Introduction
Prostate cancer (PCa) is one of the most common
cancers worldwide (1), with an
incidence rate that has increased in China in recent years
(2). Prostate specific antigen (PSA)
screening is a primary method for the surveillance of PCa. However,
PSA exhibits a low specificity, which leads to the incorrect
diagnoses and treatment of patients with PCa (3). Therefore, the discovery and
identification of new biomarkers are essential for monitoring
patients with PCa.
Citron-kinase (CIT) comprises an amino-terminal
serine/threonine kinase domain, which is highly conserved between
insects and mammals (4). It has been
revealed that CIT is critical for cytokinesis (5,6). CIT is
also involved in the cleavage of the furrow and midbody, which is
essential to cellular abscission (7–9).
Furthermore, CIT phosphorylates the regulatory light chain of
myosin II at the Ser 19/Thr 18 positions, consequently activating
myosin II, which is the primary motor protein and responsible for
cytokinesis (10).
In the current study, increased expression of CIT
was identified as an oncogene by bioinformatic analysis. This
result was verified by reverse transcription-quantitative (RT-q)PCR
and immunochemistry. The aim of the current study was to assess the
role of CIT in PCa and to determine the possibility of using CIT in
the diagnosis and therapy of patients with PCa.
Materials and methods
Dataset gene expression analysis
mRNA expression profiles and associated PCa clinical
datasets (PRAD_2015_02_24) from The Cancer Genome Atlas (TCGA) were
downloaded from the University of California Santa Cruz cancer
genome browser (https://xena.ucsc.edu/welcome-to-ucsc-xena/). The
profile contained 52 cases of normal tissue and 499 cases of
primary PCa tissue. Microarray data were normalized and compared
using Biometric Research Program (BRB) array tools developed by Dr
Richard Simon and Dr Yingdong Zhao (http://linus.nci.nih.gov/BRB-ArrayTools) (11). Differentially expressed genes (DEGs)
were filtered by comparing cancer and normal tissue, Gleason grades
≥7 and Gleason grades <7, PSA ≥10 ng/ml and PSA <10 ng/ml,
Ta-2 and T3-4, regional lymph node metastasis (N1) and no regional
lymph nodes metastasis (N0), and metastasis to distant organs (M1)
and no distant metastasis (M0). DEGs were defined as a fold-change
(FC) >1 and P<0.01. Volcano plots were established to
visualize the genes that were screened.
Patients and tissues
To determine the expression of CIT mRNA in patients
with PCa, fresh PCa tissue (n=35) and benign prostatic hyperplasia
tissue (BPH; n=20) were collected from the First Affiliated
Hospital of Chongqing Medical University (Chongqing, China). All
samples were confirmed by pathological examination and subsequently
stored in liquid nitrogen (−196°C) for mRNA analysis. Patient
characteristics are shown in Table
I.
 | Table I.Characteristics of prostate cancer
patients. |
Table I.
Characteristics of prostate cancer
patients.
| Items | N (%) |
|---|
| Sample type |
|
|
Aggressive PCa | 131 (48.34) |
| Primary
PCa | 140 (51.66) |
| Origin |
|
| The
First Affiliated Hospital of Chongqing Medical University | 156 (57.56) |
| The
Zigong No. 4 People's Hospital | 53 (19.56) |
| The
Zigong No. 1 People's Hospital | 62 (22.88) |
| Age, years |
|
|
<70 | 25 (9.23) |
|
70–79 | 148 (54.61) |
|
≥80 | 98 (36.16) |
| Gleason score |
|
|
<7 | 88 (32.47) |
| 7 | 89 (32.84) |
| ≥8 | 94 (34.69) |
| PSA level |
|
|
<4 | 34 (12.55) |
|
4-9.9 | 35 (12.92) |
|
10-19.9 | 41 (15.13) |
|
≥20 | 161 (59.40) |
| pT stage |
|
|
≤T2 | 150 (55.35) |
| T3 | 75 (27.68) |
| T4 | 46 (16.97) |
| pN stage |
|
| N0 | 255 (94.10) |
| N1 | 16 (5.90) |
| pM stage |
|
| M0 | 254 (93.73) |
| M1 | 17 (6.27) |
| Therapy |
|
|
ADT | 88 (32.47) |
| PR | 183 (67.53) |
| BCR after ADT |
|
| No | 15 (17.05) |
|
Yes | 68 (77.27) |
|
Loss | 5 (5.68) |
| CSM after RP |
|
| No | 137 (74.86) |
|
Yes | 35 (19.13) |
| Loss or
death for other cause | 11 (6.01) |
Formalin fixed paraffin embedded BPH (n=39) and PCa
(n=271) samples were retrieved from the Pathology Department of the
First Affiliated Hospital of Chongqing Medical University, Zigong
Fourth People's Hospital and Zigong First People's Hospital from
2005 to 2017. None of the patients recruited into the present study
received chemotherapy, radiation therapy androgen deprivation (ADT)
or radical prostatectomy (RP) prior to enrollment. The use of
tissue was approved by the Ethics committee of the First Affiliated
Hospital of Chongqing Medical University (approval no. 2018-69),
Zigong Fourth People's Hospital (approval no. 2018-32) and Zigong
First People's Hospital (approval no. 2018-47).
Patients were sub-divided into a high-risk group
(HR-group) when one of the following criteria was met: i) Gleason
grades ≥8; ii) T2c-T4 tumor or iii) PSA level ≥20 ng/ml (12). Patients that exhibited local invasion
and metastasis were considered to have aggressive PCa (13). The Gleason score was evaluated
according to the guidelines conducted by World Health Organization
and the International Society of Urological Pathology (14,15).
Moreover, patients with PCa were stratified into three grades
including low, middle and high grade, which determined by a Gleason
sum <5, between 5 to 7, and >7, respectively (16).
Patients who received RP were followed-up by a
telephone call and patients who received ADT were monitored via
continuous serum PSA surveillance (in 6-month intervals). The
follow-up time of patients receiving ADT was 14.6±8.2 months with
54.2% patients being followed-up for more than one year. The
follow-up time of patients receiving RP was 25±20.3 months, with
66.3% patients being followed-up for more than one year. Due to the
different therapies administered and the follow-up methods used,
follow-up outcomes were stratified to cancer-specific mortality
(CSM) for patients receiving RP and biochemical recurrence (BCR)
for patients receiving ADT. BCR was defined when patients exhibited
a PSA level ≥0.2 ng/ml on at least two consecutive postoperative
occasions, as described previously (17).
Immunohistochemistry
Tissues from the patients were fixed in 10% buffered
formalin at room temperature for 2 days, and then were transferred
to 70% ethanol overnight. The infiltrated tissues were embedded
into paraffin blocks. A single 3-µm section was cut from each
block. Immunochemistry and the assessment of immunoreactivity were
performed as described previously (18). The sections were incubated with
primary antibody (1:50; cat. no. YT0931; ImmunoWay Biotechnology
Company) at 4°C overnight. CIT immunoreactivity was scored by
multiplying the staining intensity by the percentage of area
stained. Intensity was scored as follows: 0 (no staining), 1 (weak
staining), 2 (moderate staining) and 3 (strong staining). The
percentage of area stained was defined as follows: 0 (no staining),
1 (1–25% of cells stained), 2 (26–50% of cells stained), 3 (51–75%
of cells stained), 4 (>75% of cells stained). A high expression
of CIT (H-CIT) was defined as 6–12, whereas a low expression of CIT
(L-CIT) was defined as 0–5 (18).
CIT immunohistochemical staining was scored under a light
microscope independently by two experienced pathologists (LY and
ZT) who were blinded to patient clinical information.
RT-qPCR
The isolation of total RNA and RT-qPCR were
performed as described previously (19). All samples were amplified in
triplicate. To calculate the expression of CIT mRNA in samples,
GAPDH was used as reference gene. The following primers were used
in RT-qPCR: CIT forward, 5′-ACCATAGCTGAGTTACAGGAGC-3′ and reverse,
5′-GTCCCCGGTTGCTTTCTCT-3′; GAPDH forward,
5′-TGGAAGGACTCATGACCACA-3′ and reverse,
5′-TTCAGCTCAGGGATGACCTT-3′.
Statistical analyses
Statistical analyses were performed using SPSS 20.0
software (IBM Corp.) and Prism 5.0 software (GraphPad Software,
Inc.). Comparison between groups was made by unpaired t-tests or
Kruskal-Wallis test. The association between CIT expression and the
clinicopathological parameters of patients with PCa was analyzed
using a χ2 test. Follow-up outcomes were stratified to
CSM for patients that received RP or BCR for patients that received
ADT. The Kaplan-Meier method and a log-rank test were established
to plot survival curves. Univariate and multivariate Cox regression
analysis by backward selection were used to evaluate the prognostic
significance of CIT for predicting BCR and CSM. The experiments
were repeated 3 times and the data were presented as mean ±
standard error. P<0.05 was considered to indicate a
statistically significant difference.
Results
CIT is screened as an oncogene in
PCa
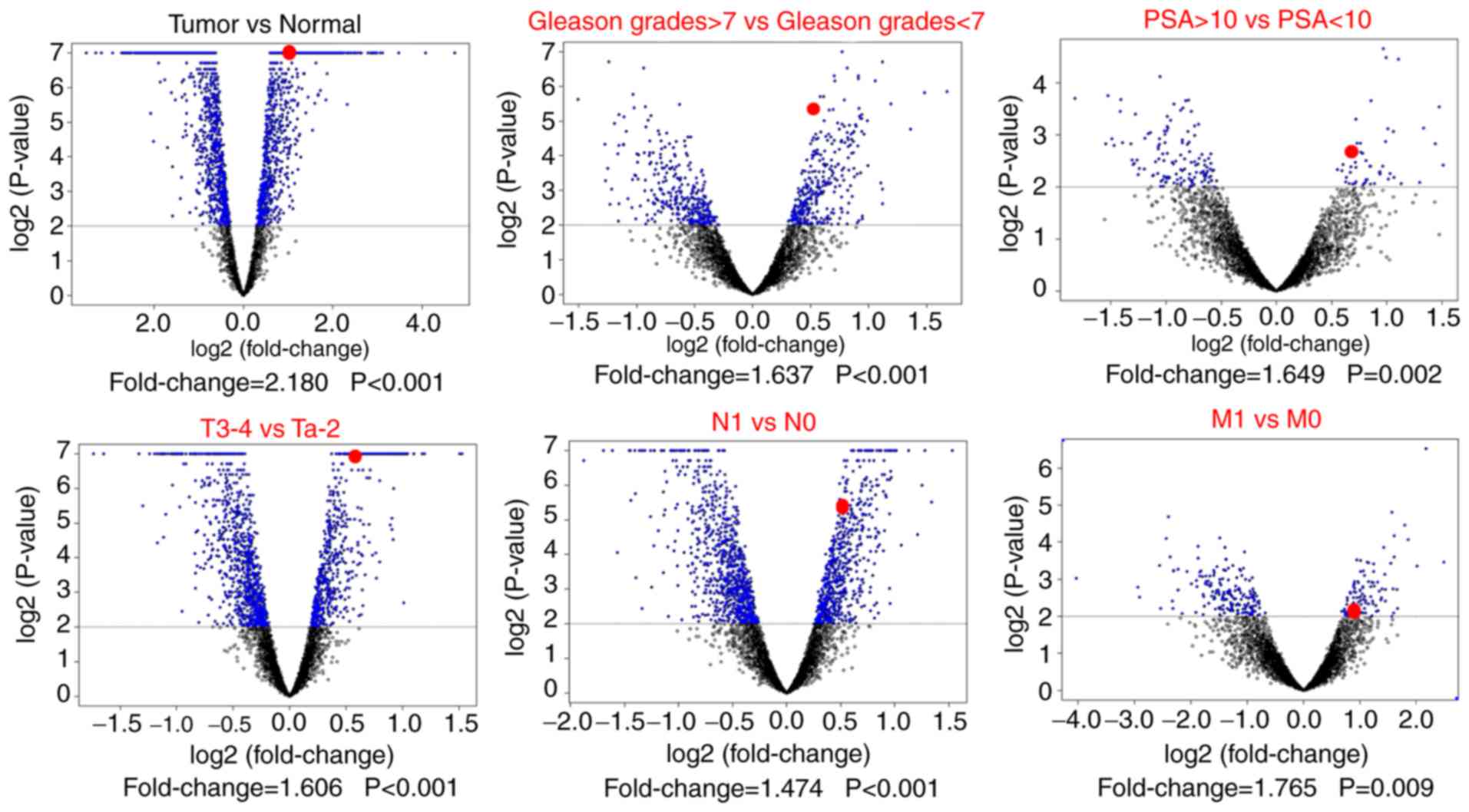
A total of 3,279 DEGs were filtered from the TCGA
profile when comparing normal prostate gland tissue with PCa
tissue. A further screening was performed by dividing groups
according to Gleason grades, serum PSA levels and tumor, node and
metastasis (TNM) stages (Fig. 1). A
total of 30 DEGs were identified to be significant in all of these
comparisons (Table II).
Significantly high expression of CIT mRNA was exhibited in PCa
samples (FC=2.180; P<0.001) and in patients with Gleason grades
≥7 (FC=1.637; P<0.001), serum PSA levels ≥10 ng/ml (FC=1.649;
P=0.002), T3-T4 (FC=1.606; P<0.001), positive lymph node
invasion (LNI; FC=1.474; P<0.001) and distant metastasis
(FC=1.765; P=0.009). The results indicate that CIT may be a
potential PCa-associated oncogene.
 | Table II.The differential expression of citron
kinase mRNA in the Cancer Genome Atlas mRNA expression profiles
(PRAD_2015_02_24). |
Table II.
The differential expression of citron
kinase mRNA in the Cancer Genome Atlas mRNA expression profiles
(PRAD_2015_02_24).
|
| Tumor vs.
normal | Gleason ≥7 vs.
Gleason <7 | PSA ≥10 vs. PSA
<10 | T3-4 vs. Ta-2 | N1 vs. N0 | M1 vs. M0 |
|---|
|
|
|
|
|
|
|
|
|---|
| Gene name | FC | P | FC | P | FC | P | FC | P | FC | P | FC | P |
|---|
| CIT | 2.18 | <0.001 | 1.64 | <0.001 | 1.65 | <0.001 | 1.61 | <0.001 | 1.47 | <0.001 | 1.77 | 0.010 |
| STAC | 1.55 | <0.001 | −1.78 | <0.001 | −2.63 | <0.001 | −1.45 | <0.001 | −1.85 | <0.001 | −3.72 | <0.001 |
| HELLS | 1.35 | <0.001 | 1.41 | <0.001 | 1.66 | <0.001 | 1.43 | <0.001 | 1.42 | <0.001 | 1.75 | 0.010 |
| RIC3 | −1.40 | <0.001 | −1.38 | <0.001 | −1.56 | 0.010 | −1.38 | <0.001 | −1.42 | <0.001 | −2.21 | <0.001 |
| C8orf46 | −1.54 | <0.001 | −1.30 | 0.010 | −1.59 | 0.010 | −1.29 | <0.001 | −1.32 | <0.001 | −1.87 | 0.010 |
| PTN | −1.89 | <0.001 | −1.75 | <0.001 | −2.04 | 0.010 | −1.73 | <0.001 | −1.89 | <0.001 | −2.65 | 0.010 |
| NTF3 | −2.00 | <0.001 | −1.26 | 0.010 | −1.49 | <0.001 | −1.17 | <0.001 | −1.24 | <0.001 | −1.73 | <0.001 |
| PAGE4 | −2.19 | <0.001 | −2.22 | <0.001 | −2.78 | <0.001 | −2.05 | <0.001 | −2.65 | <0.001 | −5.16 | <0.001 |
| BMPER | −2.25 | <0.001 | −1.91 | <0.001 | −2.67 | <0.001 | −1.46 | <0.001 | −1.84 | <0.001 | −3.01 | <0.001 |
| RSPO2 | −2.27 | <0.001 | −1.85 | <0.001 | −2.28 | <0.001 | −1.73 | <0.001 | −1.94 | <0.001 | −3.11 | <0.001 |
| FXYD1 | −2.31 | <0.001 | −1.58 | <0.001 | −1.97 | 0.010 | −1.36 | <0.001 | −1.75 | <0.001 | −2.55 | 0.010 |
| RNF112 | −2.41 | <0.001 | −1.74 | <0.001 | −2.21 | <0.001 | −1.68 | <0.001 | −1.87 | <0.001 | −3.00 | <0.001 |
| PROK1 | −2.42 | <0.001 | −2.13 | <0.001 | −2.40 | <0.001 | −2.01 | <0.001 | −2.40 | <0.001 | −3.73 | <0.001 |
|
C20orf200 | −2.55 | <0.001 | −1.67 | <0.001 | −1.83 | <0.001 | −1.55 | <0.001 | −1.68 | <0.001 | −2.32 | <0.001 |
| ANO4 | −2.82 | <0.001 | −1.72 | <0.001 | −2.23 | <0.001 | −1.79 | <0.001 | −1.99 | <0.001 | −2.64 | <0.001 |
| GSTM5 | −2.96 | <0.001 | −1.53 | <0.001 | −2.01 | <0.001 | −1.44 | <0.001 | −1.75 | <0.001 | −2.27 | 0.010 |
| B3GALT2 | −3.08 | <0.001 | −1.69 | <0.001 | −2.01 | <0.001 | −1.48 | <0.001 | −1.95 | <0.001 | −2.40 | <0.001 |
| ADRA1D | −3.19 | <0.001 | −2.05 | <0.001 | −1.98 | 0.010 | −1.63 | <0.001 | −1.97 | <0.001 | −2.86 | <0.001 |
| NDP | −3.30 | <0.001 | −1.72 | <0.001 | −2.04 | <0.001 | −1.48 | <0.001 | −1.71 | <0.001 | −2.58 | <0.001 |
| HIF3A | −3.55 | <0.001 | −1.78 | <0.001 | −2.42 | <0.001 | −1.67 | <0.001 | −2.06 | <0.001 | −2.91 | <0.001 |
| SMOC1 | −4.19 | <0.001 | −1.72 | <0.001 | −2.30 | <0.001 | −2.03 | <0.001 | −2.48 | <0.001 | −3.21 | <0.001 |
| LDB3 | −4.28 | <0.001 | −1.80 | <0.001 | −2.06 | 0.010 | −1.64 | <0.001 | −2.00 | <0.001 | −3.00 | <0.001 |
|
LOC572558 | −4.35 | <0.001 | −2.29 | <0.001 | −2.46 | <0.001 | −2.15 | <0.001 | −2.57 | <0.001 | −3.98 | <0.001 |
|
PPARGC1A | −4.42 | <0.001 | −1.60 | <0.001 | −2.08 | <0.001 | −1.60 | <0.001 | −1.98 | <0.001 | −2.43 | 0.010 |
| HRNBP3 | −4.54 | <0.001 | −2.17 | <0.001 | −2.48 | <0.001 | −2.11 | <0.001 | −2.72 | <0.001 | −5.42 | <0.001 |
| SRD5A2 | −4.57 | <0.001 | −1.98 | <0.001 | −2.17 | 0.010 | −2.18 | <0.001 | −2.74 | <0.001 | −5.26 | <0.001 |
| COL4A6 | −4.95 | <0.001 | −1.84 | <0.001 | −2.01 | 0.010 | −1.78 | <0.001 | −1.98 | <0.001 | −3.32 | <0.001 |
| LGR6 | −6.41 | <0.001 | −1.78 | <0.001 | −2.44 | <0.001 | −1.59 | <0.001 | −2.02 | <0.001 | −3.45 | <0.001 |
Expression of CIT is increased in
PCa
The expression of CIT mRNA was increased in PCa when
compared with BPH (Fig. 2A). The
immunoreactivity of CIT is presented in Fig. 2B. None and low staining were detected
in BPH and low-grade PCa, whereas moderate and strong staining was
detected in middle- and high-grade PCa. The staining scores of CIT
were significantly increased in primary and aggressive PCa,
compared with BPH (P<0.001; Fig.
2C). Additionally, compared with the non-HR-group, CIT
expression was significantly increased in the HR-group (P<0.001;
Fig. 2D). As presented in Table III, the percentage of patients with
H-CIT was significantly associated with Gleason grades (P=0.001),
serum PSA levels (P=0.001), T stages (P<0.001), lymph node
invasion (P=0.032) and metastasis (P=0.021). These results were
consistent with those of the aforementioned bioinformatic
analysis.
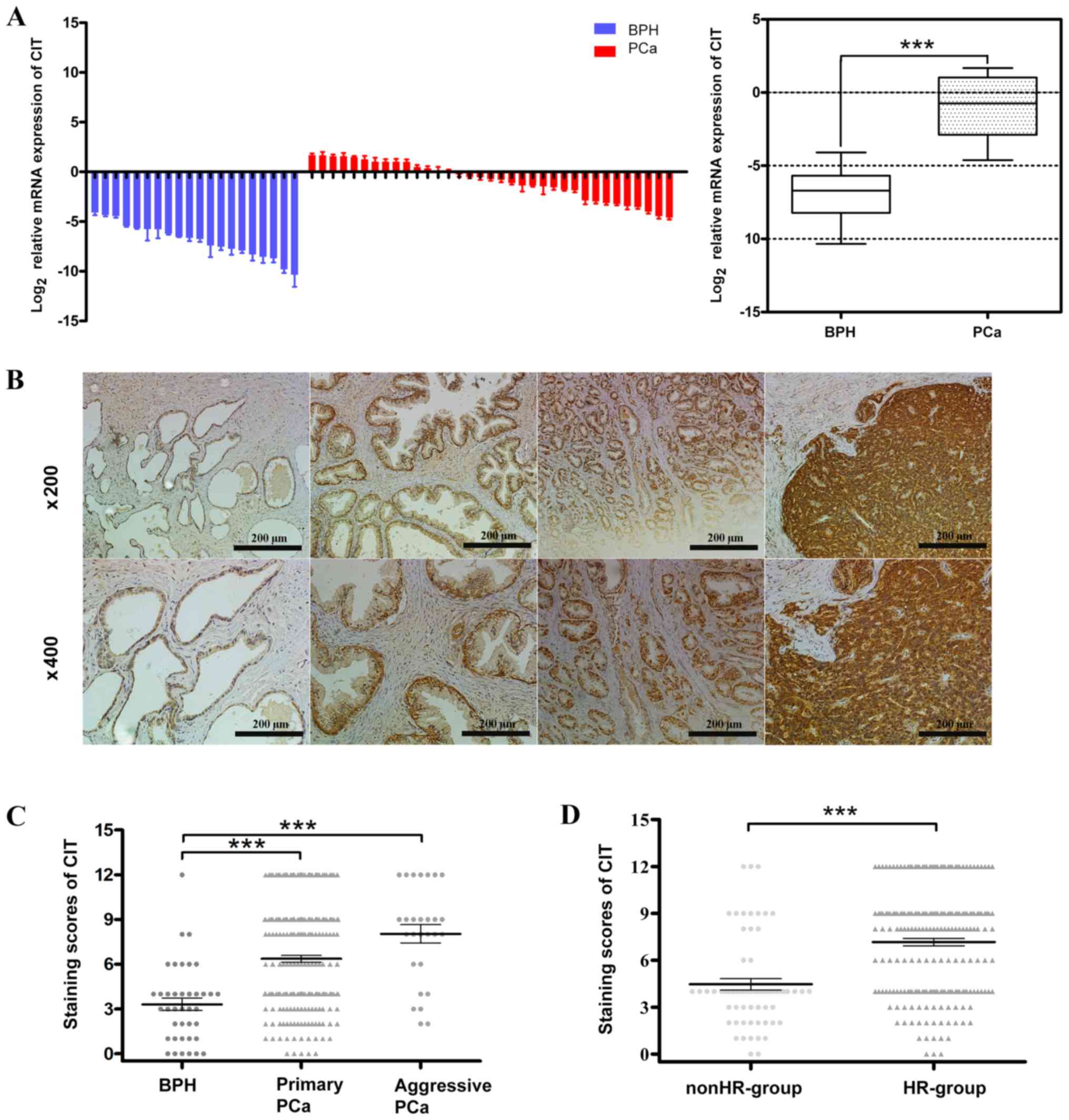 | Figure 2.CIT expression is increased in PCa.
(A) CIT mRNA was extracted from 35 cases of fresh PCa and 20 cases
of BPH. The results of reverse transcription-quantitative PCR
revealed that the mRNA expression of CIT was increased in PCa
samples. Data are presented as the SEM. The overall comparison
between PCa and BPH is presented in the box plot with the median
result, in which the bottom and top of the boxes represent the
maximum and minimum value, respectively. (B) The slides for IHC
were cut from formalin fixed paraffin embedded tissue obtained from
39 cases of BPH and 271 cases of PCa. Representative images of IHC
indicate CIT staining. No staining was present in BPH tissue; light
staining was exhibited in low-grade PCa, moderate staining was
revealed in middle-grade PCa and strong staining was indicated in
high-grade PCa. Each image was captured at a respective
magnification of ×200 and ×400, respectively. Compared with BPH,
there was a significant increase in primary and aggressive PCa,
whereas no statistically significant difference was observed
between primary and aggressive PCa (C) The expression of CIT in
HR-group was also higher than nonHR-group (D). Error bars represent
the SEM. The data in A and D were analyzed using an unpaired
t-test, and the data in C were analyzed using Kruskal-Wallis test
***P<0.001. CIT, citron kinase; PCa, prostate cancer; BPH,
benign prostatic hyperplasia; IHC, immunohistochemistry; SEM,
standard error of the mean; HR, high risk. |
 | Table III.Correlation between CIT and clinical
parameters of prostate cancer patients. |
Table III.
Correlation between CIT and clinical
parameters of prostate cancer patients.
| Parameters | No. (%) | Low CIT
expression | High CIT
expression | P-value |
|---|
| Gleason scores |
|
|
| 0.001 |
|
<7 | 88 | 52 (59.09) | 36 (40.91) |
|
| ≥7 | 183 | 69 (37.70) | 114 (62.30) |
|
| Serum PSA
(ng/ml) |
|
|
| 0.001 |
|
<10 | 69 | 43 (62.32) | 26 (37.68) |
|
|
≥10 | 202 | 78 (38.61) | 124 (61.39) |
|
| pT stage |
|
|
| <0.001 |
|
Ta-T2 | 150 | 87 (58.00) | 63 (42.00) |
|
|
T3-T4 | 121 | 34 (28.10) | 87 (71.90) |
|
| LNI |
|
|
| 0.032 |
| N0 | 255 | 118 (46.46) | 137 (53.94) |
|
| N1 | 16 | 3 (18.75) | 13 (81.25) |
|
| Metastasis |
|
|
| 0.021 |
| M0 | 254 | 118 (46.46) | 136 (53.54) |
|
| M1 | 17 | 3 (17.65) | 14 (82.35) |
|
CIT is a risk factor for poor outcomes
in patients with PCa
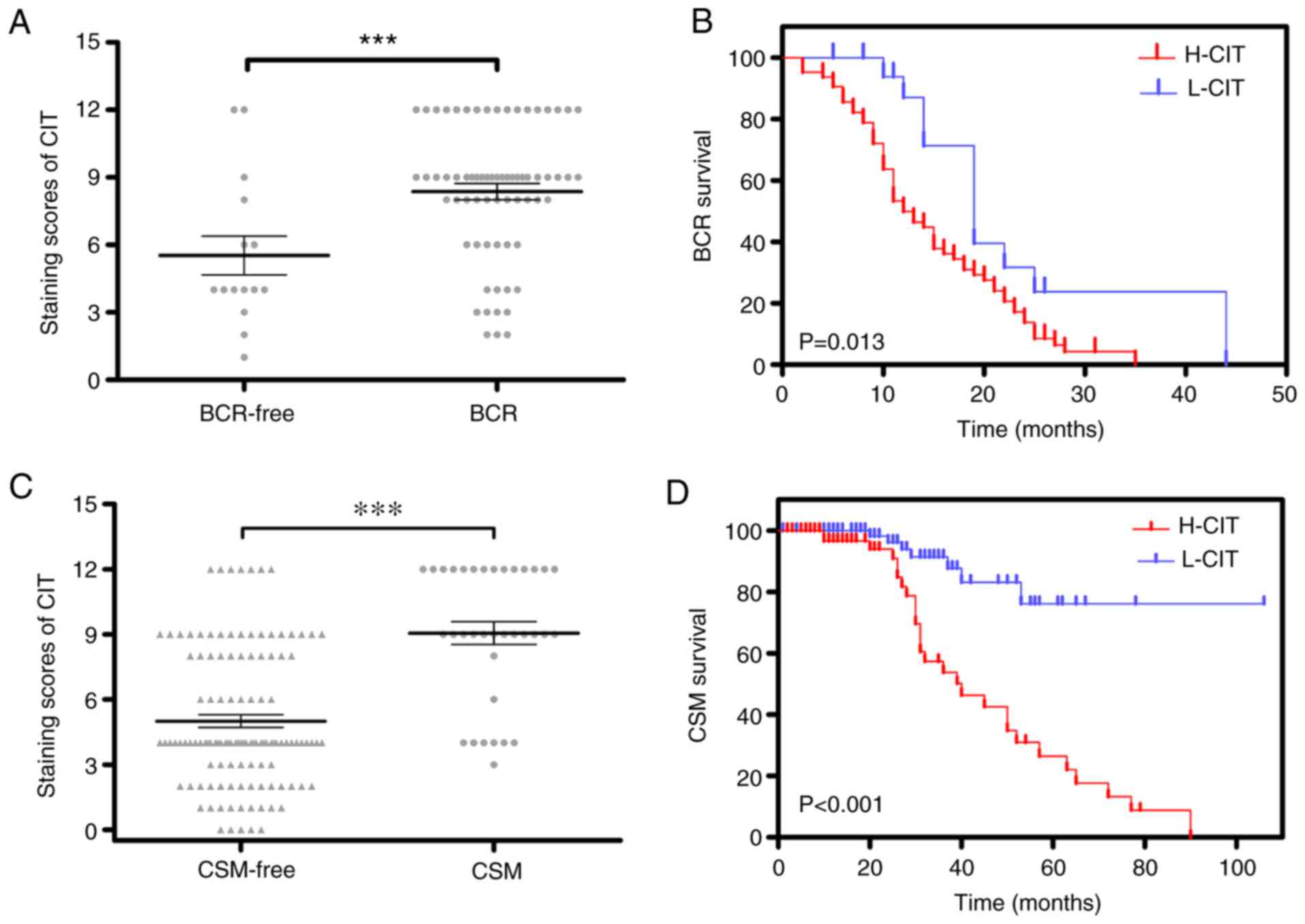
In IHC, the protein level of CIT expression was
significantly upregulated in BCR patients (P<0.001; Fig. 3A) and the recurrence time of patients
with H-CIT was significantly decreased compared with L-CIT
(P=0.013; Fig. 3B). Further
multivariate analysis demonstrated that the independent value of
H-CIT [hazard ratio (HR)=1.090–4.231; P=0.027] and LNI
(HR=1.002–4.294; P=0.049) was significant for BCR prediction
(Table IV).
 | Table IV.Univariate and Multivariate Cox
regression analysis for BCR. |
Table IV.
Univariate and Multivariate Cox
regression analysis for BCR.
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Variables | Hazard ratio (95%
CI) | P-value | Hazard ratio (95%
CI) | P-value |
|---|
| CIT (low vs.
high) | 2.231
(1.137–4.377) | 0.020 | 2.147
(1.090–4.231) | 0.027 |
| Gleason score
(<8 vs. ≥8) | 2.309
(1.148–4.644) | 0.019 | 1.561
(0.663–3.676) | 0.404 |
| Serum PSA level
(<10 vs. ≥10 ng/ml) | 2.634
(1.124–6.171) | 0.026 | 2.238
(0.949–5.277) | 0.066 |
| T stage (Ta-2 vs.
T3-4) | 1.021
(0.614–1.699) | 0.935 | 1.034
(0.601–1.782) | 0.903 |
| LNI (N0 vs.
N1) | 2.181
(1.060–4.489) | 0.034 | 2.074
(1.002–4.294) | 0.049 |
| Metastasis (M0 vs.
M1) | 1.225
(0.623–2.409) | 0.556 | 0.515
(0.237–1.118) | 0.094 |
The results also revealed that the expression of CIT
was increased in CSM patients (Fig.
3C). The Kaplan-Meier survival curve revealed that patients
with H-CIT exhibited shorter survival times compared with patients
with L-CIT (P<0.001; Fig. 3D).
Multivariate analysis also revealed that the independent risk
factors of CSM were CIT (HR=2.408–12.802; P=0.000), Gleason grades
(HR=1.148–5.068; P=0.020) and T stages (HR=1.815–8.085; P<0.001;
Table V).
 | Table V.Univariate and multivariate Cox
regression analysis for cancer-specific morality. |
Table V.
Univariate and multivariate Cox
regression analysis for cancer-specific morality.
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Variables | Hazard ratio (95%
CI) | P-value | Hazard ratio (95%
CI) | P-value |
|---|
| CIT (low vs.
high) | 5.316
(2.314–12.213) | <0.001 | 5.553
(2.408–12.802) | <0.001 |
| Gleason score
(<8 vs. ≥8) | 1.764
(0.875–3.556) | 0.108 | 2.412
(1.148–5.068) | 0.020 |
| Serum PSA, ng/ml
(<10 vs. ≥10) | 1.452
(0.628–3.355) | 0.383 | 0.869
(0.341–2.173) | 0.751 |
| T stage (Ta-2 vs.
T3-4) | 2.977
(1.504–5.895) | 0.002 | 3.831
(1.815–8.085) | 0.000 |
| LNI (N0 vs.
N1) | 4.584
(1.570–13.384) | 0.005 | 0.684
(0.181–2.586) | 0.576 |
| Metastasis (M0 vs.
N0) | 3.032
(1.570–13.384) | 0.134 | 1.134
(0.288–5.645) | 0.878 |
Discussion
A previous study of CIT in PCa demonstrated that the
loss of CIT inhibited the proliferation of LNCaP and C4-2B cells
(20), however the limited number of
cell types available and lack of investigation in a clinical
setting restricted the study. The current study screened CIT as a
potential oncogene in PCa. CIT was highly expressed in PCa samples
and was associated with Gleason scores, serum PSA levels, T stage
and risk groups. Furthermore, patients with a high CIT expression
were more likely to exhibit an increased BCR and CSM compared with
those with a low CIT expression. Additionally, the high expression
of CIT was determined to be a risk factor for BCR and CSM in
patients with PCa.
Cytokinesis is the final stage of cell division, in
which two daughter cells are separated (21). Resolving the midbody during the final
stage of abscission serves an important role in cytokinesis
(5). Failure to complete cytokinesis
may lead to tetraploidy and the presence of multiple centrosomes,
which has been proposed to promote tumorigenesis (22). Pihan et al (23) observed that centrosomes were
structurally and numerically abnormal in the majority of patients
with PCa. Furthermore, bladder cancer samples frequently contain a
number of centrosomes that are significantly increased as a result
of cytokinesis failure (24). CIT is
specifically required during the late stages of cytokinesis for the
organization and function of the midbody (7,25). The
overexpression of CIT kinase-active mutants causes the
dysregulation of cytokinesis, which results in the production of
multinucleate cells (26).
Therefore, the disrupted function of CIT may contribute to
cytokinesis failure, leading to the progression of cancer. Madhavan
et al (27) revealed that the
activation of the CIT/kinesin family member kinesin like protein
KIF14 (KIF14) axis, where CIT localizes to the central spindle via
the kinesin-3 motor, KIF14, is involved in the carcinogenesis of
retinoblastoma.
Various kinases have been demonstrated to be
intimately involved in processes and to contribute to tumor cell
proliferation and survival (28).
Certain kinases are considered to be oncogenic due to their
transforming capacity, including BRAF in colon carcinoma and ALK in
neuroblastoma (29,30). In addition, Rho-associated protein
kinase serves an essential role in the metastasis and proliferation
of breast cancer and hepatocellular carcinoma (31,32). The
knockdown of CIT directly inhibits the proliferation of breast
cancer and hepatocellular carcinoma cells (33,34).
Since a previous study determined that CIT is an essential kinase
that targets Rho-associated kinases (including ROCK and ROK)
(27), it seems likely that CIT
serves an important role in these cancers by interacting with Rho
signaling. Previous studies have also revealed that Rho signaling
factors are involved in the invasion of PCa cells (35,36),
such that CIT may also participate in the regulation of Rho
signaling, which serves a key role in the progression of PCa.
Currently, the main clinical signatures of patients
with PCa include TNM stage PSA levels and Gleason scores (37). The results of the current study
revealed that a high expression of CIT was positively associated to
a high T stage, serum PSA level and Gleason score. Furthermore, CIT
was determined to be an independent predictor of BCR and CSM. These
data indicated that CIT may serve as a potential marker of PCa and
may compensate for these clinical signatures. Currently, ADT is one
of the primary methods of treatment for patients with PCa (38). However, certain patients that receive
ADT will still advance to castration-resistant PCa and suffer from
a poor prognosis (39). Although
recent studies have determined that the glucocorticoid receptor can
be targeted to improve anti-androgen therapy (40,41), new
targets in the process of castration resistance should be explored.
In the current study, patients with a high CIT expression exhibited
shorter PSA recurrence time, which implies that CIT may serve a
role in androgen-resistant PCa.
However, the number of PCa samples was limited in
the current study and the mechanism of CIT in PCa also needs to be
further elucidated. More patient samples should therefore be
utilized in further study and the interaction between CIT and the
Rho pathway should be determined in PCa cell lines.
In conclusion, the results of the current long-term
retrospective study indicated that CIT is an independent indicator
of CSM and BCR. CIT may therefore be a potential biomarker of PCa
in the future. Although further study is required to assess the
function and mechanism of CIT in PCa, it may still serve as a
biomarker to improve the survival of patients with PCa.
Acknowledgements
The authors would like to thank Professor Fangzhou
Song (Department of Biochemistry & Molecular Biology, Molecular
Medicine & Cancer Research Center, Chongqing Medical
University, Chongqing, PR China) and Professor Xiaoni Zhong
(Department of Health Statistics and Information Management, School
of Public Health and Management, Chongqing Medical University,
Chongqing, China.) for helpful suggestions. Thanks to Dr. Yutao
Zhang (Department of Pathology, Zigong First People's Hospital) and
Dr. Yu Li (Department of Pathology, the First Affiliated Hospital
of Chongqing Medical University) for the supports of pathological
information and immunohistochemical assessment.
Funding
Funding was received from: National Natural Science
Foundation of China (grant no. 30972999); Nature Science Foundation
of Chongqing (grant no. Cstc2016shms-ztzx0054 and
cstc2015jcyjBX0045); the Science and technology planning project of
Yuzhong District (grant no. 20150111); the Health and Family
Planning Commission Foundation of Chongqing Municipal (grant no.
Cstc2012gg-yyjs10043); the Health Bureau of Chongqing (grant no.
20132082) and the Chongqing Education Commission (grant no.
CYS16125).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JD and JL analyzed the data and made major
contributions to writing the manuscript. YQ, YJL and YL performed
the experiments and wrote the initial draft of the manuscript. JH,
WW, LM and HL analyzed the data and contributed to revising the
article. DW and QY contributed to the design of the study and
provided final approval of the manuscript. WJ and YLia contributed
to the design of the study and assisted with writing the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The Ethics committee of the First Affiliated
Hospital of Chongqing Medical University, Zigong Fourth People's
Hospital and Zigong First People's Hospital approved the use of
these samples for the educational purposes of this research. The
consent from patients or patients' families was obtained
verbally.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
PCa
|
prostate cancer
|
|
BPH
|
benign prostatic hyperplasia
|
|
CIT
|
citron kinase
|
|
ADT
|
androgen deprivation therapy
|
|
PR
|
prostatectomy
|
|
PSA
|
prostate specific antigen
|
|
CSM
|
cancer specific morality
|
|
BCR
|
biochemical recurrence
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pokorny MR, de Rooij M, Duncan E, Schröder
FH, Parkinson R, Barentsz JO and Thompson LC: Prospective study of
diagnostic accuracy comparing prostate cancer detection by trans
rectal ultrasound-guided biopsy versus magnetic resonance (MR)
imaging with subsequent MR-guided biopsy in men without previous
prostate biopsies. Eur Urol. 66:22–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang W, Vazquez L, Apperson M and Kennedy
MB: Citron binds to PSD-95 at glutamatergic synapses on inhibitory
neurons in the hippocampus. J Neurosci. 19:96–108. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fujiwara T, Bandi M, Nitta M, Ivanova EV,
Bronson RT and Pellman D: Cytokinesis failure generating
tetraploids promotes tumorigenesis in p53-null cells. Nature.
437:1043–1047. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ganem NJ, Storchova Z and Pellman D:
Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev.
17:157–162. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
McKenzie C, Bassi ZI, Debski J, Gottardo
M, Callaini G, Dadlez M and D'Avino PP: Cross-regulation between
aurora B and citron kinase controls midbody architecture in
cytokinesis. Open Biol. 6:1600192016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
D'Avino PP and Capalbo L: Regulation of
midbody formation and function by mitotic kinases. Semin Cell Dev
Biol. 53:57–63. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gai M, Camera P, Dema A, Bianchi F, Berto
G, Scarpa E, Germena G and Di Cunto F: Citron kinase controls
abscission through RhoA and anillin. Mol Biol Cell. 22:3768–3778.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yamashiro S, Totsukawa G, Yamakita Y,
Sasaki Y, Madaule P, Ishizaki T, Narumiya S and Matsumura F: Citron
kinase, a Rho-dependent kinase, induces di-phosphorylation of
regulatory light chain of myosin II. Mol Biol Cell. 14:1745–1756.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao Y and Simon R: BRB-ArrayTools data
archive for human cancer gene expression: A unique and efficient
data sharing resource. Cancer Inform. 6:9–15. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mottet N, Bellmunt J, Bolla M, Briers E,
Cumberbatch MG, De Santis M, Fossati N, Gross T, Henry AM, Joniau
S, et al: EAU-ESTRO-SIOG guidelines on prostate cancer Part 1:
Screening, diagnosis, and local treatment with curative intent. Eur
Urol. 71:618–629. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu L, Toriseva M, Tuomala M, Seikkula H,
Elo T, Tuomela J, Kallajoki M, Mirtti T, Taimen P, Boström PJ, et
al: Increased expression of fibroblast growth factor 13 in prostate
cancer is associated with shortened time to biochemical recurrence
after radical prostatectomy. Int J Cancer. 139:140–152. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Eble JN, Sauter G, Epstein JE and
Sesterhenn IA: World Health Organization Classification of Tumours.
Pathology and genetics of the urinary system and male genital
organs. IARC Press; Lyon: 2004. pp. 159–215. 2004
|
|
15
|
Epstein JI, Allsbrook WC Jr, Amin MB and
Egevad LL; ISUP Grading Committee, : The 2005 International Society
of Urological Pathology (ISUP) consensus conference on Gleason
grading of prostatic carcinoma. Am J Surg Pathol. 29:1228–1242.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Goktas S, Yilmaz MI, Caglar K, Sonmez A,
Kilic S and Bedir S: Prostate cancer and adiponectin. Urology.
65:1168–1172. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu J, Xiao M, Li J, Wang D, He Y, He J,
Gao F, Mai L, Li Y, Liang Y, et al: Activation of UPR signaling
pathway is associated with the malignant progression and poor
prognosis in prostate cancer. Prostate. 77:274–281. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Roudier MP, Winters BR, Coleman I, Lam HM,
Zhang X, Coleman R, Chéry L, True LD, Higano CS, Montgomery B, et
al: Characterizing the molecular features of ERG-Positive tumors in
primary and castration resistant prostate cancer. Prostate.
76:810–822. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sheng X, Li WB, Wang DL, Chen KH, Cao JJ,
Luo Z, He J, Li MC, Liu WJ and Yu C: YAP is closely correlated with
castration-resistant prostate cancer, and downregulation of YAP
reduces proliferation and induces apoptosis of PC-3 cells. Mol Med
Rep. 12:4867–4876. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Whitworth H, Bhadel S, Ivey M, Conaway M,
Spencer A, Hernan R, Holemon H and Gioeli D: Identification of
kinases regulating prostate cancer cell growth using an RNAi
phenotypic screen. PLoS One. 7:e389502012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Glotzer M: The molecular requirements for
cytokinesis. Science. 307:1735–1739. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Boveri T: Concerning the origin of
malignant tumors by Theodor Boveri. Translated and annotated by
Henry Harris. J Cell Sci. 121:1–84. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pihan GA, Purohit A, Wallace J, Malhotra
R, Liotta L and Doxsey SJ: Centrosome defects can account for
cellular and genetic changes that characterize prostate cancer
progression. Cancer Res. 61:2212–2219. 2001.PubMed/NCBI
|
|
24
|
Yamamoto Y, Eguchi S, Junpei A, Nagao K,
Sakano S, Furuya T, Oga A, Kawauchi S, Sasaki K and Matsuyama H:
Intercellular centrosome number is correlated with the copy number
of chromosomes in bladder cancer. Cancer Genet Cytogenet.
191:38–42. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bassi ZI, Audusseau M, Riparbelli MG,
Callaini G and D'Avino PP: Citron kinase controls a molecular
network required for midbody formation in cytokinesis. Proc Natl
Acad Sci USA. 110:9782–9787. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Madaule P, Eda M, Watanabe N, Fujisawa K,
Matsuoka T, Bito H, Ishizaki T and Narumiya S: Role of citron
kinase as a target of the small GTPase Rho in cytokinesis. Nature.
394:491–494. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Madhavan J, Mitra M, Mallikarjuna K,
Pranav O, Srinivasan R, Nagpal A, Venkatesan P and Kumaramanickavel
G: KIF14 and E2F3 mRNA expression in human retinoblastoma and its
phenotype association. Mol Vis. 15:235–240. 2009.PubMed/NCBI
|
|
28
|
Zhang J, Yang PL and Gray NS: Targeting
cancer with small molecule kinase inhibitors. Nat Rev Cancer.
9:28–39. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Garnett MJ and Marais R: Guilty as
charged: B-RAF is a human oncogene. Cancer Cell. 6:313–319. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mossé YP, Laudenslager M, Longo L, Cole
KA, Wood A, Attiyeh EF, Laquaglia MJ, Sennett R, Lynch JE, Perri P,
et al: Identification of ALK as a major familial neuroblastoma
predisposition gene. Nature. 455:930–935. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Grise F, Bidaud A and Moreau V: Rho
GTPases in hepatocellular carcinoma. Biochim Biophys Acta.
1795:137–151. 2009.PubMed/NCBI
|
|
32
|
Tang Y, Olufemi L, Wang MT and Nie D: Role
of Rho GTPases in breast cancer. Front Biosci. 13:759–776. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fu Y, Huang J, Wang KS, Zhang X and Han
ZG: RNA interference targeting CITRON can significantly inhibit the
proliferation of hepatocellular carcinoma cells. Mol Biol Rep.
38:693–702. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
McKenzie C and D'Avino PP: Investigating
cytokinesis failure as a strategy in cancer therapy. Oncotarget.
7:87323–87341. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Somlyo AV, Bradshaw D, Ramos S, Murphy C,
Myers CE and Somlyo AP: Rho-kinase inhibitor retards migration and
in vivo dissemination of human prostate cancer cells. Biochem
Biophys Res Commun. 269:652–659. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu Y, He L, Zhang L, Chen J, Yi Z, Zhang
J, Liu M and Pang X: Anacardic acid (6-pentadecylsalicylic acid)
inhibits tumor angiogenesis by targeting Src/FAK/Rho GTPases
signaling pathway. J Pharmacol Exp Ther. 339:403–411. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Porten SP, Whitson JM, Cowan JE,
Cooperberg MR, Shinohara K, Perez N, Greene KL, Meng MV and Carroll
PR: Changes in prostate cancer grade on serial biopsy in men
undergoing active surveillance. J Clin Oncol. 29:2795–800. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sharifi N, Gulley JL and Dahut WL:
Androgen deprivation therapy for prostate cancer. JAMA.
294:238–244. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kirby M, Hirst C and Crawford ED:
Characterising the castration-resistant prostate cancer population:
A systematic review. Int J Clin Pract. 65:1180–1192. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Puhr M, Hoefer J, Eigentler A, Ploner C,
Handle F, Schaefer G, Kroon J, Leo A, Heidegger I, Eder I, et al:
The Glucocorticoid receptor is a key player for prostate cancer
cell survival and a target for improved Antiandrogen therapy. Clin
Cancer Res. 24:927–938. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Arora VK, Schenkein E, Murali R, Subudhi
SK, Wongvipat J, Balbas MD, Shah N, Cai L, Efstathiou E, Logothetis
C, et al: Glucocorticoid receptor confers resistance to
antiandrogens by bypassing androgen receptor blockade. Cell.
155:1309–1322. 2013. View Article : Google Scholar : PubMed/NCBI
|