Introduction
Glioma is the most common type of primary brain
tumor; however, the diagnosis and treatment of glioma remain the
most challenging in adults and children (1). Primary glioma is divided into four
grades according to the histologic criteria of the World Health
Organization (WHO) (2); low-grade (I
and II) glioma tumors have a relatively good prognosis, and
high-grade tumors (III and IV) are associated with poor clinical
outcomes and tumor recurrence (2).
In glioblastoma (GBM), the most common and malignant type of
glioma, the 5-year survival rate from first diagnosis remains at
<5%, despite progress in multidisciplinary and comprehensive
treatment (3). At present, tumor
hypoxia has been identified as the primary cause of poor prognosis
in patients with GBM (4). Hypoxia is
the most important characteristic of aggressive tumors, since the
tissues or organs receiving inadequate oxygen, which can lead to
rapid tumor growth; the hypoxic tumor microenvironment is involved
in different tumor biology processes, including initiation,
metabolism, development and metastasis. A previous study has
suggested that hypoxic conditions lead to abnormal activation of
hypoxia-inducible factors (HIFs), which serve an important role in
the regulation of tumor signaling (5). HIF-1α, one of two subunits composing
HIF-1, is considered a necessary oxygen-sensitive protein in the
regulation of hypoxic signaling pathways (6). The degradation pathway of HIF-1α is
dependent on oxygen; thus, hypoxia in tumor tissues leads to the
elevation of HIF-1α levels (7). It
has been reported that HIF-1α is positively enhanced in most
malignant types of cancer and serves an essential role in mediating
the hypoxic effects by regulating numerous target genes, including
microRNAs (miRNAs/miRs), to adjust the adaptation of tumor cells to
hypoxic conditions (8).
miRNAs are a series of evolutionarily conserved
small non-coding RNAs that can cause mRNA destabilization and/or
translational inhibition by base-pairing with the 3′-untranslated
regions of encoding mRNAs (9). A
range of miRNAs has been reported as tumor suppressors or oncogenes
for the irreplaceable mechanism in tumor initiation and
progression, and as therapeutic targets (10,11).
Increasing evidence supports the idea that aberrant miRNA
expression serves important roles in tumor hypoxia and is
associated with HIF-1α (12,13). HIF-1α is involved in multidrug
resistance by regulating miR-27 in gastric cancer cells (14). Overexpression of miR-210 induced by
hypoxia is associated with tumor progression and prognosis in upper
tract urothelial carcinoma (15).
miR-224, which is regulated by hypoxia, promotes the growth,
migration and invasion of gastric cancer cells (16). HIF-1α-inducible miR-421 increases
metastasis, suppresses apoptosis and results in cisplatin
resistance in gastric cancer by targeting E-cadherin and caspase-3
(17).
One of the main HIF-1-dependent genes is carbonic
anhydrase 9 (CA9). CA9 is overexpressed in a number of tumor types
and serves as a prognostic factor for hypoxic, aggressive and
malignant types of cancer (18). CA9
is a transmembrane glycoprotein with enzymatic activity that is
located on the cell surface and is inducible by hypoxia (19). Therefore, CA9 has been identified as
a potential target for cancer treatment, as it is enriched in
hypoxic niches of neoplastic tissues (12).
Tumor hypoxia causes a number of biological
alterations in tumor viability, including abnormal secretion and
release of exosomes into the tumor microenvironment (20). Exosomes are 30–140 nm nanovesicles
carrying genetic materials secreted from various cell types,
including cancer cells, released into the tumor microenvironment
and maybe taken in by other types of cells (21). miRNAs packaged by exosomes are stable
and specific, and thus, appropriate as potential biomarkers to
diagnose and distinguish diseases, including various malignancies
(22,23). The present study aimed to investigate
exosomal-(exo-) miR-210 levels in the serum, which may reflect
hypoxic conditions, as a potential diagnostic and prognostic
biomarker in patients with glioma.
Materials and methods
Patients and clinical samples
The present study was approved by the Medical School
of Chinese PLA (Beijing, China) and Tianjin Huanhu Hospital
(Tianjin, China). In total, 91 patients diagnosed with glioma by
clinicopathology and 50 healthy volunteers were recruited from the
Medical School of Chinese PLA and Tianjin Huanhu Hospital between
October 2011 and July 2014. The study included 52 male and 39
female patients. The median age of the patients was 45 years
(range, 19–72 years). The Karnofsky Performance Status (KPS)
(24) scores ranged between 70 and
100, and the median score was 90 (Table
I). The inclusion criteria were: i) Histological diagnosis of
glioma; ii) age between 18 and 80 years; and iii) glioma presented
on MRI or PET/CT scans. The exclusion criteria were: i) Severe
systemic infection and organ failure; ii) pregnancy; iii) severe
immunological disorders (autoimmune disease, immunosuppression);
iv) multiple cancers; and v) anaphylaxis induced by synthetic
peptides. None of the patients were treated with chemotherapy or
radiotherapy prior to surgery. All procedures in the present study
involving human participants were performed in accordance with
relevant ethical standards. Blood samples were collected at the
specific time points; pre-operation, post-operation and recurrence,
to examine the expression levels of exo-miR-210. Patients with
co-morbidities receiving other medication were excluded from the
present study. All participants provided written informed consent
prior to recruitment. All patients with glioma were followed up at
regular intervals of 3 months for up to 5 years. Although the
terminal point of the x-axis is 70 months, the survival curve was
limited to 60 months. Survival times were calculated from the first
diagnosis to the date of death caused by glioma or the last date of
follow-up, and cases of mortality unrelated to glioma were excluded
from the present study.
 | Table I.Association between clinical
characteristics and the exo-miR-210 level. |
Table I.
Association between clinical
characteristics and the exo-miR-210 level.
|
|
| Exo-miR-210 |
|---|
|
|
|
|
|---|
| Parameter | Patients (n) | Mean
expression | P-value |
|---|
| Sex |
|
| 0.724 |
|
Male | 52 | 0.0030 |
|
|
Female | 39 | 0.0034 |
|
| Age, years |
|
| 0.479 |
|
≤45 | 46 | 0.0029 |
|
|
>45 | 45 | 0.0035 |
|
| Tumor size, cm |
|
| 0.539 |
| ≤5 | 28 | 0.0030 |
|
|
>5 | 63 | 0.0032 |
|
| Relapse |
|
| 0.023a |
| No | 42 | 0.0023 |
|
|
Yes | 49 | 0.0040 |
|
| WHO grade |
|
| 0.001a |
|
I–II | 32 | 0.0010 |
|
|
III–IV | 59 | 0.0044 |
|
| KPS score |
|
| 0.332 |
|
≤90 | 41 | 0.0033 |
|
|
>90 | 50 | 0.0031 |
|
Isolation of exosomes from serum
The extraction and purification of exosomes from
blood serum and were performed as previously described (9). The serum was extracted from blood
samples immediately. The collected serum was aliquoted and stored
in liquid nitrogen. The samples were centrifuged to remove cells
and other debris in the serum, first at 3,000 × g for 30 min at
room temperature, then at 10,000 × g for 20 min at room
temperature. Subsequently, microvesicles that were larger than
exosomes were removed from supernatants by centrifuging at 100,000
× g for 30 min at 4°C. Subsequently, the supernatants were
harvested, and again centrifuged at 10,000 × g for 70 min at 4°C.
Finally, the supernatants were gently decanted, and the exosome
sediments were resuspended in PBS. The extracted exosomal samples
were stored at −80°C until further experiments.
Exosome characterization
The morphology and the size of the exosomes were
imaged by Transmission Electron Microscopy (TEM). For TEM,
collected exosomes were resuspended in 2% paraformaldehyde
(Sigma-Aldrich; Merck KGaA) and spread onto carbon/Formvar-coated
grid (Ted Pella, Inc.) for 30 min at room temperature. The grid was
then washed with DPBS and fixed with 1% glutaraldehyde
(Sigma-Aldrich; Merck KGaA). TEM images were captured with a JEOL
JEM-2100F microscope (JEOL, Ltd). Magnification, ×10,000.
miRNA isolation from exosomes and
quantitative (q)PCR
The MirVana microRNA isolation kit (Thermo Fishes
Scientific, Inc.) was used to isolate miRNAs from exosomes in
accordance with the manufacturer's protocol. The isolated RNA was
reverse transcribed and amplified using the TaqMan MicroRNA Reserve
Transcription kit (Applied Biosystem; Thermo Fishes Scientific,
Inc.). For the synthesis of cDNA, the reaction mixtures were
incubated at 37.8°C for 60 min and 95°C for 5 min, and then held at
4°C. The cDNA specimens were stored at −20°C until polymerase chain
reaction (PCR). qPCR was performed using the QuantiTect SYBR-Green
PCR mixture (Invitrogen; Thermo Fisher Scientific, Inc.) as
described previously (9,25). The reaction mixtures were incubated
at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec, 56°C
for 30 sec and 72°C for 35 sec. miR-16, which is stably expressed
across samples, was used as an endogenous control. The following
primers were used: miR-210 forward, 5′-TTGACCTGTGCGTGTGACA-3′ and
reverse, 5′-TATGGTTGTTCTGCTCTCTGTCTC-3′; miR-16-1 forward,
5′-CGCCTGTAGCAGCACGTAA-3′ and reverse, 5′-CAGAGCAGGGTCCGAGGTA-3′.
The expression levels of the miRNAs were normalized to U6, and
quantified using the 2−ΔΔCq method (25).
Immunohistochemistry and analysis
Immunohistochemical staining was used to evaluate
the expression levels of HIF-1α and CA9. Briefly, paraffin-embedded
and formalin-fixed samples were cut into 4-µm sections, which were
then processed for immunohistochemistry. The sections were
de-waxed, rehydrated, blocked with hydrogen peroxide, and the
antigens were retrieved in a microwave in 10 mM citrate buffer (pH
6.0) for 10 min and cooled to room temperature. In order to block
non-specific staining the tissue was incubated in blocking buffer,
1 ml Protein blocking buffer (cat. no. ab126587; Abcam) diluted in
9 ml PBS, for 30 min at room temperature. Following antigen
retrieval for 20 min in citrate buffer (0.01 M citric acid, pH 6.0)
at room temperature, sections were incubated overnight at 4°C with
the primary antibody (1:100). HIF-1α (cat. no. ab51608) and CA9
(cat. no. ab216021) antibodies were purchased from Abcam.
Anti-mouse immunoglobulin G (IgG)-horseradish peroxidase (HRP) and
anti-goat IgG-HRP goat anti-rabbit secondary antibodies (1:200;
cat. no. ab205718; Santa Cruz Biotechnology, Inc.) was incubated
with the sections for 1 h at room temperature, and
3,3′-diaminobenzidine buffer was used for signal detection. The
sections were scored blindly by two pathologists using a light
microscope at ×200 magnification.
Either citrate buffer pH 6 or EDTA buffer pH 9 was
used to perform antigen retrieval prior to cooling at 4°C for 20
min. Subsequently, 3% hydrogen peroxide was applied to block
endogenous peroxidase activity for 10 min. Primary antibodies (1:20
dilution) were incubated with samples for 15 min at room
temperature. 3,3′-diaminobenzidine substrate was added for 5 min at
room temperature until color developed prior to washing in running
water for 10 min. Slides were then counterstained in 10%
hematoxylin at room temperature for 60 sec and blued with Scotts'
tap water before being dehydrated through a series of graded
alcohols. Cover slips were applied using distrene, plasticizer and
xylene. H-score was used to determine the expression of HIF-1α and
CA9: A combination of the intensity (0–3) and percentage of cells stained, with a
range of 0–300. HIF-1α scores only included nuclear expression and
CA9 scores contained the staining of the nucleus and cytoplasm, as
previously described (26).
Statistical analysis
SPSS version 13.0 (SPSS, Inc.) was used to assess
and analyze statistical data. All quantitative data are presented
as the mean ± standard deviation (SD). Univariate and multivariate
analysis of variance were performed. ANOVA was performed to analyze
the significance between two groups (such as low-grade vs.
high-grade glioma), and Student-Newman-Keuls test was performed to
calculate the P-values. Kaplan-Meier analysis, a log-rank test and
Cox regression analysis were used to calculate survival rates and
predict factors associated with survival. P<0.05 was considered
to indicate a statistically significant difference. Technical and
biological triplicates of each experiment were performed.
Results
Clinical characteristics of
patients
Exosomal samples from the serum were collected from
91 patients with glioma and 50 healthy volunteers as a control.
Patients with glioma included 12 patients with WHO grade I
pilocytic astrocytoma, 20 patients with WHO grade II diffuse
astrocytoma, 22 patients with WHO grade III anaplastic astrocytoma
and 37 patients with WHO grade IV primary GBM. A total of 58
patients received radio-chemotherapy after surgery. At the last
follow-up, 72 patients had succumbed to primary glioma, including 3
patients in the WHO grade I, 14 patients in WHO grade II, 20
patients in WHO grade III and 35 patients in WHO grade IV groups.
The 1- and 2-year overall survival (OS) rates were 91.2 and 57.1%,
respectively.
Exo-miR-210 in the serum may serve as
a diagnostic biomarker in glioma
A previous study demonstrated that serum miR-210 was
overexpressed in patients with glioma, and high levels of serum
miR-210 have been identified to be associated with worse overall
survival (27). The present study
aimed to further explore whether serum exo-miR-210 may serve as a
diagnostic or prognostic biomarker for patients with glioma. Data
from exosome characterization are shown in Fig. S1. Initially, levels of serum
exo-miR-210 were analyzed by qPCR to compare the differences
between patients with glioma (n=91) and healthy controls (n=50).
Following normalization to miR-16, there was a significant
difference in the expression levels of serum exo-miR-210 between
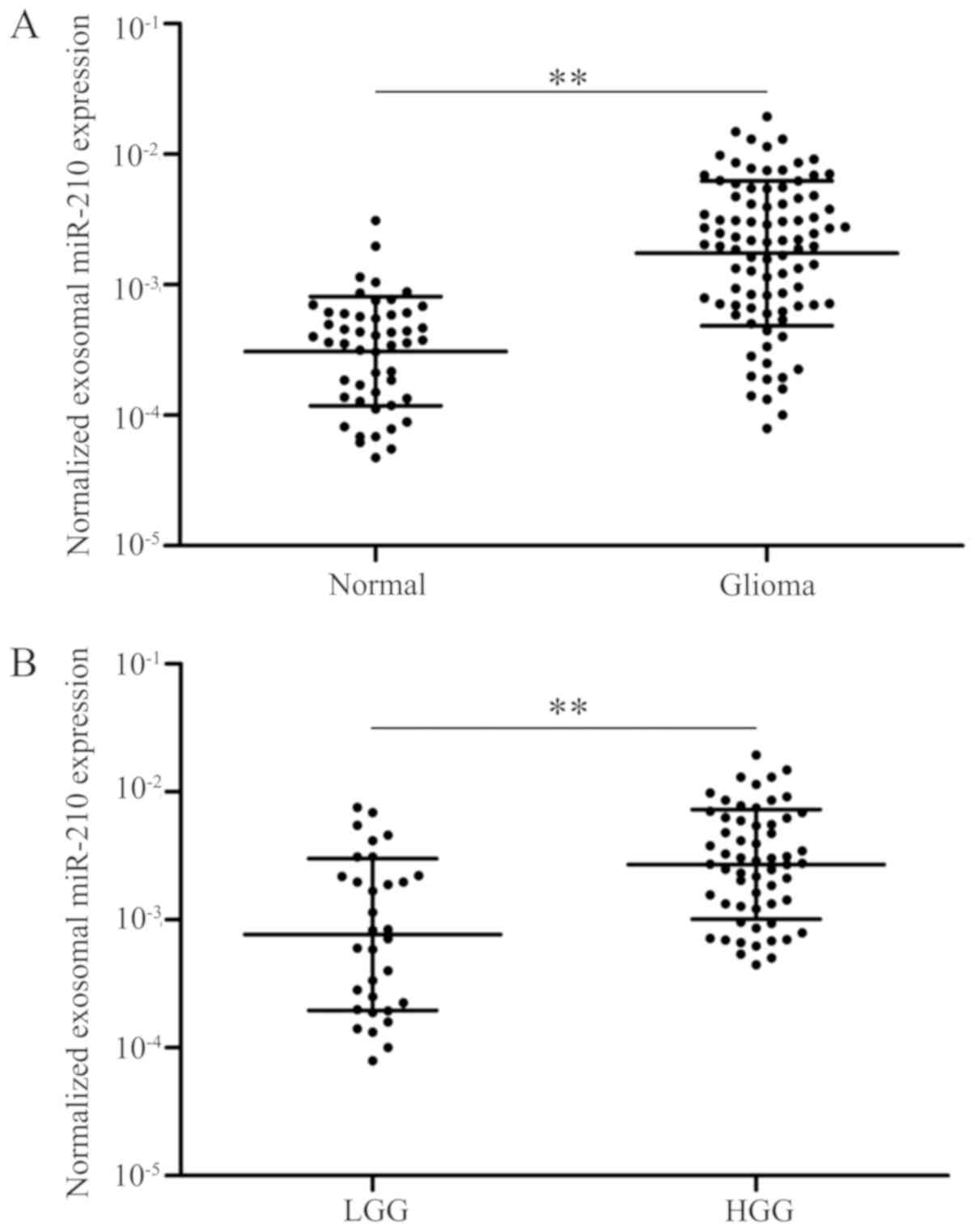
patients with glioma and healthy controls (P<0.01; Fig. 1A). Notably, patients with low-grade
(I–II) glioma exhibited relatively low levels of serum exo-miR-210
compared with patients with high-grade (III–IV) glioma (P<0.01;
Fig. 1B). Collectively, the data
suggested that serum exo-miR-210 was abnormally overexpressed in
the serum of patients with glioma and increased with increasing
grades of glioma; therefore, it maybe used as a biomarker for the
diagnosis of glioma.
Expression levels of exo-miR-210 in
the serum are associated with specific clinicopathological
features
The putative associations between expression levels
of exo-miR-210 and different clinicopathological features were
explored. All patients were separated into two groups (low and high
exo-miR-210 expression) according to the median concentration of
exo-miR-210. The percentages of patients with glioma included in
the high expression group of exo-miR-210 increased with grade, with
16.7, 20.0, 59.1 and 73.0% of patients with grade I, II, II and IV,
respectively, being included in the high expression group. There
was no statistically significant association identified between the
level of serum exo-miR-210 and other clinicopathological
parameters, including sex, KPS and age at diagnosis (P>0.05;
Table I). Advanced pathological
grades were statistically significantly associated with serum
exo-miR-210 expression (P<0.01; Table
I).
Serum exo-miR-210 levels reflect the
dynamics of GBM
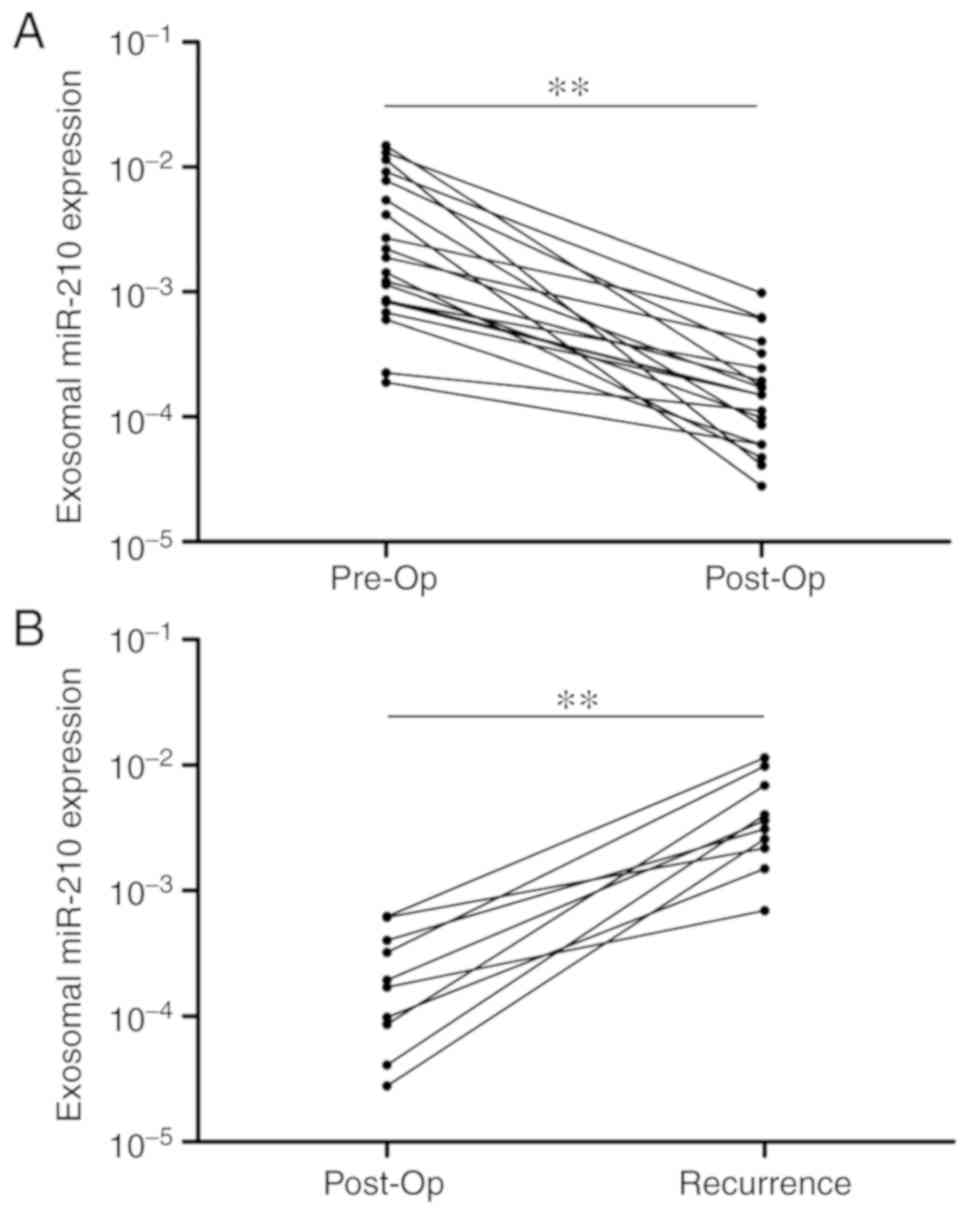
Subsequently, 20 patients with GBM from the original
group were selected, and the expression levels of serum exo-miR-210
were detected pre- and post-operation. The gap period between the
pre-and post-operation period was fixed at 1–2 weeks. Notably,
serum exo-miR-210 levels were markedly decreased following curative
surgical resection (Fig. 2A).
Furthermore, the expression levels of serum exo-miR-210 were
compared after surgery and at tumor recurrence in 10 patients with
recurring GBM. Although relatively low levels of exo-miR-210 were
found at the post-operation time point, a significant increase was
detected at the time of GBM recurrence (Fig. 2B). Overall, serum exo-miR-210
secreted and released by GBM cancer cells could reflect tumor
dynamics during treatment.
Expression of serum exo-miR-210 as a
potential diagnostic and prognostic biomarker for glioma
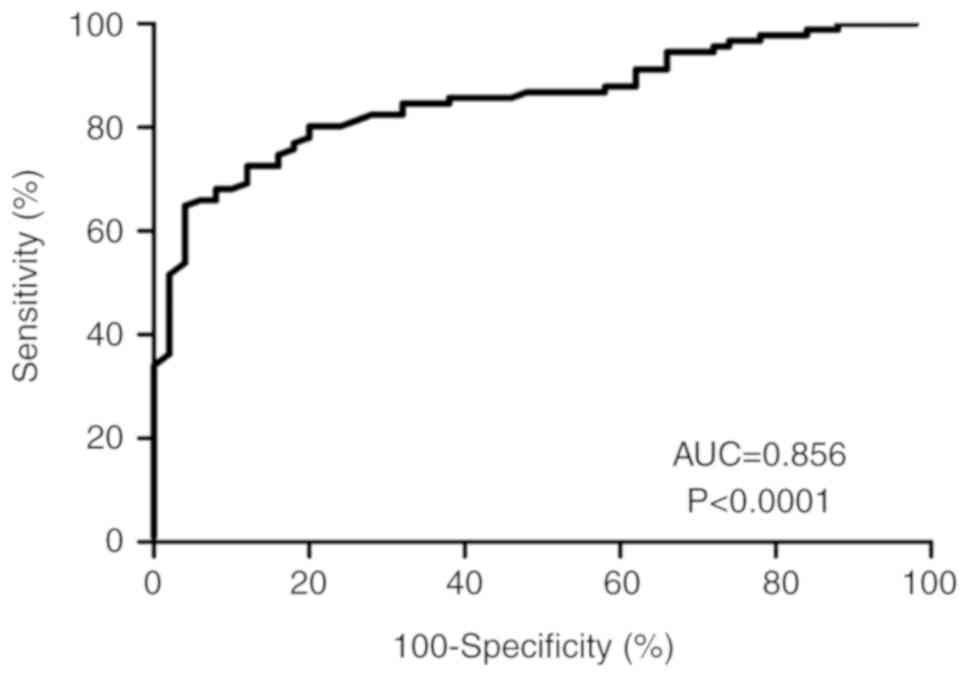
To evaluate the diagnostic value of exo-miR-210
expression in patients with glioma, the present study applied
receiver operating characteristic curve analysis and calculated the
corresponding area under the curve (AUC) values to verify the
potential diagnosis and discriminatory accuracy of exo-miR-210. As
shown in Fig. 3, serum exo-miR-210
levels were robust in distinguishing patients with glioma from
healthy controls, with an AUC value of 0.856 (95% CI, 0.795–0.917),
whereas the sensitivity, specificity, and positive and negative
predictive values to discriminate patients with glioma were 83.2,
94.3, 93.5 and 70.3%, respectively.
Our previous study demonstrated that exo-miR-210
expression is associated with glioma grades, which suggests that
exo-miR-210 may be an important predictor of poor prognosis in
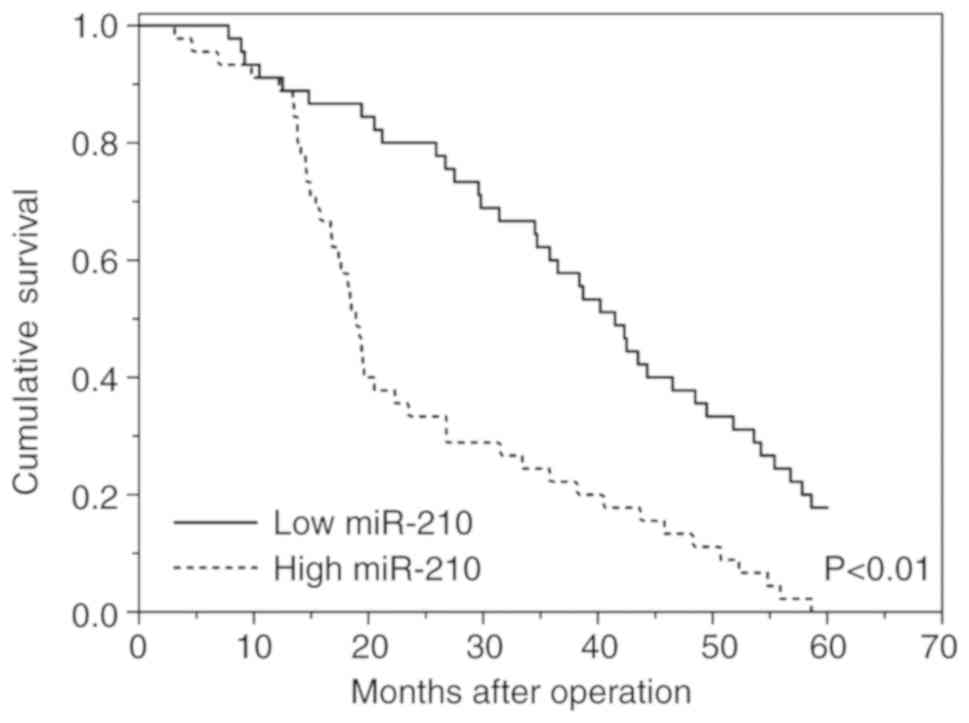
higher-grade compared with lower-grade gliomas (27). In this cohort, patients with high
levels of exo-miR-210 exhibited a poor survival rate in contrast to
patients with low expression levels of exo-miR-210 according to
Kaplan-Meier survival analysis (Fig.
4). Subsequently, Cox regression analyses were carried out to
assess the prognostic value of serum exo-miR-210 levels for OS. The
results showed a significant association between serum exo-miR-210
levels and OS in patients with glioma (P<0.01; hazard ratio,
3.63; 95% CI, 1.54–5.67; Table II).
Statistically significant associations were also observed in
pathological grades. No significant association was observed in any
of the other clinicopathological parameters tested, including sex,
KPS and age at diagnosis (Table
II). Overall, levels of serum exo-miR-210 secreted by glioma
cells could be used as excellent indexes to differentiate patients
with glioma from non-glioma patients, and as a valuable and novel
biomarker in the diagnosis and prognosis of human glioma.
 | Table II.Univariate and multivariate analyses
of prognostic parameters in patients with glioma. |
Table II.
Univariate and multivariate analyses
of prognostic parameters in patients with glioma.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Parameter | P-value | HR (95% CI) | P-value | HR (95% CI) |
|---|
| Age (≤45 vs.
>45) | 0.77 | 1.26
(0.64–1.91) | 0.48 | 1.03
(0.51–1.67) |
| Sex (male vs.
female) | 0.64 | 1.09
(0.45–1.56) | 0.59 | 1.13
(0.57–1.78) |
| KPS (≤90
vs.>90) | 0.19 | 1.37
(0.84–1.98) | 0.43 | 0.98
(0.60–1.37) |
| WHO grade (I–II vs.
III–IV) | <0.01 | 7.88
(3.54–12.53) | <0.01 | 11.25
(5.34–19.67) |
| Exosomal
miR-210 | <0.01 | 3.63
(1.54–5.67) | <0.01 | 4.31
(1.94–8.13) |
Expression levels of serum exo-miR-210
are associated with hypoxia
A number of studies (28,29) have
demonstrated that the overexpression of miR-210 is regulated in a
HIF-1α-dependent manner in numerous types of cancer. Integrated
analysis of microRNA, mRNA and chromatin
immunoprecipitation-sequencing data has revealed that miR-210
expression under hypoxia is regulated at the transcriptional and
post-transcriptional levels, with the presence of HIF-1α binding
sites (30). Previous studies have
demonstrated that miR-210 is associated with tumor hypoxia and
predicts benefit from hypoxic modification (31,32).
Based on these previous studies, the present study examined the
association between serum exo-miR-210 expression and HIF-1α and CA9
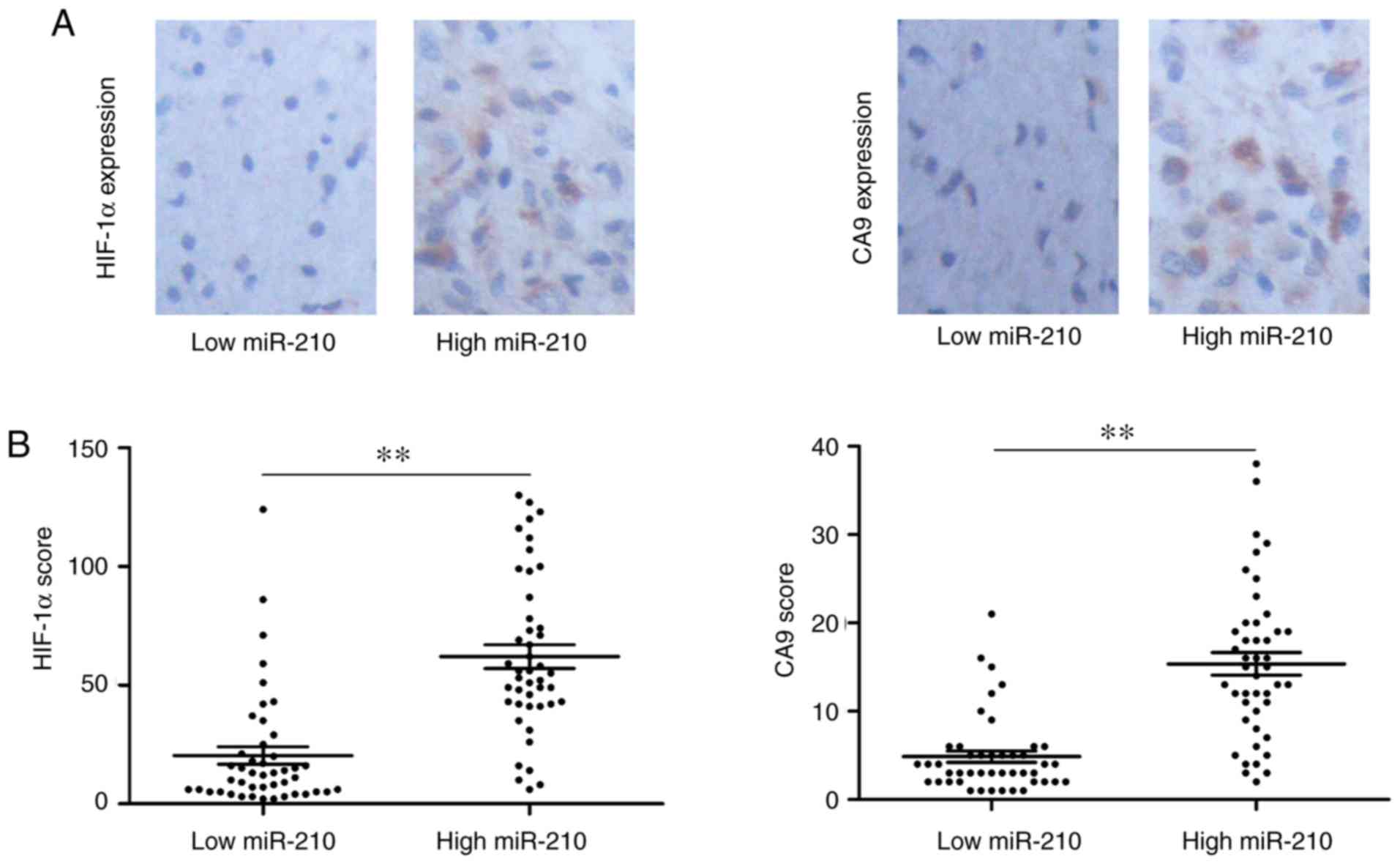
levels. Representative images of immunohistochemistry of HIF-1α and
CA9 are shown in Fig. 5A. When
exo-miR-210 expression was compared with conventional hypoxic
markers, higher serum exo-miR-210 expression was accompanied by
high HIF-1α and CA9 protein levels (Fig.
5B). Based on the results of the present study, it can be
concluded that serum exo-miR-210 could be used as a potential
biomarker of hypoxia.
Discussion
Biomarkers could provide a more accurate assessment
to predict the diagnosis and prognosis of various types of cancer
according to tumor biology. Exosomes are a reservoir of valuable
and novel biomarkers for cancer diagnosis and prognosis. Exosomes
secreted to the extracellular microenvironment are involved in the
cell-cell communication to neighboring and distant cells, favoring
secretion of growth factors, cytokines and angiopoietic factors,
which can induct tumor proliferation, metastasis, therapeutic
efficacy and immune responses (33).
Exosomes can be isolated from blood or other bodily fluids to
reveal progression of cancer occurring in the body (34). miRNA can be horizontally transferred
into the extracellular microenvironment by exosomes to modify the
microenvironment. Exosomal miRNAs are emerging as an identified
group of messengers and effectors in intercellular communication
(35).
Serum exo-miRNAs can function as novel non-invasive
biomarkers for the detection and monitoring of various types of
cancer and putatively as therapeutic targets. Levels of serum
exo-miR-21 are increased and associated with the progression and
aggressiveness of esophageal squamous cell cancer (36), whereas levels of serum exo-miR-19a
have been identified to serve as prognostic biomarkers for
recurrence in patients with colorectal cancer (37). Exo-miR-21 extracted from
cerebrospinal fluids is associated with a poor prognosis and tumor
recurrence in patients with glioma (38). Levels of serum exo-miR-1290 and
exo-miR-375 are associated with castration resistance in prostate
cancer (39). miR-223-3p
encapsulated by exosomes has been identified as an important
biomarker for the early detection of invasive breast cancer
(40). Exo-miR-423-5p has been
reported as a minimally invasive biomarker in the diagnosis of
gastric cancer, and can promote tumor growth and metastasis by
targeting SUFU negative regulator of hedgehog signaling (41). The present study obtained data, which
suggested that the exo-miR-210 level in serum was a diagnostic,
prognostic and predictive biomarker for glioma, and associated with
hypoxic conditions. miR-210, which is a regulator of several
cellular functions, dependent on or independent of hypoxia
signaling, is involved in numerous biological processes of numerous
tumors throughout the human body (42). miR-210 has been identified as an
activator of tumor initiation and development, and its high levels
are associated with a negative clinical outcome (43). It has been reported to be involved in
cell survival, differentiation, angiogenesis, metabolism and cell
cycle control (15,44). In addition, elevated miR-210 levels
increase the proliferation and decrease apoptosis of GBM cells by
targeting polypyrimidine tract binding protein 3 (45). miR-210 extracted from the peripheral
blood is a promising and minimally invasive biomarker that can be
employed to diagnose and predict the outcome of glioma (27). miR-210, which has been reported as
one of the abnormally upregulated miRNAs in hypoxic cells, is
highly conserved and required for cell survival (46). In addition, since other studies have
reported that the level of miR-210 is dependent on HIF-1α
regulation, increased expression of miR-210 in tissues and serum
has become a signature for tumor hypoxia (47,48).
The aim of the present study was to examine whether
exo-miR-210 is a potential non-invasive biomarker for the diagnosis
and prognosis of glioma and associated with hypoxic conditions.
Firstly, a significant difference was identified in the expression
levels of serum exo-miR-210 between patients with glioma and
healthy controls. Importantly, serum exo-miR-210 exhibited a
relatively low level in low-grade (I–II) compared with high grade
(III–IV) glioma. Additionally, serum exo-miR-210 levels were
markedly decreased following curative surgical resection and
significantly increased at the time of GBM recurrence. Notably,
high levels of exo-miR-210 were associated with a poor survival
rate compared with low expression levels of exo-miR-210 according
to Kaplan-Meier survival analysis. Subsequently, Cox regression
analyses revealed that serum exo-miR-210 levels were associated
with the OS of patients with glioma. Finally, overexpression of
exo-miR-210 was significantly associated with high protein levels
of HIF-1a and CA9 and reflected hypoxia in patients with glioma.
Although the present study discussed the potential use of
exo-miR-210 as a biomarker in glioma, the underlying mechanism of
exo-miR-210 in hypoxic conditions has not been identified. Future
studies are needed to explore the unique properties of exo-miR-210
and allow the development of highly sensitive strategies to rapidly
and non-invasively monitor the pathological condition of patients
with glioma.
In summary, serum exo-miR-210 expression is
abnormally elevated in glioma, particularly GBM, and may function
as a novel biomarker for predicting diagnosis and progression of
patients with glioma. Further larger independent studies are
required prior to applying the results of the present study to
clinical practice.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the China
National Natural Scientific Fund (grant nos. 81772690, 81502654 and
81872468).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
TX conceived and designed the study. FL and XY
performed the experiments.
Ethics approval and consent to
participate
The present study was approved by the Medical School
of Chinese PLA (Beijing, China) and Tianjin Huanhu Hospital
(Tianjin, China). All participants provided written informed
consent prior to recruitment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Savage N: Searching for the roots of brain
cancer. Nature. 561:S50–S51. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 world health organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ahmed SU, Carruthers R, Gilmour L,
Yildirim S, Watts C and Chalmers AJ: Selective inhibition of
parallel DNA damage response pathways optimizes radiosensitization
of glioblastoma stem-like cells. Cancer Res. 75:4416–4428. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Colwell N, Larion M, Giles AJ, Seldomridge
AN, Sizdahkhani S, Gilbert MR and Park DM: Hypoxia in the
glioblastoma microenvironment: Shaping the phenotype of cancer
stem-like cells. Neuro Oncol. 19:887–896. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Majmundar AJ, Wong WJ and Simon MC:
Hypoxia-inducible factors and the response to hypoxic stress. Mol
Cell. 40:294–309. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu D, Potluri N, Lu J, Kim Y and
Rastinejad F: Structural integration in hypoxia-inducible factors.
Nature. 524:303–308. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Semenza GL: Hypoxia-inducible factors:
Mediators of cancer progression and targets for cancer therapy.
Trends Pharmacol Sci. 33:207–214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sadri N and Zhang PJ: Hypoxia-inducible
factors: Mediators of cancer progression; Prognostic and
therapeutic targets in soft tissue sarcomas. Cancers (Basel).
5:320–333. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lan F, Qing Q, Pan Q, Hu M, Yu H and Yue
X: Serum exosomal miR-301a as a potential diagnostic and prognostic
biomarker for human glioma. Cell Oncol (Dordr). 41:25–33. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yue X, Cao D, Lan F, Pan Q, Xia T and Yu
H: MiR-301a is activated by the Wnt/β-catenin pathway and promotes
glioma cell invasion by suppressing SEPT7. Neuro Oncol.
18:1288–1296. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lan F, Yu H, Hu M, Xia T and Yue X:
miR-144-3p exerts anti-tumor effects in glioblastoma by targeting
c-Met. J Neurochem. 135:274–286. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu K, Zhan Y, Yuan Z, Qiu Y, Wang H, Fan
G, Wang J, Li W, Cao Y, Shen X, et al: Hypoxia induces drug
resistance in colorectal cancer through the HIF-1α/miR-338-5p/IL-6
feedback loop. Mol Ther. 2019.(Epub ahead of print). View Article : Google Scholar
|
|
13
|
Zhou Y, Xu Q, Shang J, Lu L and Chen G:
Crocin inhibits the migration, invasion, and epithelial-mesenchymal
transition of gastric cancer cells via miR-320/KLF5/HIF-1α
signaling. J Cell Physiol. 234:17876–17885. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao Q, Li Y, Tan BB, Fan LQ, Yang PG and
Tian Y: HIF-1α induces multidrug resistance in gastric cancer cells
by inducing MiR-27a. PLoS One. 10:e01327462015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ke HL, Li WM, Lin HH, Hsu WC, Hsu YL,
Chang LL, Huang CN, Li CC, Chang HP, Yeh HC, et al:
Hypoxia-regulated MicroRNA-210 overexpression is associated with
tumor development and progression in upper tract urothelial
carcinoma. Int J Med Sci. 14:578–584. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
He C, Wang L, Zhang J and Xu H:
Hypoxia-inducible microRNA-224 promotes the cell growth, migration
and invasion by directly targeting RASSF8 in gastric cancer. Mol
Cancer. 16:352017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ge X, Liu X, Lin F, Li P, Liu K, Geng R,
Dai C, Lin Y, Tang W, Wu Z, et al: MicroRNA-421 regulated by HIF-1α
promotes metastasis, inhibits apoptosis, and induces cisplatin
resistance by targeting E-cadherin and caspase-3 in gastric cancer.
Oncotarget. 7:24466–24482. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Benej M, Pastorekova S and Pastorek J:
Carbonic anhydrase IX: Regulation and role in cancer. Subcell
Biochem. 75:199–219. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ziffels B, Stringhini M, Probst P, Fugmann
T, Sturm T and Neri D: Antibody-based delivery of cytokine payloads
to carbonic anhydrase IX leads to cancer cures in immunocompetent
tumor-bearing mice. Mol Cancer Ther. 18:1544–1554. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Meng W, Hao Y, He C, Li L and Zhu G:
Exosome-orchestrated hypoxic tumor microenvironment. Mol Cancer.
18:572019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hoshino A, Costa-Silva B, Shen TL,
Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di
Giannatale A, Ceder S, et al: Tumour exosome integrins determine
organotropic metastasis. Nature. 527:329–335. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nouraee N, Khazaei S, Vasei M, Razavipour
SF, Sadeghizadeh M and Mowla SJ: MicroRNAs contribution in tumor
microenvironment of esophageal cancer. Cancer Biomarkers.
16:367–376. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xiao GY, Cheng CC, Chiang YS, Cheng WT,
Liu IH and Wu SC: Exosomal miR-10a derived from amniotic fluid stem
cells preserves ovarian follicles after chemotherapy. Sci Rep.
6:231202016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Khalid MA, Achakzai IK, Ahmed Khan S,
Majid Z, Hanif FM, Iqbal J, Laeeq SM and Luck NH: The use of
karnofsky performance status (KPS) as a predictor of 3 month post
discharge mortality in cirrhotic patients. Gastroenterol Hepatol
Bed Bench. 11:301–305. 2018.PubMed/NCBI
|
|
25
|
Yue X, Lan F, Hu M, Pan Q, Wang Q and Wang
J: Downregulation of serum microRNA-205 as a potential diagnostic
and prognostic biomarker for human glioma. J Neurosurg.
124:122–128. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Eustace A, Irlam JJ, Taylor J, Denley H,
Agrawal S, Choudhury A, Ryder D, Ord JJ, Harris AL, Rojas AM, et
al: Necrosis predicts benefit from hypoxia-modifying therapy in
patients with high risk bladder cancer enrolled in a phase III
randomised trial. Radiother Oncol. 108:40–47. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lai NS, Wu DG, Fang XG, Lin YC, Chen SS,
Li ZB and Xu SS: Serum microRNA-210 as a potential noninvasive
biomarker for the diagnosis and prognosis of glioma. Br J Cancer.
112:1241–1246. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dang K and Myers KA: The role of
hypoxia-induced miR-210 in cancer progression. Int J Mol Sci.
16:6353–6372. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang W, Ma J, Zhou W, Zhou X, Cao B, Fan D
and Hong L: Biological implications and clinical value of mir-210
in gastrointestinal cancer. Expert Rev Gastroenterol Hepatol.
11:539–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Camps C, Saini HK, Mole DR, Choudhry H,
Reczko M, Guerra-Assunção JA, Tian YM, Buffa FM, Harris AL,
Hatzigeorgiou AG, et al: Integrated analysis of microRNA and mRNA
expression and association with HIF binding reveals the complexity
of microRNA expression regulation under hypoxia. Mol Cancer.
13:282014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Agrawal R, Pandey P, Jha P, Dwivedi V,
Sarkar C and Kulshreshtha R: Hypoxic signature of microRNAs in
glioblastoma: Insights from small RNA deep sequencing. BMC
Genomics. 15:6862014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Irlam-Jones JJ, Eustace A, Denley H,
Choudhury A, Harris AL, Hoskin PJ and West CM: Expression of
miR-210 in relation to other measures of hypoxia and prediction of
benefit from hypoxia modification in patients with bladder cancer.
Br J Cancer. 115:571–578. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li D, Liu J, Guo B, Liang C, Dang L, Lu C,
He X, Cheung HY, Xu L, Lu C, et al: Osteoclast-derived exosomal
miR-214-3p inhibits osteoblastic bone formation. Nat Commun.
7:108722016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Munagala R, Aqil F and Gupta RC: Exosomal
miRNAs as biomarkers of recurrent lung cancer. Tumour Biol.
37:10703–10714. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhou W, Fong MY, Min Y, Somlo G, Liu L,
Palomares MR, Yu Y, Chow A, O'Connor ST, Chin AR, et al:
Cancer-secreted miR-105 destroys vascular endothelial barriers to
promote metastasis. Cancer Cell. 25:501–515. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tanaka Y, Kamohara H, Kinoshita K,
Kurashige J, Ishimoto T, Iwatsuki M, Watanabe M and Baba H:
Clinical impact of serum exosomal microRNA-21 as a clinical
biomarker in human esophageal squamous cell carcinoma. Cancer.
119:1159–1167. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Matsumura T, Sugimachi K, Iinuma H,
Takahashi Y, Kurashige J, Sawada G, Ueda M, Uchi R, Ueo H, Takano
Y, et al: Exosomal microRNA in serum is a novel biomarker of
recurrence in human colorectal cancer. Br J Cancer. 113:275–281.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shi R, Wang PY, Li XY, Chen JX, Li Y,
Zhang XZ, Zhang CG, Jiang T, Li WB, Ding W and Cheng SJ: Exosomal
levels of miRNA-21 from cerebrospinal fluids associated with poor
prognosis and tumor recurrence of glioma patients. Oncotarget.
6:26971–26981. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Huang X, Yuan T, Liang M, Du M, Xia S,
Dittmar R, Wang D, See W, Costello BA, Quevedo F, et al: Exosomal
miR-1290 and miR-375 as prognostic markers in castration-resistant
prostate cancer. Eur Urol. 67:33–41. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yoshikawa M, Iinuma H, Umemoto Y,
Yanagisawa T, Matsumoto A and Jinno H: Exosome-encapsulated
microRNA-223-3p as a minimally invasive biomarker for the early
detection of invasive breast cancer. Oncol Lett. 15:9584–9592.
2018.PubMed/NCBI
|
|
41
|
Yang H, Fu H, Wang B, Zhang X, Mao J, Li
X, Wang M, Sun Z, Qian H and Xu W: Exosomal miR-423-5p targets SUFU
to promote cancer growth and metastasis and serves as a novel
marker for gastric cancer. Mol Carcinog. 57:1223–1236. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bavelloni A, Ramazzotti G, Poli A, Piazzi
M, Focaccia E, Blalock W and Faenza I: MiRNA-210: A current
overview. Anticancer Res. 37:6511–6521. 2017.PubMed/NCBI
|
|
43
|
Ren CX, Leng RX, Fan YG, Pan HF, Wu CH and
Ye DQ: MicroRNA-210 and its theranostic potential. Expert Opin Ther
Targets. 20:1325–1338. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bonora M, Wieckowsk MR, Chinopoulos C,
Kepp O, Kroemer G, Galluzzi L and Pinton P: Molecular mechanisms of
cell death: Central implication of ATP synthase in mitochondrial
permeability transition. Oncogene. 34:16082015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang S, Lai N, Liao K, Sun J and Lin Y:
MicroRNA-210 regulates cell proliferation and apoptosis by
targeting regulator of differentiation 1 in glioblastoma cells.
Folia Neuropathol. 53:236–244. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kulshreshtha R, Ferracin M, Wojcik SE,
Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM,
Negrini M, et al: A microRNA signature of hypoxia. Mol Cell Biol.
27:1859–1867. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ying Q, Liang L, Guo W, Zha R, Tian Q,
Huang S, Yao J, Ding J, Bao M, Ge C, et al: Hypoxia-inducible
microRNA-210 augments the metastatic potential of tumor cells by
targeting vacuole membrane protein 1 in hepatocellular carcinoma.
Hepatology. 54:2064–2075. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cheng HH, Mitchell PS, Kroh EM, Dowell AE,
Chéry L, Siddiqui J, Nelson PS, Vessella RL, Knudsen BS, Chinnaiyan
AM, et al: Circulating microRNA profiling identifies a subset of
metastatic prostate cancer patients with evidence of
cancer-associated hypoxia. PLoS One. 8:e692392013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Martin RC, Gerstenecker A, Nabors LB,
Marson DC and Triebel KL: Impairment of medical decisional capacity
in relation to karnofsky performance status in adults with
malignant brain tumor. Neurooncol Pract. 2:13–19. 2015.PubMed/NCBI
|