Introduction
In developing countries, stomach cancer is the third
most frequently diagnosed cancer and is one of the leading causes
of cancer-related death (1). The
annual incidence and mortality of gastric cancer (GC) in China is
estimated to ~679,100 and 498,000 cases, respectively (2). Despite recent advances in chemotherapy
for GC, the outcomes of anticancer therapy remain
unsatisfactory.
Tyrosine-protein phosphatase non-receptor type
11 (PTPN11) encodes Src homology 2 domain-containing
protein tyrosine phosphatase (SHP-2), which participates in
multiple intracellular signaling pathways and plays an important
role in tumor cell proliferation, apoptosis, invasion, metastasis
and drug resistance (3,4).
H. pylori (Hp) infection is the primary risk
factor for GC (5). Previously,
evidence suggests that the well-known carcinogenic protein Cag-A of
Hp is associated with SHP-2 expression in gastric mucosal
epithelial cells. The Cag-A protein is released by Hp, enters
epithelial cells via the type IV secretion system and is activated
by tyrosine phosphorylation, which enables this protein to acquire
the ability to interact with the tumor promoting enzyme tyrosine
phosphorylase SHP-2. This process is regulated by host cell
phosphatase and affects a large number of downstream pathways
ultimately leading to decreased adhesion and polarity of epithelial
cells (3,6,7).
Therefore, SHP-2 is considered one of the key proteins that links
Cag-A with gastric cancer. However, only a limited number of
patients with Hp-positive chronic gastritis or a peptic ulcer
eventually develop into GC (8). This
suggests that specific differences may appear between Hp-infected
hosts, such as genetic or epigenetic changes associated with the
PTPN11 gene, or the differences noted in SHP-2 protein
expression.
PTPN11 mutations have been extensively
investigated in the past years. Germline mutations in PTPN11
cause Noonan syndrome (9–11) and its clinically related Leopard
syndrome (12), whereas somatic
mutations of PTPN11 contribute to leukemogenesis (13–17), as
well as in the development of specific solid tumors, including
neuroblastoma (18,19), metachondromatosis (20,21),
brain tumors (22–24), neurofibromatosis (25), optic nerve pilomyxoid astrocytoma
(26), breast carcinoma (27,28),
colorectal cancer (29,30) and Ewing sarcoma (31). However, oncogenic mutations of
PTPN11 are rare in the majority of solid tumors including GC
(32,33).
Previous studies have detected the presence of
PTPN11 polymorphisms in GC (34–36).
However, these PTPN11 polymorphisms were shown to be
associated with gastric atrophy instead of GC in Chinese and
Japanese subjects (37,38). These findings indicated that with the
exception of mutations and polymorphisms, additional abnormal
expression changes in the PTPN11 gene were involved in the
development of GC.
Previously, the role of DNA methylation in the study
of GC has received increasing attention in the identification of
the mechanisms responsible for GC formation (39). However, the association between
PTPN11 methylation and GC has not been reported to date.
Therefore, the current study aimed to investigate the contribution
of PTPN11 methylation in GC.
Materials and methods
Study subjects
A total of 112 GC patients (mean, 56.56; range,
21–83 years), including 76 male and 36 female patients, were
recruited in the First Affiliated Hospital of Soochow University
between December 2010 and April 2014. Gastric mucosa tissues of the
primary tumor site and the corresponding adjacent normal tissues (5
cm away from the tumor) were collected from the patients. During
this period, the GC patients were followed up and their survival
data was collected. The patients were diagnosed by pathological
examination and none of them received radiotherapy or chemotherapy
prior to surgical resection. All patients who participated in the
present study had signed an informed consent form. The study was
approved by the Ethics Committee of Ningbo University.
DNA extraction, bisulphite conversion
and sequencing
Total DNA was extracted from the tissue samples by
the EZNA™ Tissue DNA kit (Omega Bio-Tek, Inc.) and its
concentration was determined using a Nanodrop 2000
spectrophotometer (Thermo Fisher Scientific, Inc.). Bisulphite
treatment was achieved using the EZ DNA Methylation-Gold kit™ (Zymo
Research Corp.). Typically, 500 ng of the original DNA sample was
denatured by NaOH and bisulphite was used to convert the
unmethylated cytosine to uracil, while the methylated cytosine
remained unchanged (40). The total
volume of the reaction was 30 µl. In addition, a part of the
bisulphite-converted products were randomly selected for Sanger
sequencing to verify the specificity of the quantitative
methylation specific PCR (qMSP) assay.
SYBR green-based qMSP
The SYBR green-based qMSP used β-actin (ACTB)
as an internal control. The qMSP reaction consisted of 10 µl SYBR
Green I Master mix (Roche Diagnostics), 1 µl primers and 1.0 µl
bisulphite-modified DNA template (10 ng/µl). The reaction volume
was made up to 20 µl by addition of ddH2O. The primer
sequences used in the qMSP assays were the following:
5′-GAGGTTCGGAGATAGTAGGTAAT-3′ for the PTPN11 forward primer,
5′-GATTTCATTCATTTCGTTCCACAA-3′ for the PTPN11 reverse
primer, 5′-TGGTGATGGAGGAGGTTTAGTAAGT-3′ for the ACTB forward
primer and 5′-AACCAATAAAACCTACTCCTCCCTTAA-3′ for the ACTB
reverse primer. The primers used in the present study were designed
by the Primer Premier 5.0 software (PREMIER Biosoft Inc.). The
designed primers were evaluated using Oligo 6.0 software (DBA Oligo
Inc.) and NCBI primer-blast tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/).
The PCR reactions were initiated at 95°C for 10 min, followed by 45
cycles at 95°C for 30 sec and 58°C and 72°C for 30 sec. A melting
curve analysis was used at 95°C for 15 sec and at 58°C for 1 min.
Subsequently, the temperature was increased every sec for 0.11°C
until it reached 95°C. The percentage of methylated reference (PMR)
was calculated so as to represent the PTPN11 methylation
levels. The equation used was as follows:
[PMR=2−ΔΔCqx100%, ΔΔCq=sample DNA
(CqPTPN11-CqACTB)-fully methylated DNA
(CqPTPN11-CqACTB)] (41).
Bioinformatic analysis
The methylation levels of PTPN11 and the
expression profiles of this gene [Stomach Adenocarcinoma, The
Cancer Genome Atlas (TCGA), Provisional] were available from
cBioPortal (http://www.cbioportal.org/). The data comprised 478 GC
samples from which DNA was extracted. The samples were used to
evaluate the correlation between PTPN11 methylation and mRNA
expression levels. A total of 177 samples were derived from
subjects with Hp infection. The PTPN11 methylation levels
were obtained from the TCGA gastric cancer database, including
three tumor Hp(+), 17 tumor Hp(−), 58 non-tumor Hp(+) and 99
non-tumor Hp(−) samples.
Statistical analysis
SPSS 16.0 software (SPSS Inc.) was used for
statistical analysis. The normal distributed data were described as
mean ± standard deviation, and the non-normal distributed data were
described as median with interquartile ranges. The paired sample t
test was used to assess the differences in the methylation levels
between tumor and adjacent normal tissues. The data were analyzed
following subgroup analysis. The spearman rank test was used to
assess the association between PTPN11 methylation levels and
PTPN11 expression levels. Kaplan-Meier and log-rank test
analyses were applied to assess patient survival. A non-parametric
Mann-Whitney U test and two independent sample t-tests were used to
calculate differences in PTPN11 methylation in Hp-infected
samples and non-Hp-infected samples. A two-sided P<0.05 was
considered to indicate a statistically significant difference.
Results
Verification of experimental
reliability
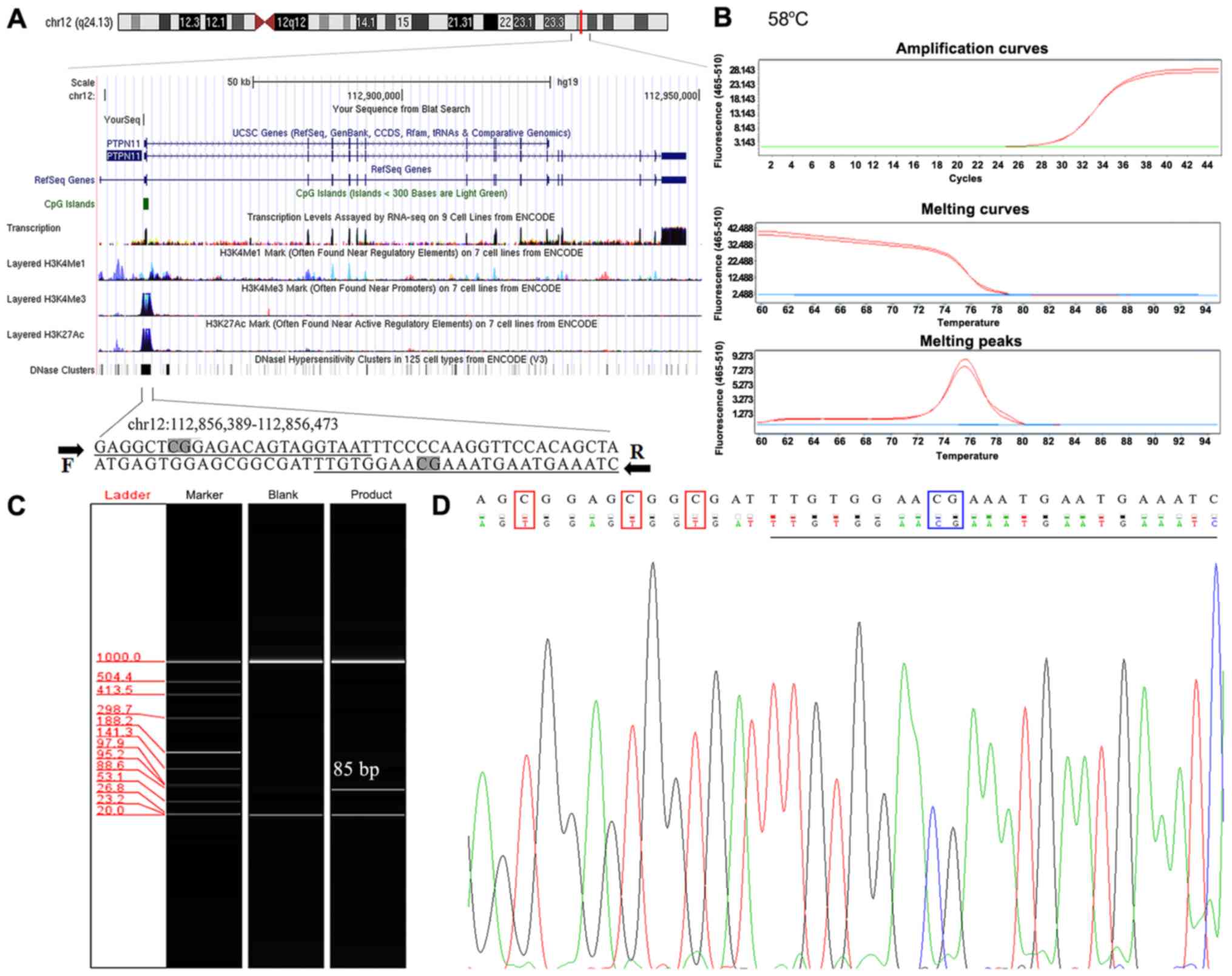
The specificity of primers in this experiment was
verified by Oligo 6.0 software and NCBI primer-blast tool. The
evaluation indicated that the primers were methylation-specific.
Each of the upstream and downstream primers used in the present
study contained one CpG cytosine site and multiple non-CpG cytosine
sites (Fig. 1A). The SYBR-green qMSP
product formed a single dissolution curve at ~76°C, suggesting that
the qMSP product exhibited a uniform melting temperature (Fig. 1B). In addition, the qMSP product was
further analyzed by automated capillary electrophoresis and the
results indicated a single band of 85 bp, confirming that the
amplified qMSP products were homogenous (Fig. 1C). To further verify the specificity
of the primers, random qMSP products were selected for Sanger
sequencing and the results confirmed that the bisulfite conversion
of the DNA template was complete (Fig.
1D). Therefore, all the aforementioned quality control results
indicated that the qMSP process was unlikely to amplify fragments
that were incompletely converted.
PTPN11 hypomethylation exists in GC
and upregulates PTPN11 expression
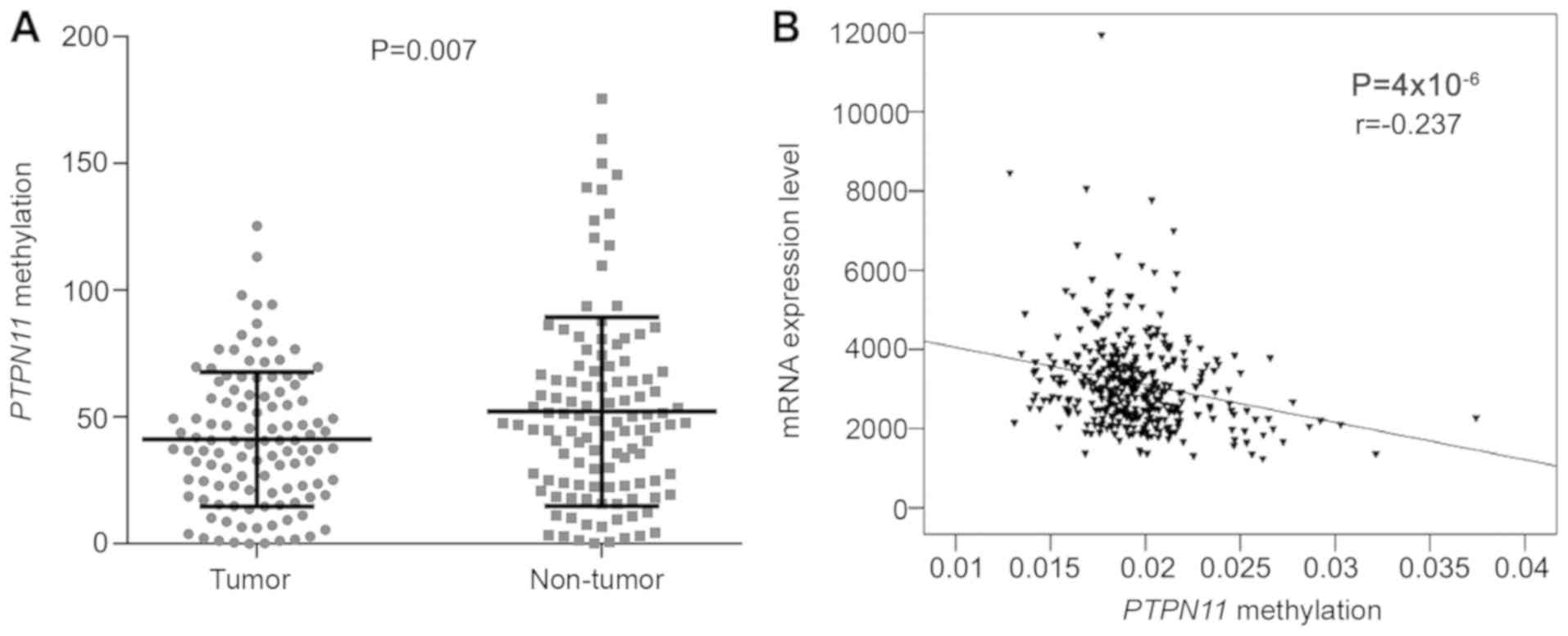
The results indicated that the PTPN11
promoter was significantly hypomethylated in GC tissues compared
with its corresponding methylation levels in the adjacent normal
tissues [mean with standard deviation (SD): 40.91±26.33 vs.
51.99±37.37, P=0.007, Fig. 2A]. In
addition, PTPN11 expression data was extracted from 478 GC
samples present in the TCGA database of Stomach Adenocarcinoma. The
results indicated an inverse correlation between PTPN11
methylation and PTPN11 expression (P=4×10−6,
r=−0.237, Fig. 2B).
Results of the subgroup analyses
Subgroup analyses by different clinical phenotypes
were performed to compare PTPN11 methylation levels between
the tumor and adjacent normal samples. The data demonstrated that
PTPN11 hypomethylation was specific to male subjects
(39.44±25.93 vs. 52.06±37.10, P=0.015) and the patients with
history of heavy drinking (36.01±21.16 vs. 60.07±46.66, P=0.019,
Fig. 3A). In addition, the
association of PTPN11 hypomethylation with GC was specific
to patients with low/no tumor differentiation (39.97±26.01 vs.
50.96±35.90, P=0.010), positive lymphatic metastasis [LN (+),
39.45±26.20 vs. 53.34±38.49, P=0.002] and tumor, node and
metastasis (TNM) stage III+IV (38.62±25.72 vs. 49.91±36.00,
P=0.008; Fig. 3B).
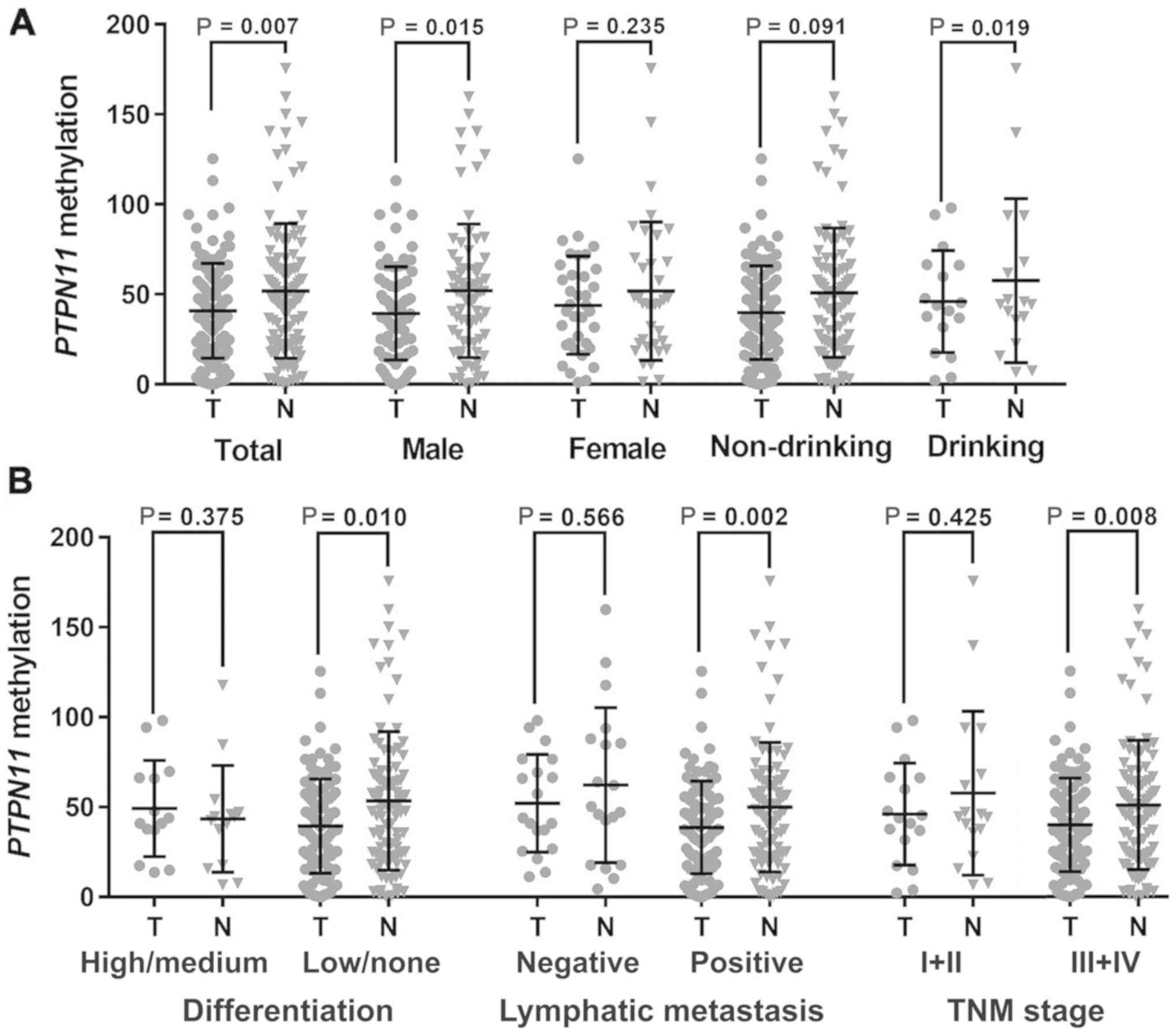 | Figure 3.Subgroup analysis by clinical
characteristics. (A) Subgroup tests by gender and drinking history.
(B) Subgroup tests by gastric cancer differentiation, LN and TNM
stage. The plots are presented as mean with standard deviation. The
P-value was calculated by the paired-samples t test. A significant
P-value was indicated in the following subgroups: Male subjects
(39.44±25.93 vs. 52.06±37.10, P=0.015), heavy drinking subjects
(36.01±21.16 vs. 60.07±46.66, P=0. 019), low/no differentiation
(39.97±26.01 vs. 50.96±35.90, P=0.010), positive LN (39.45±26.20
vs. 53.34±38.49, P=0.002) and TNM stage III+IV (38.62±25.72 vs.
49.91±36.00, P=0.008). TNM, tumor, node, metastasis; LN, lymphatic
metastasis; T, tumor tissue; N, adjacent normal tissue. |
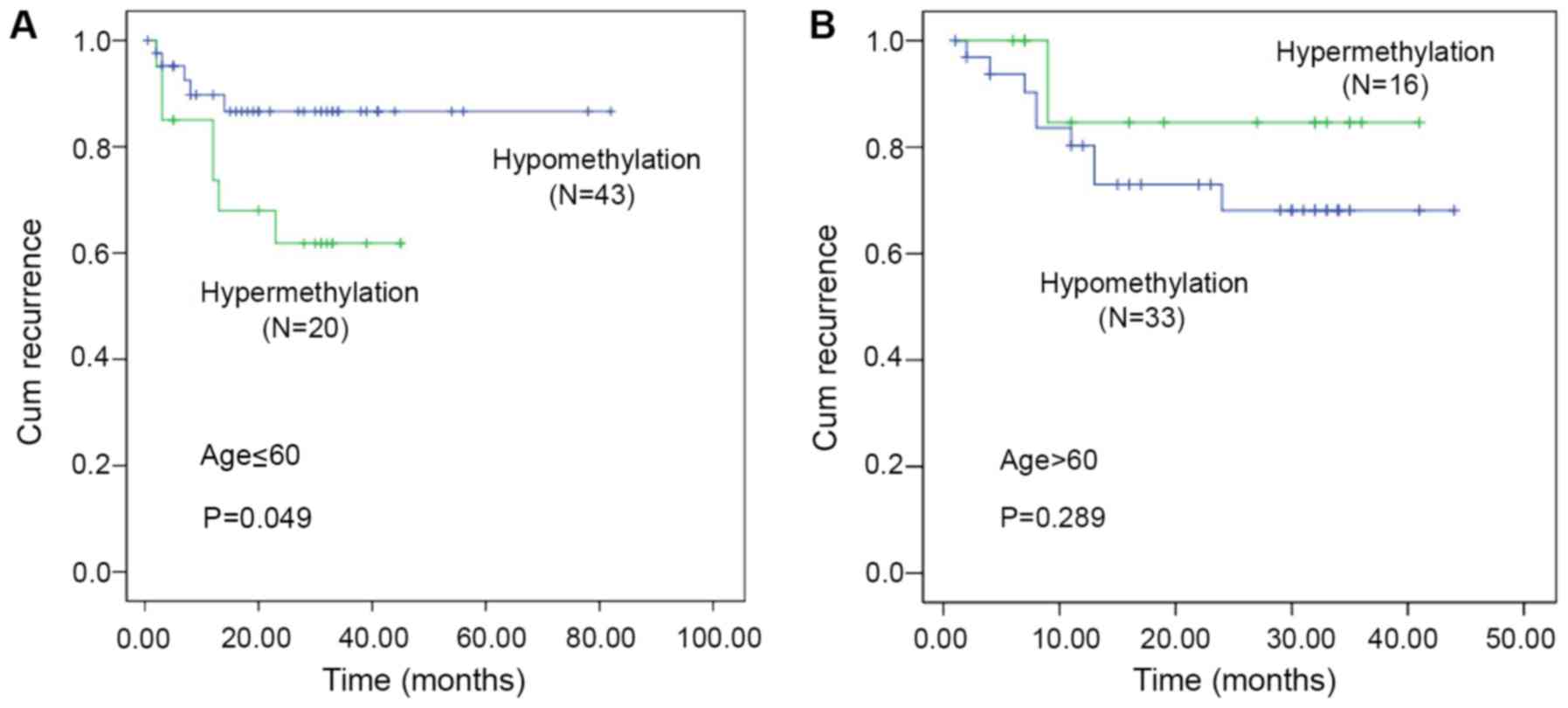
Hypomethylation cohort aged ≤60 tends
to have a higher recurrence rate
During a seven-year follow-up of 112 GC patients,
five patients were lost to follow-up and 34 patients did not
survive. The groups that exhibited higher tumor methylation levels
compared with those noted in the adjacent normal tumors were
defined as the hypermethylation cohort. The subjects that exhibited
lower methylation levels in tumor vs. normal tissues were defined
as the hypomethylation cohort. Kaplan-Meier analysis indicated no
statistical significance between PTPN11 methylation levels
and overall survival (P=0.484) or tumor recurrence (P=0.485).
However, when stratified by age, the hypomethylation cohort aged
≤60 years demonstrated a higher recurrence rate of GC (mean
recurrence: 25.03 vs. 22.25 months, P=0.049; Fig. 4A). In addition, no significant
association was noted between PTPN11 methylation levels and
GC recurrence in different methylation cohorts aged >60 years
(mean recurrence: 22.19 months vs. 22.76 months, P=0.289, Fig. 4B).
Association between Hp infection and
PTPN11 hypomethylation
To further investigate the association between Hp
infection and PTPN11 hypomethylation, the data from the
samples and those from the TCGA database were analyzed (eight CpGs,
Fig. 5A). In China, the detection of
Hp is not a routine assay used in the screening of gastric cancer.
In the present study, the samples were isolated from 9 patients
with Hp, including one Hp(+) patient and eight Hp(−) patients. The
data indicated that the tumor Hp(+) tissues exhibited decreased
PTPN11 methylation levels compared with those noted in the
non-tumor Hp(+) tissues (PMR: 46.88 vs. 53.62). Similarly, the
tumor Hp(−) tissues exhibited lower PTPN11 methylation
levels than the non-tumor Hp(−) tissues although the difference was
not significant (PMR: 43.59±27.90 vs. 50.76±31.21, P=0.664,
Fig. 5B). In addition, the
methylation levels of eight CpG sites from the TCGA database with
Hp infection were compared (Fig.
5A). The analysis demonstrated that the Hp infection status in
the tumor samples was not associated with the levels of
PTPN11 methylation. However, Hp infection was associated
with hypermethylation of 2 PTPN11 CG sites in non-tumor
tissues (cg09337511: P=0.027, cg24032304: P=0.016). A total of four
PTPN11 CG sites were noted that exhibited significant
hypomethylation in the tumor Hp(−) compared with the corresponding
non-tumor Hp(−) tissues, (cg10069827: P=0.003, cg08573574: P=0.011,
cg09337511: P=0.023, cg27541540: P=0.0003).
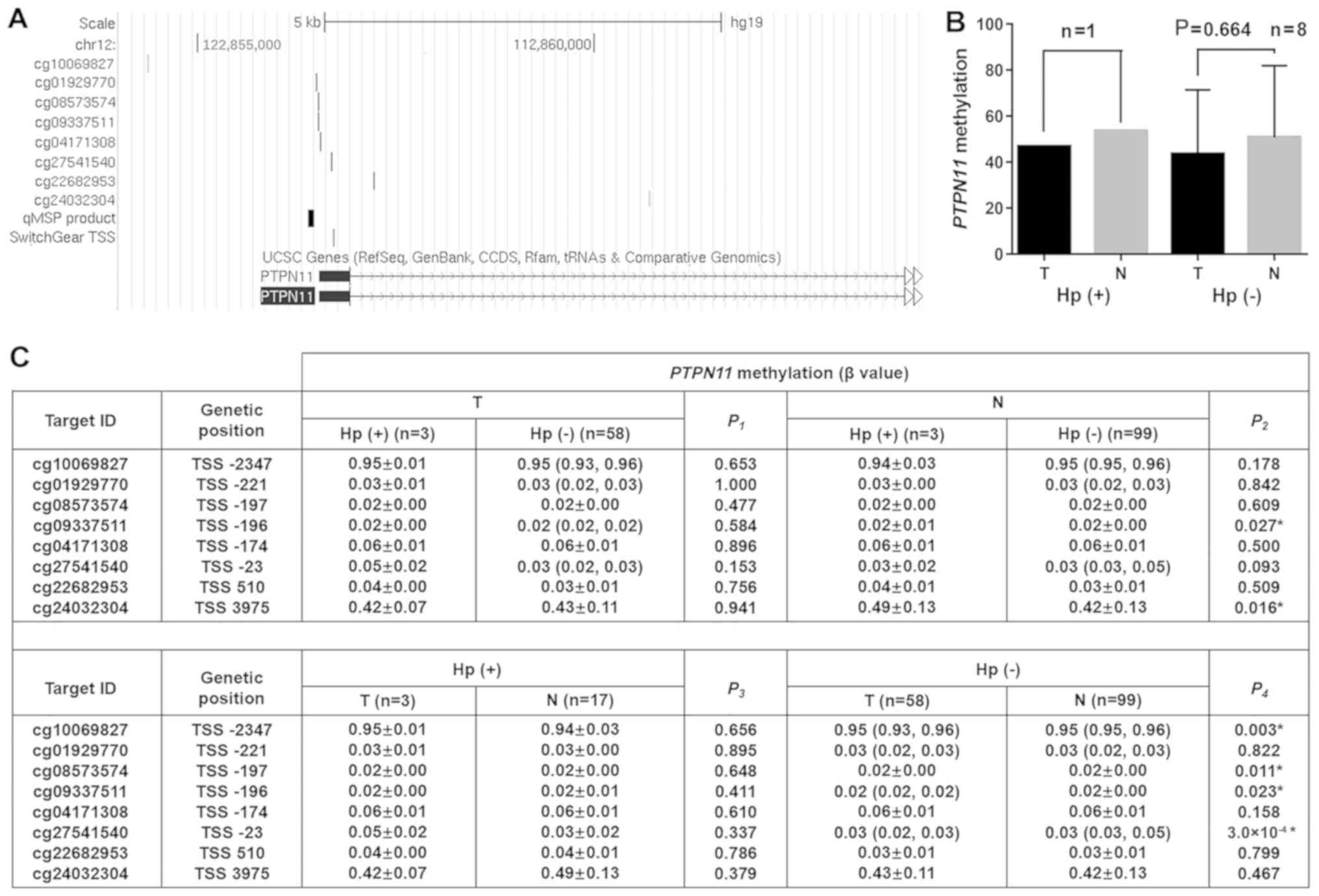 | Figure 5.Association between Hp infection and
PTPN11 methylation. (A) Genomic locations of the qMSP
product, CpG sites in TCGA and SwitchGear TSS at PTPN11
locus (Hg19). (B) The samples were compared according to the Hp
status and PTPN11 methylation levels of the tumor and the
adjacent non-tumor tissues. The comparisons were performed using
the paired sample t test. (C) TCGA samples were grouped according
to tumor status and Hp infection, and the association between
PTPN11 methylation and Hp infection was analyzed. The normal
distributed data were described as mean ± standard deviation, and
the non-normal distributed data were described as median with
interquartile ranges. When the two sets of data were normally
distributed, the P-value was calculated by the two independent
sample t tests. In any other case, a non-parametric Mann-Whitney U
test was used. *P<0.05. Hp, Helicobacter pylori; TGCA,
The Cancer Genome Atlas; PTPN11, Tyrosine-Protein Phosphatase
Non-Receptor Type 11; TSS, transcriptional start site; T, tumor
tissue; N, non-tumor tissue; n, number; qMSP, quantitative
methylation specific PCR. |
Discussion
Deregulation of SHP-2 (a protein encoded by
PTPN11) by the Hp-related protein Cag-A can lead to GC risk
(3). Although ~70% of patients with
GC are Hp-positive, only 1–2% of patients with chronic gastritis or
Hp-positive peptic ulcer eventually develop GC (42). The expression levels of the SHP-2
protein were increased in Hp(+) GC patients, although the
differences noted were not significant (43). In the current study, it was
demonstrated that PTPN11 was hypomethylated in GC, which has
not been previously reported. Since abnormal gene methylation
always interferes with expression (44), it was hypothesized that
hypomethylation of PTPN11 may be one of the mechanisms
involved in the development of GC.
PTPN11 is a specific gene, which exhibits a
two-sided effect in cancer progression (45). PTPN11 has been characterized
as a tumor suppressor gene (46,47) in
liver cancer and as a proto-oncogene in leukemia (45). PTPN11 expression was increased
in leukemia (48), breast cancer
(49) and in thyroid tumors
(50), whereas it was decreased in
colon (51) and liver cancer
(46,52). Previous studies have shown an
increase in PTPN11 mRNA and protein levels in GC (43,53,54),
suggesting that PTPN11 may be a proto-oncogene in GC. In the
present study, the results indicated that PTPN11 was
hypomethylated in GC. In addition, TCGA data analysis revealed an
inverse correlation between PTPN11 methylation and
PTPN11 expression. These findings may explain the decreased
methylation pattern and the high expression profile of
PTPN11 in GC.
In addition, subgroup analysis indicated that
PTPN11 hypomethylation was specific for male subjects and GC
patients with a history of heavy drinking. Chinese men were more
likely to suffer from GC and the male mortality rate in China was
~twice that noted in Chinese women (2). Heavy drinking is a risk factor for GC
(55). Therefore, whether
PTPN11 hypomethylation occurs only in male subjects and
heavy drinkers requires further studies. In addition, it was found
that PTPN11 hypomethylation was specific for poorly
differentiated GC patients and TNM III+IV GC patients. Patients
with advanced TNM staging exhibited a poor prognosis (56). Poorly differentiated cancer cells are
also a feature of advanced GC. Therefore, the present study
hypothesized that PTPN11 hypomethylation may be associated
with the progression of GC, which can be further studied in the
future.
Previous studies have shown that PTPN11
overexpression indicates poor prognosis in liver cancer patients
(57). The PTPN11 rs2301756
polymorphism has been shown to be associated with decreased risk of
GC and with an improved response to chemotherapy (34). In addition, the gene panel containing
PTPN11 in colorectal cancer and oral squamous cell carcinoma
has a high prognostic value (58,59). The
3-year survival rate of GC patients with high SHP-2 expression was
significantly decreased compared with patients with low SHP-2
expression and the postoperative recurrence mortality of high SHP-2
expression was also significantly increased compared with patients
with low SHP-2 expression (53). The
correlation between the methylation status of the PTPN11
gene and the prognosis of GC has not been reported previously.
Therefore, a 7-year follow-up of GC patients was performed in the
current study and the parameters survival time and postoperative
recurrence time were assessed. Although the current analysis
indicated that PTPN11 methylation exhibited no prognostic
value on the survival and recurrence of patients with GC, following
stratification by age, it was shown that the hypomethylation cohort
with an average age ≤60 years exhibited a higher recurrence rate of
GC. SHP-2 abnormalities were associated with tumor cell
proliferation, invasion and metastasis (3). Young cancer patients may be more prone
to tumor progression due to increased body metabolism compared with
elderly cancer patients. Therefore, PTPN11 hypomethylation
may be a prognostic indicator for postoperative recurrence of GC
patients under 60 years of age.
Several studies have indicated that the SHP-2
protein, which is encoded by PTPN11 is an intracellular
target of Cag-A (3,6,8). This
protein is a virulence factor of Hp (3,6,8). Recently, Jiang et al (43) demonstrated that although the
expression levels of SHP-2 in the gastric cancer Hp(+) group were
increased compared with those noted in the gastric cancer Hp(−)
group, the differences noted were not statistically significant.
The present study revealed that both Hp infection and tumor status
may change the methylation levels of specific PTPN11 CpG
sites. However, only one and three tumor Hp (+) samples were found
in the samples used in the present study and in those derived from
the TCGA database. Therefore, the present findings require
verification in the future with larger sample sizes of subjects
with Hp infection.
The present study exhibits certain limitations.
Firstly, the samples used and those derived from the TCGA database
were not sufficient to ensure a plausible association of GC
incidence and PTPN11 methylation with the status of Hp
(positive or negative). Therefore, the correlation between Hp
infection and PTPN11 methylation should be further tested in
larger datasets with known Hp infection status. Secondly, although
an inverse association between PTPN11 methylation and mRNA
expression was noted by TCGA data analysis, future work is required
to evaluate whether PTPN11 hypomethylation can lead to
elevated SHP-2 expression in GC patients.
In summary, the present study indicated that
PTPN11 was hypomethylated in GC and that this could be
associated with SHP-2 overexpression in GC. Future study is
required to verify this hypothesis. Hypomethylation of
PTPN11 may be specific for men, patients with a history of
heavy drinking, patients with poor tumor differentiation and
patients with TNM III+IV stage GC. In addition, the present study
further demonstrated that PTPN11 hypomethylation could
predict recurrence of GC in patients aged ≤60 years.
Acknowledgements
Not applicable.
Funding
The present study was supported by the K. C. Wong
Magna Fund in Ningbo University.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
SD, CX and LX cenceived and designed the study and
gave the final approval of the submitted version. CZ, RP, JT, JW,
BL and TH performed the data analyses and conducted the
experiments. CZ contributed to figure preparation. LX collected
samples and wrote the paper. All the authors have read and approved
the final manuscript.
Ethics approval and consent to
participate
All patients who participated in the present study
had signed an informed consent form. The study was approved by the
Ethics Committee of Ningbo University.
Patient consent for publication
Written informed consent was obtained from each
patient.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yamazaki S, Yamakawa A, Ito Y, Ohtani M,
Higashi H, Hatakeyama M and Azuma T: The CagA protein of
Helicobacter pylori is translocated into epithelial cells and binds
to SHP-2 in human gastric mucosa. J Infect Dis. 187:334–337. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang J, Zhang F and Niu R: Functions of
Shp2 in cancer. J Cell Mol Med. 19:2075–2083. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Inoue K: Gastric cancer risk
classification (ABC classification). Nihon Rinsho. 71:1472–1478.
2013.(In Japanese). PubMed/NCBI
|
|
6
|
Hatakeyama M: H. pylori oncoprotein CagA
and gastric cancer. Nihon Rinsho. 70:1699–1704. 2012.(In Japanese).
PubMed/NCBI
|
|
7
|
Hatakeyama M: Deregulation of SHP-2
tyrosine phosphatase by the Helicobacter pylori virulence factor
CagA. Keio J Med. 51 (Suppl 2):S26–S32. 2002. View Article : Google Scholar
|
|
8
|
Uemura N, Okamoto S, Yamamoto S, Matsumura
N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N and Schlemper RJ:
Helicobacter pylori infection and the development of gastric
cancer. N Engl J Med. 345:784–789. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Müller PJ, Rigbolt KT, Paterok D, Piehler
J, Vanselow J, Lasonder E, Andersen JS, Schaper F and Sobota RM:
Protein tyrosine phosphatase SHP2/PTPN11 mistargeting as a
consequence of SH2-domain point mutations associated with Noonan
Syndrome and leukemia. J Proteomics. 84:132–147. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mutesa L, Pierquin G, Janin N, Segers K,
Thomée C, Provenzi M and Bours V: Germline PTPN11 missense mutation
in a case of Noonan syndrome associated with mediastinal and
retroperitoneal neuroblastic tumors. Cancer Genet Cytogenet.
182:40–42. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee KA, Williams B, Roza K, Ferguson H,
David K, Eddleman K, Stone J, Edelmann L, Richard G, Gelb BD and
Kornreich R: PTPN11 analysis for the prenatal diagnosis of Noonan
syndrome in fetuses with abnormal ultrasound findings. Clin Genet.
75:190–194. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Motegi S, Yokoyama Y, Ogino S, Yamada K,
Uchiyama A, Perera B, Takeuchi Y, Ohnishi H and Ishikawa O:
Pathogenesis of multiple lentigines in LEOPARD syndrome with PTPN11
gene mutation. Acta Derm Venereol. 95:978–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tartaglia M, Niemeyer CM, Shannon KM and
Loh ML: SHP-2 and myeloid malignancies. Curr Opin Hematol.
11:44–50. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ibáñez M, Carbonell-Caballero J,
García-Alonso L, Such E, Jiménez-Almazán J, Vidal E, Barragán E,
López-Pavía M, LLop M, Martín I, et al: The mutational landscape of
acute promyelocytic leukemia reveals an interacting network of
co-occurrences and recurrent mutations. PLoS One. 11:e01483462016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen L, Chen W, Mysliwski M, Serio J, Ropa
J, Abulwerdi FA, Chan RJ, Patel JP, Tallman MS, Paietta E, et al:
Mutated Ptpn11 alters leukemic stem cell frequency and reduces the
sensitivity of acute myeloid leukemia cells to Mcl1 inhibition.
Leukemia. 29:1290–1300. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yuan L, Lu L, Yang Y, Sun H, Chen X, Huang
Y, Wang X, Zou L and Bao L: Genetic mutational profiling analysis
of T cell acute lymphoblastic leukemia reveal mutant FBXW7 as a
prognostic indicator for inferior survival. Ann Hematol.
94:1817–1828. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang WY, Chen XJ, Wang SC, Guo Y, Liu TF,
Chang LX, Liu F and Zhu XF: Gene mutations and clinical
characteristics in children with juvenile myelomonocytic leukemia.
Zhongguo Dang Dai Er Ke Za Zhi. 17:1–5. 2015.(In Chinese).
PubMed/NCBI
|
|
18
|
Pugh TJ, Morozova O, Attiyeh EF,
Asgharzadeh S, Wei JS, Auclair D, Carter SL, Cibulskis K, Hanna M,
Kiezun A, et al: The genetic landscape of high-risk neuroblastoma.
Nat Genet. 45:279–284. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Obasaju P, Brondon J, Mir S, Fordham LA,
Lee S and Blatt J: Somatic PTPN11 mutation in a child with
neuroblastoma and protein losing enteropathy. J Pediatr Hematol
Oncol. 40:328–330. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
McFarlane J, Knight T, Sinha A, Cole T,
Kiely N and Freeman R: Exostoses, enchondromatosis and
metachondromatosis; diagnosis and management. Acta Orthop Belg.
82:102–105. 2016.PubMed/NCBI
|
|
21
|
Bowen ME, Boyden ED, Holm IA,
Campos-Xavier B, Bonafé L, Superti-Furga A, Ikegawa S,
Cormier-Daire V, Bovée JV, Pansuriya TC, et al: Loss-of-function
mutations in PTPN11 cause metachondromatosis, but not Ollier
disease or Maffucci syndrome. PLoS Genet. 7:e10020502011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Siegfried A, Cances C, Denuelle M, Loukh
N, Tauber M, Cavé H and Delisle MB: Noonan syndrome, PTPN11
mutations, and brain tumors. A clinical report and review of the
literature. Am J Med Genet A. 173:1061–1065. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rankin J, Short J, Turnpenny P, Castle B
and Hanemann CO: Medulloblastoma in a patient with the PTPN11
p.Thr468Met mutation. Am J Med Genet A. 161A:2027–2029. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Roccograndi L, Binder ZA, Zhang L, Aceto
N, Zhang Z, Bentires-Alj M, Nakano I, Dahmane N and O'Rourke DM:
SHP2 regulates proliferation and tumorigenicity of glioma stem
cells. J Neurooncol. 135:487–496. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Thiel C, Wilken M, Zenker M, Sticht H,
Fahsold R, Gusek-Schneider GC and Rauch A: Independent NF1 and
PTPN11 mutations in a family with neurofibromatosis-Noonan
syndrome. Am J Med Genet A. 149A:1263–1267. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nair S, Fort JA, Yachnis AT and Williams
CA: Optic nerve pilomyxoid astrocytoma in a patient with Noonan
syndrome. Pediatr Blood Cancer. 62:1084–1086. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hu Z, Wang X, Fang H, Liu Y, Chen D, Zhang
Q, Liu X, Wei D, Qu C and Wang S: A tyrosine phosphatase SHP2
gain-of-function mutation enhances malignancy of breast carcinoma.
Oncotarget. 7:5664–5676. 2016.PubMed/NCBI
|
|
28
|
Beigbeder A, Chartier FJM and Bisson N:
MPZL1 forms a signalling complex with GRB2 adaptor and PTPN11
phosphatase in HER2-positive breast cancer cells. Sci Rep.
7:115142017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang Q, Li Y, Zhao R, Wang X, Fan C, Xu
Y, Liu Y, Li J and Wang S: The gain-of-function mutation E76K in
SHP2 promotes CAC tumorigenesis and induces EMT via the
Wnt/β-catenin signaling pathway. Mol Carcinog. 57:619–628. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gagné-Sansfaçon J, Coulombe G, Langlois
MJ, Langlois A, Paquet M, Carrier J, Feng GS, Qu CK and Rivard N:
SHP-2 phosphatase contributes to KRAS-driven intestinal oncogenesis
but prevents colitis-associated cancer development. Oncotarget.
7:65676–65695. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang N, Liu H, Yue G, Zhang Y, You J and
Wang H: Molecular Heterogeneity of Ewing Sarcoma as Detected by Ion
Torrent Sequencing. PLoS One. 11:e01535462016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Je EM, Choi YJ, Yoo NJ and Lee SH:
Oncogenic PTPN11 mutations are rare in solid tumors. Pathol Oncol
Res. 21:225–227. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Martinelli S, Carta C, Flex E, Binni F,
Cordisco EL, Moretti S, Puxeddu E, Tonacchera M, Pinchera A,
McDowell HP, et al: Activating PTPN11 mutations play a minor role
in pediatric and adult solid tumors. Cancer Genet Cytogenet.
166:124–129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhuo C, Shao M, Chen C, Lin C, Jiang D,
Chen G, Tian H, Wang L, Li J and Lin X: Chemotherapy effectiveness
and prognosis of gastric cancer influenced by PTPN11 polymorphisms.
Cell Physiol Biochem. 39:1537–1552. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
He C, Tu H, Sun L, Xu Q, Li P, Gong Y,
Dong N and Yuan Y: Helicobacter pylori-related host gene
polymorphisms associated with susceptibility of gastric
carcinogenesis: A two-stage case-control study in Chinese.
Carcinogenesis. 34:1450–1457. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhu F, Loh M, Hill J, Lee S, Koh KX, Lai
KW, Salto-Tellez M, Iacopetta B, Yeoh KG and Soong R; Singapore
Gastric Cancer Consortium, : Genetic factors associated with
intestinal metaplasia in a high risk Singapore-Chinese population:
A cohort study. BMC Gastroenterol. 9:762009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jiang J, Jia ZF, Kong F, Jin MS, Wang YP,
Tian S, Suo J and Cao X: Association of polymorphism of PTPN 11
encoding SHP-2 with gastric atrophy but not gastric cancer in
Helicobacter pylori seropositive Chinese population. BMC
Gastroenterol. 12:892012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hishida A, Matsuo K, Goto Y, Naito M,
Wakai K, Tajima K and Hamajima N: Associations of a PTPN11 G/A
polymorphism at intron 3 with Helicobactor pylori seropositivity,
gastric atrophy and gastric cancer in Japanese. BMC Gastroenterol.
9:512009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sapari NS, Loh M, Vaithilingam A and Soong
R: Clinical potential of DNA methylation in gastric cancer: A
meta-analysis. PLoS One. 7:e362752012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li J, Chen C, Bi X, Zhou C, Huang T, Ni C,
Yang P, Chen S, Ye M and Duan S: DNA methylation of CMTM3, SSTR2,
and MDFI genes in colorectal cancer. Gene. 630:1–7. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kristensen LS, Mikeska T, Krypuy M and
Dobrovic A: Sensitive Melting Analysis after Real Time-Methylation
Specific PCR (SMART-MSP): High-throughput and probe-free
quantitative DNA methylation detection. Nucleic Acids Res.
36:e422008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Testino G, Testino R and Ancarani AO:
Helicobacter pylori and gastric carcinogenesis. Recenti Prog Med.
92:573–577. 2001.(In Italian). PubMed/NCBI
|
|
43
|
Jiang J, Jin MS, Kong F, Wang YP, Jia ZF,
Cao DH, Ma HX, Suo J and Cao XY: Increased expression of tyrosine
phosphatase SHP-2 in Helicobacter pylori-infected gastric cancer.
World J Gastroenterol. 19:575–580. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fukushige S and Horii A: DNA methylation
in cancer: A gene silencing mechanism and the clinical potential of
its biomarkers. Tohoku J Exp Med. 229:173–185. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li S, Hsu DD, Wang H and Feng GS: Dual
faces of SH2-containing protein-tyrosine phosphatase Shp2/PTPN11 in
tumorigenesis. Front Med. 6:275–279. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bard-Chapeau EA, Li S, Ding J, Zhang SS,
Zhu HH, Princen F, Fang DD, Han T, Bailly-Maitre B, Poli V, et al:
Ptpn11/Shp2 acts as a tumor suppressor in hepatocellular
carcinogenesis. Cancer Cell. 19:629–639. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jiang C, Hu F, Tai Y, Du J, Mao B, Yuan Z,
Wang Y and Wei L: The tumor suppressor role of Src homology
phosphotyrosine phosphatase 2 in hepatocellular carcinoma. J Cancer
Res Clin Oncol. 138:637–646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xu R, Yu Y, Zheng S, Zhao X, Dong Q, He Z,
Liang Y, Lu Q, Fang Y, Gan X, et al: Overexpression of Shp2
tyrosine phosphatase is implicated in leukemogenesis in adult human
leukemia. Blood. 106:3142–3149. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lucci MA, Orlandi R, Triulzi T, Tagliabue
E, Balsari A and Villa-Moruzzi E: Expression profile of tyrosine
phosphatases in HER2 breast cancer cells and tumors. Cell Oncol.
32:361–372. 2010.PubMed/NCBI
|
|
50
|
Hu ZQ, Ma R, Zhang CM, Li J, Li L, Hu ZT,
Gao QI and Li WM: Expression and clinical significance of tyrosine
phosphatase SHP2 in thyroid carcinoma. Oncol Lett. 10:1507–1512.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cai P, Guo W, Yuan H, Li Q, Wang W, Sun Y,
Li X and Gu Y: Expression and clinical significance of tyrosine
phosphatase SHP-2 in colon cancer. Biomed Pharmacother. 68:285–290.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Luo X, Liao R, Hanley KL, Zhu HH, Malo KN,
Hernandez C, Wei X, Varki NM, Alderson N, Chu C, et al: Dual Shp2
and Pten deficiencies promote non-alcoholic steatohepatitis and
genesis of liver tumor-initiating cells. Cell Rep. 17:2979–2993.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Dong S, Li FQ, Zhang Q, Lv KZ, Yang HL,
Gao Y and Yu JR: Expression and clinical significance of SHP2 in
gastric cancer. J Int Med Res. 40:2083–2089. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kim JS, Shin OR, Kim HK, Cho YS, An CH,
Lim KW and Kim SS: Overexpression of protein phosphatase
non-receptor type 11 (PTPN11) in gastric carcinomas. Dig Dis Sci.
55:1565–1569. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tramacere I, Negri E, Pelucchi C, Bagnardi
V, Rota M, Scotti L, Islami F, Corrao G, La Vecchia C and Boffetta
P: A meta-analysis on alcohol drinking and gastric cancer risk. Ann
Oncol. 23:28–36. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang D, Li C, Xu Y, Xing Y, Qu L, Guo Y,
Zhang Y, Sun X and Suo J: Clinicopathological characteristics and
prognosis of alpha-fetoprotein positive gastric cancer in Chinese
patients. Int J Clin Exp Pathol. 8:6345–6355. 2015.PubMed/NCBI
|
|
57
|
Han T, Xiang DM, Sun W, Liu N, Sun HL, Wen
W, Shen WF, Wang RY, Chen C, Wang X, et al: PTPN11/Shp2
overexpression enhances liver cancer progression and predicts poor
prognosis of patients. J Hepatol. 63:651–660. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Chang W, Gao X, Han Y, Du Y, Liu Q, Wang
L, Tan X, Zhang Q, Liu Y, Zhu Y, et al: Gene expression
profiling-derived immunohistochemistry signature with high
prognostic value in colorectal carcinoma. Gut. 63:1457–1467. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Chen SJ, Liu H, Liao CT, Huang PJ, Huang
Y, Hsu A, Tang P, Chang YS, Chen HC and Yen TC: Ultra-deep targeted
sequencing of advanced oral squamous cell carcinoma identifies a
mutation-based prognostic gene signature. Oncotarget.
6:18066–18080. 2015.PubMed/NCBI
|