Introduction
Colorectal cancer (CRC) is the third most prevalent
malignancy, with an estimated 1.4 million new cases and 693,900
deaths worldwide in 2012 (1). Tumor
sidedness has emerged as an important prognostic and predictive
factor in the treatment of patients with CRC (2). Multiple studies have demonstrated that
proximal colon cancer exhibits significantly different clinical and
biological features compared with distal colon or rectal cancer
(3). From a molecular point of view,
the former is generally diploid and exhibits higher rates of
microsatellite instability (MSI), whereas chromosomal instability
(CIN) is more frequent in the latter (4). Anatomically, they have a different
embryological origin, the proximal colon is derived from the midgut
and the distal colon and rectum are derived from the hindgut
(3). Therefore, traditionally,
patients with distal colon and rectal cancer have frequently been
grouped together in clinical or scientific research. However, there
is increasing evidence that distal colon and rectal cancer are
related to each other but are distinct in regard to their clinical
behavior, including the patterns of metastasis, response to
treatment and clinical outcome (5–7).
However, to the best of our knowledge, the underlying biological
carcinogenic backgrounds of the two types of cancer have not been
investigated.
CRC is a highly complex and heterogeneous disease
involving somatic mutation events associated with the interplay and
crosstalk between critical oncogenic pathways (8,9). Tie
et al (10) reported that
distal colon cancer exhibited a higher B-Raf proto-oncogene
serine/threonine kinase (BRAF) mutation frequency compared with
rectal cancer, and this may explain the different responses to
BRAF-targeting agents. Salem et al (11) demonstrated that catenin β1 (CTNNB1)
mutations were significantly increased in distal colon cancer
compared with rectal cancer, and a further study revealed that
tumors containing CTNNB1-mutations were frequently non-polyploid
and showed signs of immediate invasive growth (12). Improved understanding of these
mutational events and their role in the evolutionary process of
cancer may provide insight into the different clinical behaviors of
distal colon and rectal cancer.
The Memorial Sloan Kettering-Integrated Mutation
Profiling of Actionable Cancer Targets (MSK-IMPACT) is a
hybridization capture-based next-generation sequencing (NGS)
clinical assay for solid tumor molecular oncology (13). In the present study, using the
MSK-IMPACT data from cBioPortal, a systematic comparison of
molecular alterations between distal colon and rectal cancer was
performed. The results of the present study suggested that the
mutation profiles of distal colon and rectal cancer were largely
similar, but distinct in specific key genetic events, including APC
regulator of WNT signaling pathway (APC) R876*, SMAD4 R361 and BRAF
mutations.
The findings of the present study may contribute to
an improved understanding of the biology of CRC and provide
valuable information for improving management of patients with the
disease.
Materials and methods
Data and tumor samples
Data were downloaded from cBioPortal for Cancer
Genomics (cbioportal.org/msk-impact). A total of 12,670 tumors
from 11,369 unique patients were submitted for MSK-IMPACT
sequencing at the Memorial Sloan Kettering Cancer Center (MSKCC)
between January 2014 and May 2016 (14). Blood from the same patients was also
obtained to serve as a source of matched normal (germline) DNA
expression profile. Among the 1,007 CRC samples, 518 were primary
tumor samples, although four of these had no clearly annotated
tumor origins. Proximal, transverse and rectosigmoid colon cancer
were excluded, and 137 distal colon and 125 rectal tumor samples
were retained for further analysis.
MSK-IMPACT sequencing workflow
MSK-IMPACT is a comprehensive molecular profiling
assay that involves hybridization capture and deep sequencing of
all genes that are druggable by approved therapies or are targets
of experimental therapies being investigated in clinical trials at
MSKCC, as well as frequently mutated genes in human cancer (somatic
and germline mutations) (13). Two
different panels containing 341 (version 1) and 410 genes (version
2) were used, and all genes from the former panel were included in
the latter expanded panel (14). DNA
was extracted from tumor and matched normal blood samples using the
Chemagic STAR DNA Tissue-10 and Chemagic STAR DNA Blood-400 kits
(PerkinElmer, Inc.), respectively. Patient-matched blood DNA was
used to identify germline variants. Following sequencing, paired
reads were analyzed through a custom bioinformatics pipeline, and
the germline variants were filtered out. Each somatic variant
identified by the pipeline was manually reviewed to prevent
false-positive results (13,14). The alterations were described as
suggested in the Human Genome Variation Society (www.hgvs.org/mutnomen). All sequencing work was
performed at the MSKCC and reported in the original study (14).
Somatic mutation analysis
Mutation density across the tumors was expressed as
number of genetic alterations found in cancer genes present in the
MSK-IMPACT panel. Tumor mutational burden (TMB) was calculated as
the total number of non-synonymous mutations per megabase (Mb) of
the coding region target territory of the assay (0.98 Mb for
version 1 and 1.12 Mb for version 2), and further categorized as
low (0–10) or high (≥10). Following
the bioinformatics filtering, somatic point mutations were
classified as missense, truncating or in-frame mutations according
to the predicted protein sequence. Somatic gene mutation rates in
distal colon and rectal cancers were calculated, and a frequency
>5% was considered as significant. The frequencies and hotspot
density of specific driver mutations between distal colon and
rectal cancer were compared. Mutation plots were generated through
adaptation of cBioPortal visualization plots.
Statistical analysis
Statistical analyses were performed using SPSS
software (version 22.0; IBM Corp.). Continuous data were described
as either the mean ± standard deviation or median ± interquartile
range (IQR), and categorical variables as counts and frequencies.
To compare the differences in patient characteristics and the
distribution of gene mutations, Fisher's exact test, χ2
test, paired t-test, or Mann-Whitney U test were used, as
appropriate. P<0.05 was considered to indicate a statistically
significant difference.
Results
Tumor characteristics
The mutational profiles of distal colon and rectal
cancer were compared using 262 CRC samples, and the
clinicopathological features of the patients are summarized in
Table I. In the distal colon and
rectal cancer groups, 76 (53.9%) and 74 (53.0%) patients were male,
respectively (P>0.05). In addition, no significant difference in
smoking history was observed between the two groups (P>0.05).
MSK-IMPACT, an NGS platform for targeted sequencing of
cancer-related genes, was performed on all the samples. The average
depth of sample coverage for the distal colon and rectal tumors
were 740× and 743×, respectively (P>0.05). Two types of
MSK-IMPACT panels were employed for NGS throughout the study, but
there was no apparent distribution difference between the
groups.
 | Table I.Clinicopathological features of the
262 patients the colorectal tumor samples were obtained from. |
Table I.
Clinicopathological features of the
262 patients the colorectal tumor samples were obtained from.
| Clinicopathological
feature | Distal colon cancer
(n=137) | Rectal cancer
(n=125) | P-value |
|---|
| Sex |
|
|
|
| Male | 76 (53.9%) | 74 (53.0%) |
|
|
Female | 61 (46.1%) | 51 (47.0%) | 0.617 |
| Smoking history |
|
|
|
|
Previous/Current | 48 (37.3%) | 54 (41.3%) |
|
|
Never | 72 (51.3%) | 53 (50.9%) |
|
|
Unknown | 17 (11.4%) | 18 (7.8%) | 0.259 |
| MSK-IMPACT
panel |
|
|
|
| IM3_341
genes | 40 (30.5%) | 24 (18.6%) |
|
| IM5_410
genes | 97 (76.6%) | 101 (81.4%) | 0.063 |
| Sample coverage
(x) |
|
|
|
| Mean ±
SD | 740±236 | 743±228 | 0.529 |
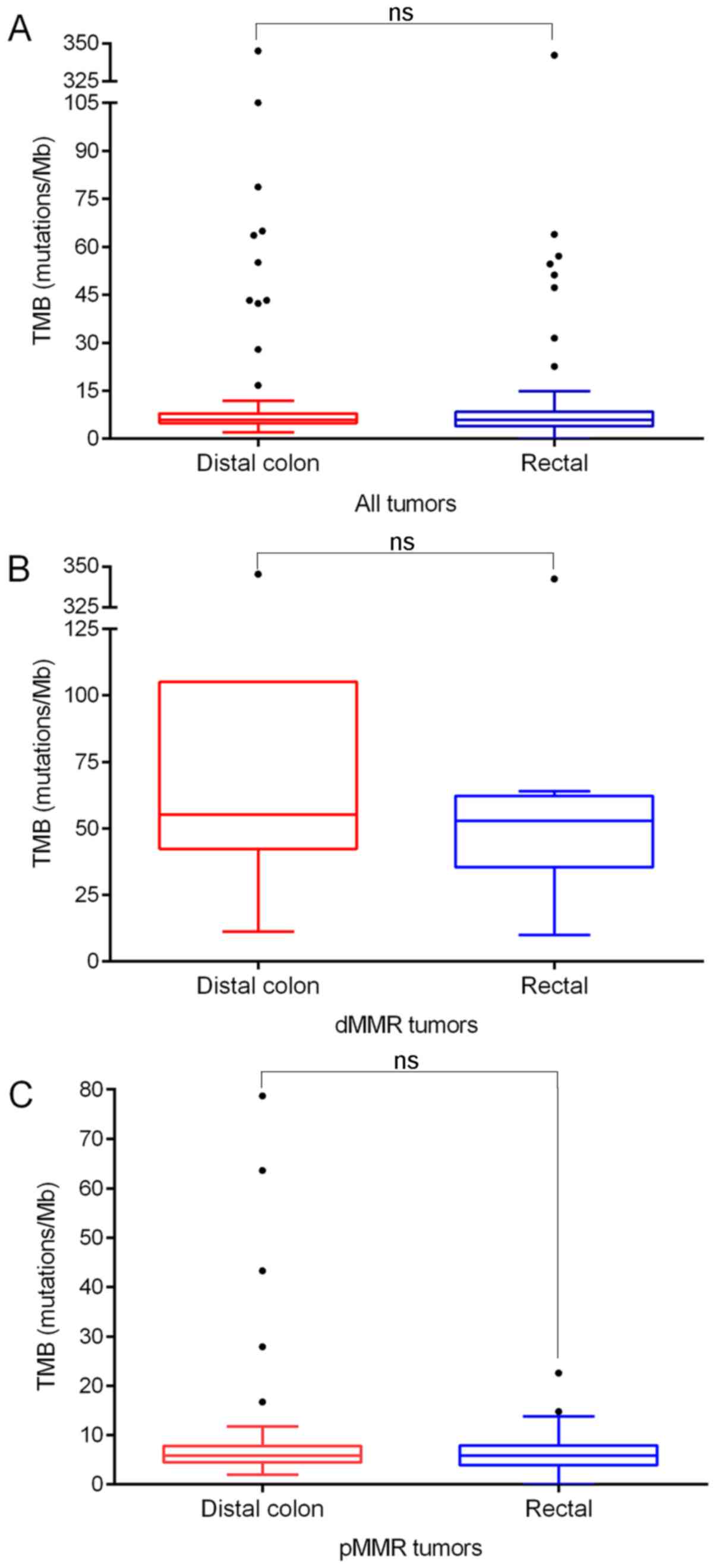
TMB analysis
TMB was calculated for each sample sequenced for
341/410 genes by MSK-IMPACT. Distal colon tumors had a median of
5.9 mutations/Mb (IQR, 3.0), which was similar to that in the
rectal tumors (median ± IQR, 5.9±4.5; P>0.05). It is worth
noting that seven cases (5.1%) in the distal colon group and eight
cases (6.4%) in the rectal group were tumors with defects in
mismatch repair (dMMR) genes (mutL homolog 1, mutS homolog 2, mutS
homolog 6 and PMS1 homolog 2 mismatch repair system component),
which showed a disproportionately higher number of mutations (55.1
and 52.9 mutations/Mb, respectively). When TMB was calculated for
proficient MMR (pMMR) tumors only, the median TMB was 5.9
mutations/Mb in both distal colon and rectal tumors, with no
significant difference (Mann-Whitney U test, both P>0.05;
Fig. 1). The association between TMB
and the clinicopathological features of CRC were examined. TMB
showed no significant association with sex, smoking history, panel
type or sample coverage (all P>0.05; Table II). Additionally, the associations
remained insignificant after removing dMMR tumors (all P>0.05;
Table II).
 | Table II.Association between TMB and the
clinicopathological features of the patients the colorectal tumor
samples were obtained from. |
Table II.
Association between TMB and the
clinicopathological features of the patients the colorectal tumor
samples were obtained from.
|
| All colorectal
tumors | pMMR colorectal
tumors |
|---|
|
|
|
|
|---|
| Clinicopathological
feature | Low TMB
(n=218) | High TMB
(n=44) | P-value | Low TMB
(n=218) | High TMB
(n=29) | P-value |
|---|
| Sex |
|
|
|
|
|
|
|
Male | 127 (58.3%) | 23 (52.3%) |
| 127 (58.3%) | 12 (41.4%) |
|
|
Female | 91 (41.7%) | 21 (47.7%) | 0.506 | 91 (41.7%) | 17 (58.6%) | 0.111 |
| Smoking
history |
|
|
|
|
|
|
|
Previous/Current | 87 (39.9%) | 15 (34.1%) |
| 87 (39.9%) | 8 (27.6%) |
|
|
Never | 102 (46.8%) | 23 (52.3%) |
| 102 (46.8%) | 16 (55.2%) |
|
|
Unknown | 29 (13.3%) | 6 (13.5%) | 0.742 | 29 (13.3%) | 5 (17.2%) | 0.412 |
| MSK-IMPACT
panel |
|
|
|
|
|
|
| IM3_341
genes | 50 (22.9%) | 14 (31.8%) |
| 50 (22.9%) | 11 (37.9%) |
|
| IM5_410
genes | 168 (77.1%) | 30 (68.2%) | 0.248 | 168 (77.1%) | 18 (62.1%) | 0.106 |
| Sample coverage
(x) |
|
|
|
|
|
|
| Mean ±
SD | 745±238 | 700±218 | 0.073 | 745±238 | 722±234 | 0.268 |
Driver mutation analysis
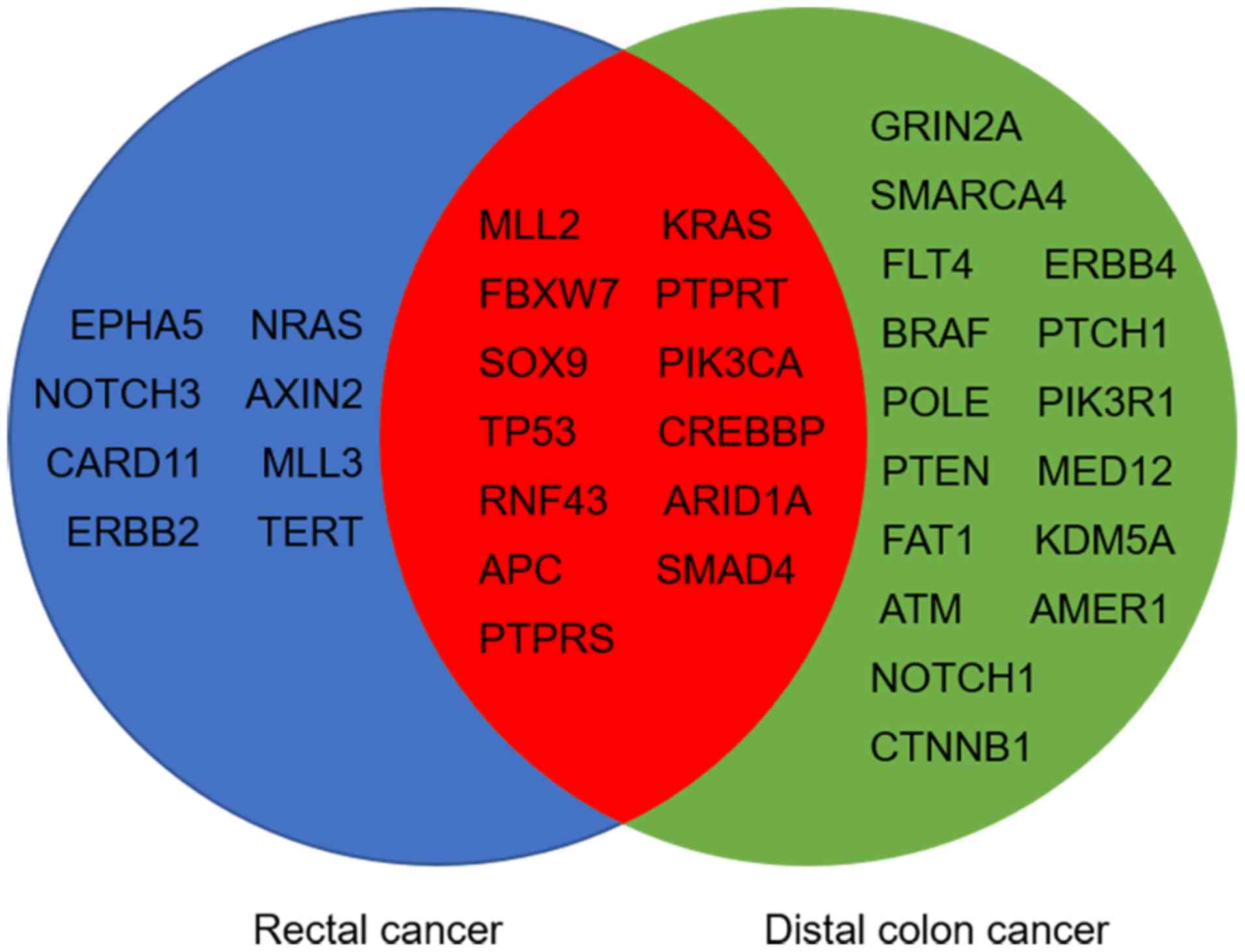
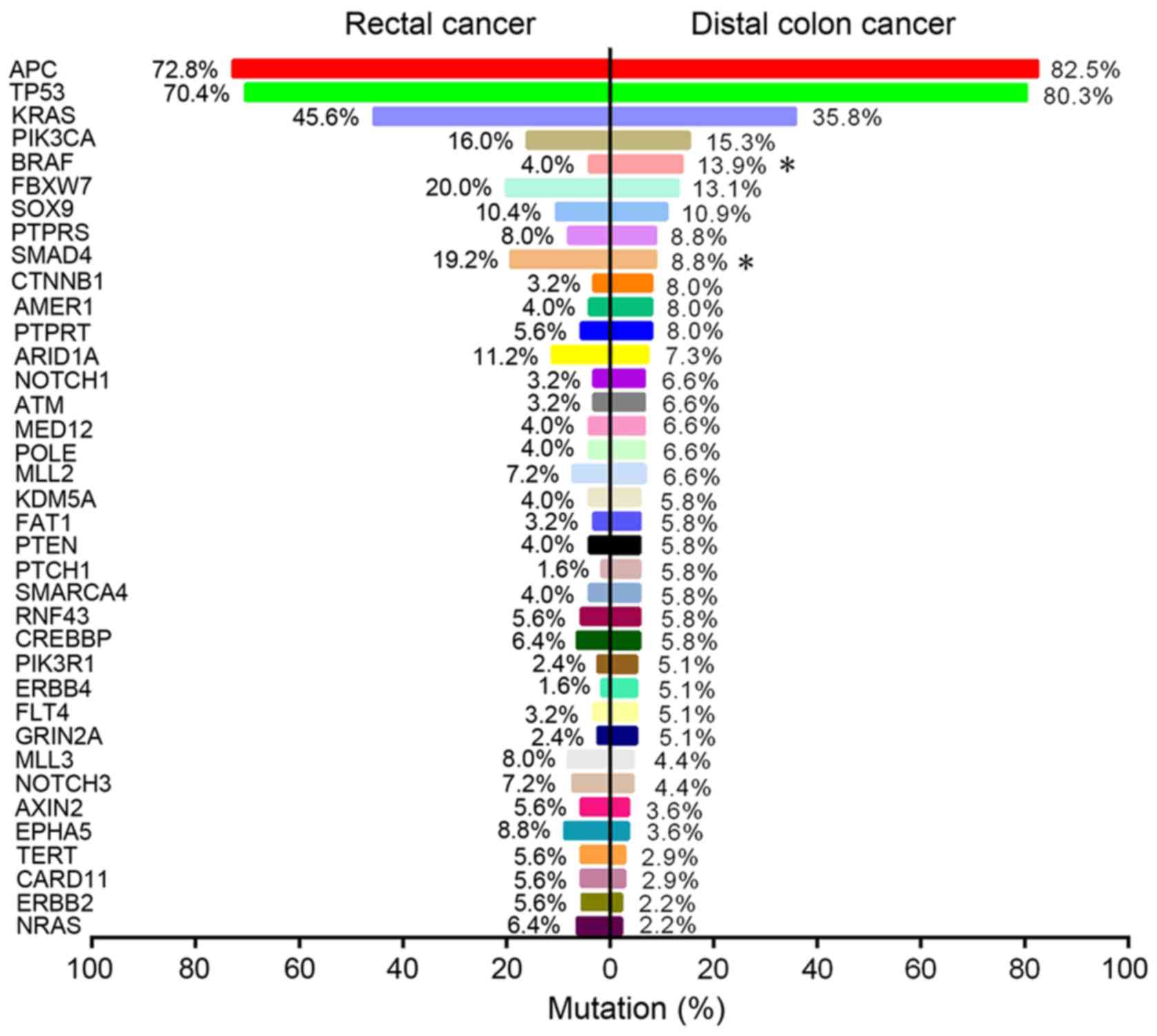
Mutational analysis showed that 29 and 21 genes were
significantly mutated in distal colon and rectal tumors,
respectively (>5% of tumor samples). Among these genes, 13
significantly mutated genes (SMGs) were shared between the two
groups (Fig. 2), including APC,
tumor protein p53 (TP53), KRAS proto-oncogene GTPase (KRAS),
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α
(PIK3CA) and SMAD4. Comparison of SMGs between the groups revealed
that BRAF mutations were significantly enriched in distal colon
cancer (13.9 vs. 4.0%; P=0.009; Fig.
3), whilst SMAD4 mutations were significantly more common in
rectal cancer (19.2 vs. 8.8%; P=0.019; Fig. 3). Despite there being no significant
difference in the frequencies of KRAS or NRAS proto-oncogene GTPase
mutations between the two groups (both P>0.05; Fig. 3), RAS was significantly more
frequently mutated in rectal cancer compared with distal colon
cancer (52.0 vs. 38.0%; P=0.025; Table
III). In addition, the data showed that KRAS and BRAF mutations
were predominantly, but not completely, exclusive, with only three
cases of distal colon and one case of rectal tumor samples carrying
both mutations concomitantly. The mutational landscape in the
subgroup of pMMR tumors was further examined, and it was
demonstrated that these differences in mutational frequencies of
BRAF, SMAD4 and RAS between distal colon and rectal tumors remained
significant (all P<0.05; Table
III).
 | Table III.Enrichment analysis of BRAF, SMAD4
and RAS mutations between distal colon and rectal cancer. |
Table III.
Enrichment analysis of BRAF, SMAD4
and RAS mutations between distal colon and rectal cancer.
|
| All colorectal
tumors | pMMR colorectal
tumors |
|---|
|
|
|
|
|---|
| Gene | Distal colon
(n=137) (%) | Rectal (n=125)
(%) | P-value | Distal colon
(n=130) (%) | Rectal (n=117)
(%) | P-value |
|---|
| BRAF |
|
|
|
|
|
|
|
Mutant | 19 (13.9) | 5 (4.0) |
| 16 (12.3) | 4 (3.4) |
|
|
Wild-type | 118 (86.1) | 120 (96.0) | 0.009 | 114 (87.7) | 113 (96.6) | 0.018 |
| SMAD4 |
|
|
|
|
|
|
|
Mutant | 12 (8.8) | 24 (19.2) |
| 12 (9.2) | 24 (20.5) |
|
|
Wild-type | 125 (91.2) | 101 (80.8) | 0.019 | 118 (90.8) | 93 (79.5) | 0.018 |
| RAS
(KRAS/NRAS) |
|
|
|
|
|
|
|
Mutant | 52 (38.0) | 65 (52.0) |
| 49 (37.7) | 60 (51.3) |
|
|
Wild-type | 85 (62.0) | 60 (48.0) | 0.025 | 81 (62.3) | 57 (48.7) | 0.040 |
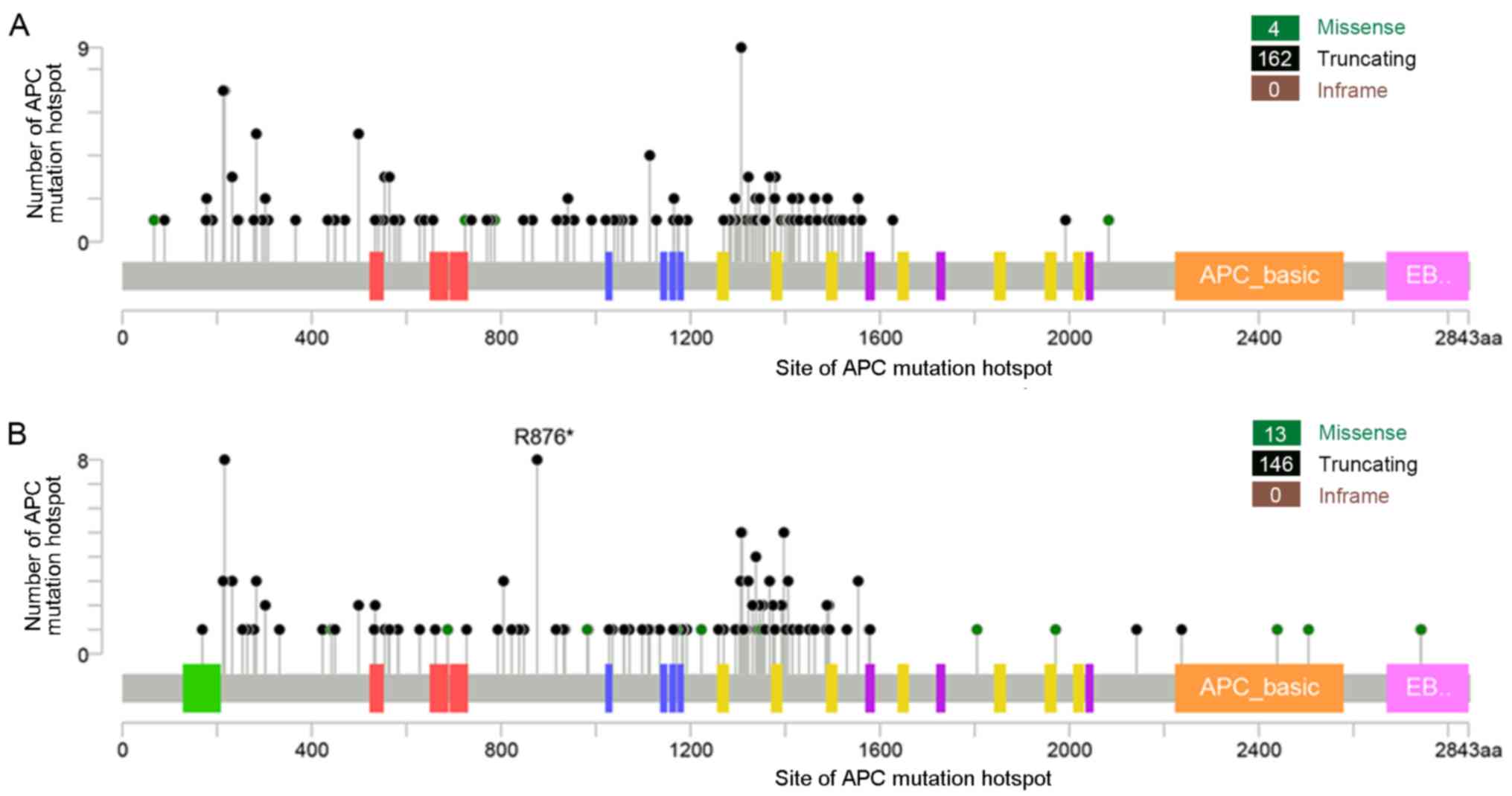
Mutation hotspot analysis
Mutation hotspot analysis of several key driver
genes was performed and it was demonstrated that in both distal
colon and rectal cancer, missense mutations were the most common
type of point mutations in TP53, KRAS, BRAF, PIK3CA and SMAD4
genes, while truncations were the predominant type of mutations in
the APC gene. APC mutations were the most frequent genetic
alterations in CRC, and codons 1,286-1,513 (mutation cluster
region) were the most commonly mutated loci, covering ~40% of APC
mutations in both groups. Additionally, APC R876* was a significant
mutation hotspot in rectal cancer compared with distal colon cancer
(seen in eight rectal and no distal colon tumor samples; P=0.002;
Fig. 4). TP53 mutations were found
scattered throughout the coding sequence, but ~25% of the mutations
were clustered at codons R175, R248 and R273 in both groups. For
KRAS, G12 and G13 were the predominant hotspots, accounting for 84
and 78% of KRAS mutations in distal colon and rectal tumors,
respectively. For BRAF, >50% of the mutations were found
clustered at codon V600 in both groups. In PIK3CA, 52 and 35% of
its mutations in distal colon and rectal tumors, respectively, were
located at codons R542, R545 and H1047. In addition, similar to APC
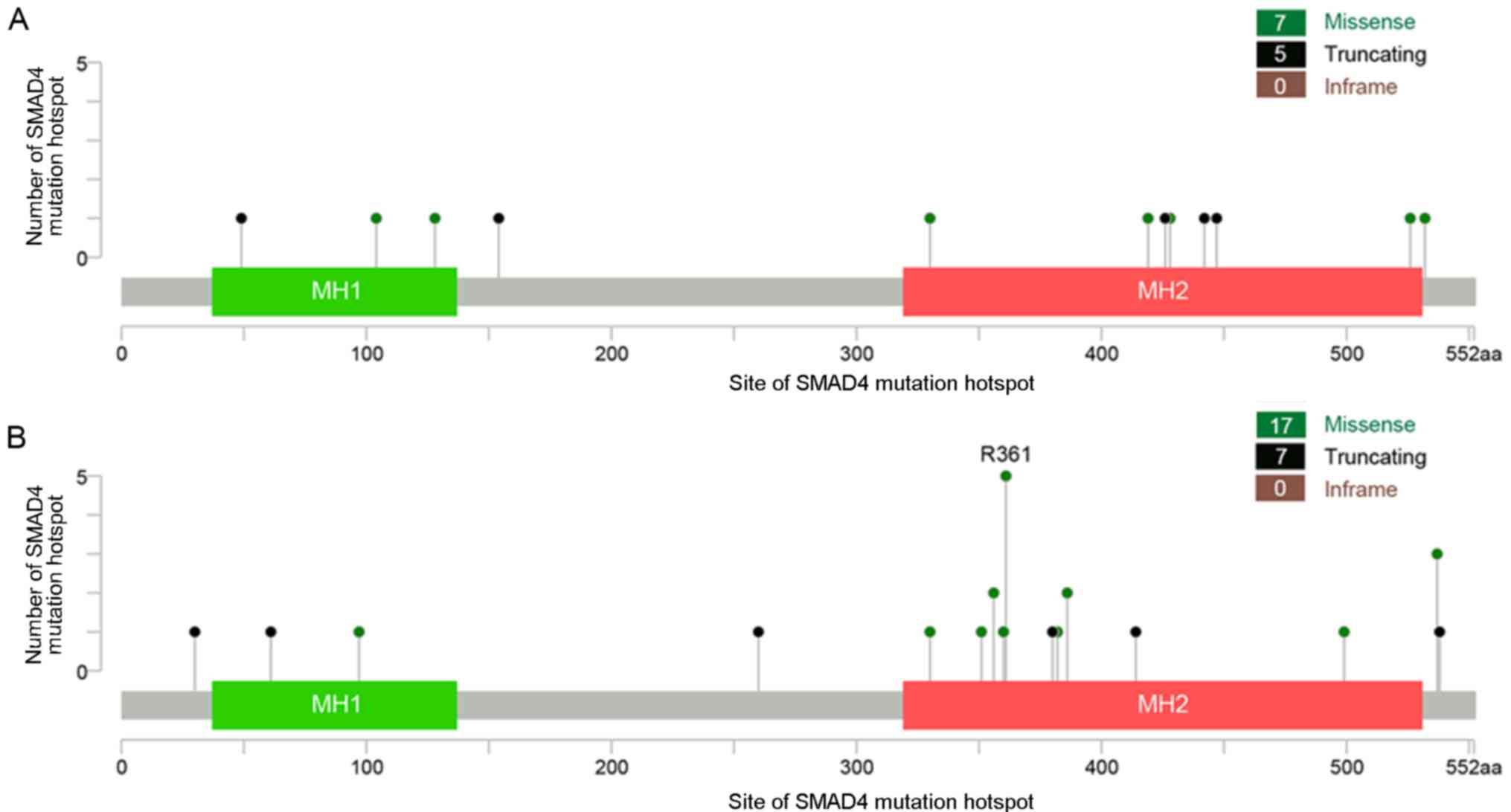
R876*, SMAD4 R361 missense mutations appeared to be present
exclusively in rectal cancer (seen in five rectal and no distal
colon tumor samples, respectively; P=0.024; Fig. 5).
Discussion
Various studies have indicated that CRC is a complex
disease with multiple genetic alterations and variable clinical
outcomes (9,15). Molecular genotyping of patients with
CRC is of vital importance in clinical decision-making regarding
diagnostic and therapeutic interventions. In the present study, by
comparing the mutational profiles of distal colon and rectal cancer
in 262 tumor samples, it was demonstrated that the genetic
differences between the two types of cancer were clinically
relevant, which emphasized the importance of the location of the
primary tumor in the management of CRC and the implications for
future clinical and scientific research.
In the present study, analysis was performed using
MSK-IMPACT data with high depth of coverage for improving the
understanding of the mutational landscape of distal colon and
rectal cancer. TMB analysis showed that the two anatomical
locations exhibited similar mutational burdens, and a high-TMB
status was present in 14.6% of distal colon cancer cases and 19.2%
of rectal cancer cases, with no significant difference. MMR-mutated
tumors showed a hypermutator phenotype and were most likely to
benefit from immune checkpoint blockade therapy (16). After removal of dMMR tumors from
analyses, the TMB level in the distal colon and rectal groups
remained similar. This finding was in agreement with the result of
a previous study (11). In addition,
the association of TMB with CRC clinicopathological characteristics
was examined, including smoking history, which was reported to be
significantly associated with a higher TMB level in lung cancer
(17), but no similar association
was identified in all the CRC cases in the present study. Previous
studies have also suggested that smoking was an independent risk
factor for the development of MSI-high CRC (18,19).
Therefore, further studies are required to validate the results
obtained.
CRC arises through a series of well-characterized
histopathological changes as the result of specific genetic ‘hits’
at certain oncogenes and tumor suppressor genes (8,20). The
present study suggested that despite sharing the same critical
genomic events, including APC, TP53, KRAS, PIK3CA and SMAD4, there
were differences in the frequencies, hotspots and significance of
these SMGs in the development of distal colon and rectal cancer.
APC and TP53 mutations are the most common genetic alterations in
both distal colon and rectal cancer and contribute functionally to
various stages of tumor progression (21,22). The
present study identified a novel, potentially targetable hotspot
mutation in APC R876* that was enriched in rectal cancer compared
with distal colon cancer. Ficari et al (23) indicated that the truncation mutation
at APC codon 876, which affected the β-catenin binding domain, was
associated with the density of adenomas of a certain mild
colorectal pathophenotype. SMAD4 is an essential mediator in the
transforming growth factor-β signaling pathway (24), and is associated with CRC metastasis,
resistance to 5-fluorouracil chemotherapy and poor outcome
(25,26). A study by Mehrvarz et al
(27) found that SMAD4 mutations
were more frequently detected in colon rather than rectal cancer,
and may be associated with the response of CRC to anti-epidermal
growth factor receptor (EGFR) therapy. However, the present study
observed that SMAD4-mutated tumors were more likely to be located
in the rectum than in the distal colon. Furthermore, the SMAD4 R361
mutation was found almost exclusively in rectal cancer and not in
distal colon cancer, suggesting that it may be involved in the
different clinical and biological behaviors associated with the two
different types of CRC, and thus may provide a potential diagnostic
or therapeutic target for rectal cancer.
Currently, RAS and BRAF mutation testing has been
incorporated into routine clinical practice for patients with CRC
receiving anti-EGFR therapy. There is also emerging evidence that
PIK3CA mutations are associated with resistance to anti-EGFR
therapy (28,29). Sartore-Bianchi et al (30) suggested that a combined mutational
analysis of the KRAS and PIK3CA/phosphatase and tensin homolog
pathways could identify up to 70% of patients with advanced CRC who
were unlikely to respond to anti-EGFR agents. The results of the
present study showed that distal colon and rectal cancer had
similar KRAS and PIK3CA mutational status, whereas BRAF and RAS
mutations were significantly enriched in distal colon and rectal
cancer, respectively. Furthermore, these differences remained
significant in the subgroup analysis of pMMR tumors. Similar to the
findings of the present study, Salem et al (11) observed that there was a significant
decrease in the frequency of BRAF mutations when moving from
proximal colon to distal colon to the rectum, suggesting that
different underlying mechanisms may be involved in rectal cancer
and distal colon cancer. In addition, in the present study it was
observed that mutations in KRAS and BRAF were primarily, but not
completely, mutually exclusive in both distal colon and rectal
cancer, thus differing from the majority of previous reports
(31,32). However, the exclusivity of the
mutational status of KRAS and BRAF may be largely due to the
high-depth sequencing coverage of the MSK-IMPACT assay, which can
detect mutations that appear only in a minority of cells in a
sample (14).
In conclusion, despite the limitation that the
present study was primarily computational and requires further
experimental validation, the results suggested that the mutation
profiles of distal colon and rectal cancer are similar in
principle, but distinct in specific key genetic events, including
APC R876*, SMAD4 R361 and BRAF mutations. Therefore, the findings
of the present study may contribute to understanding the
differences in tumor biology and clinical behavior between distal
colon and rectal cancer. The present study highlighted the
necessity to consider distal colon and rectal cancer in the context
of genetic background when selecting treatment regimens, designing
research trials and analyzing clinical outcomes.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural
Science Foundation of China (grant no. 81874201) and Shanghai
Science and Technology Commission (grant no. 19411971500).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available from cBioPortal for Cancer Genomics at
cbioportal.org/msk-impact.
Authors' contributions
ZZ and HJ designed the study. ZZ, AW and XT
performed the research. YC, ET and HJ contributed to the data
analysis. HJ supervised the study. ZZ and AW drafted the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sugai T, Habano W, Jiao YF, Tsukahara M,
Takeda Y, Otsuka K and Nakamura S: Analysis of molecular
alterations in left- and right-sided colorectal carcinomas reveals
distinct pathways of carcinogenesis: Proposal for new molecular
profile of colorectal carcinomas. J Mol Diagn. 8:193–201. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee GH, Malietzis G, Askari A, Bernardo D,
Al-Hassi HO and Clark SK: Is right-sided colon cancer different to
left-sided colorectal cancer? A systematic review. Eur J Surg
Oncol. 41:300–308. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Takahashi Y, Sugai T, Habano W, Ishida K,
Eizuka M, Otsuka K, Sasaki A, Takayuki M, Morikawa T, Unno M and
Suzuki H: Molecular differences in the microsatellite stable
phenotype between left-sided and right-sided colorectal cancer. Int
J Cancer. 139:2493–2501. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Minoo P, Zlobec I, Peterson M, Terracciano
L and Lugli A: Characterization of rectal, proximal and distal
colon cancers based on clinicopathological, molecular and protein
profiles. Int J Oncol. 37:707–718. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gao XH, Yu GY, Gong HF, Liu LJ, Xu Y, Hao
LQ, Liu P, Liu ZH, Bai CG and Zhang W: Differences of protein
expression profiles, KRAS and BRAF mutation, and prognosis in
right-sided colon, left-sided colon and rectal cancer. Sci Rep.
7:78822017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Salem ME, Yin J, Weinberg BA, Renfro LA,
Pederson LD, Maughan TS, Adams RA, Van Cutsem E, Falcone A, Tebbutt
NC, et al: Clinicopathological differences and survival outcomes
with first-line therapy in patients with left-sided colon cancer
and rectal cancer: Pooled analysis of 2,879 patients from AGITG
(MAX), COIN, FOCUS2, OPUS, CRYSTAL and COIN-B trials in the ARCAD
database. Eur J Cancer. 103:205–213. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bogaert J and Prenen H: Molecular genetics
of colorectal cancer. Ann Gastroenterol. 27:9–14. 2014.PubMed/NCBI
|
|
9
|
Worthley DL and Leggett BA: Colorectal
cancer: Molecular features and clinical opportunities. Clin Biochem
Rev. 31:31–38. 2010.PubMed/NCBI
|
|
10
|
Tie J, Gibbs P, Lipton L, Christie M,
Jorissen RN, Burgess AW, Croxford M, Jones I, Langland R, Kosmider
S, et al: Optimizing targeted therapeutic development: Analysis of
a colorectal cancer patient population with the BRAF(V600E)
mutation. Int J Cancer. 128:2075–2084. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Salem ME, Weinberg BA, Xiu J, El-Deiry WS,
Hwang JJ, Gatalica Z, Philip PA, Shields AF, Lenz HJ and Marshall
JL: Comparative molecular analyses of left-sided colon, right-sided
colon, and rectal cancers. Oncotarget. 8:86356–86368. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ahadova A, von Knebel Doeberitz M, Bläker
H and Kloor M: CTNNB1-mutant colorectal carcinomas with immediate
invasive growth: A model of interval cancers in Lynch syndrome. Fam
Cancer. 15:579–586. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cheng DT, Mitchell TN, Zehir A, Shah RH,
Benayed R, Syed A, Chandramohan R, Liu ZY, Won HH, Scott SN, et al:
Memorial Sloan Kettering-integrated mutation profiling of
actionable cancer targets (MSK-IMPACT): A hybridization
capture-based next-generation sequencing clinical assay for solid
tumor molecular oncology. J Mol Diagn. 17:251–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zehir A, Benayed R, Shah RH, Syed A,
Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, et
al: Mutational landscape of metastatic cancer revealed from
prospective clinical sequencing of 10,000 patients. Nat Med.
23:703–713. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Phipps AI, Limburg PJ, Baron JA,
Burnett-Hartman AN, Weisenberger DJ, Laird PW, Sinicrope FA, Rosty
C, Buchanan DD, Potter JD and Newcomb PA: Association between
molecular subtypes of colorectal cancer and patient survival.
Gastroenterology. 148:77–87.e2. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Le DT, Uram JN, Wang H, Bartlett BR,
Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et
al: PD-1 blockade in tumors with mismatch-repair deficiency. N Engl
J Med. 372:2509–2520. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Govindan R, Ding L, Griffith M,
Subramanian J, Dees ND, Kanchi KL, Maher CA, Fulton R, Fulton L,
Wallis J, et al: Genomic landscape of non-small cell lung cancer in
smokers and never-smokers. Cell. 150:1121–1134. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Slattery ML, Curtin K, Anderson K, Ma KN,
Ballard L, Edwards S, Schaffer D, Potter J, Leppert M and Samowitz
WS: Associations between cigarette smoking, lifestyle factors, and
microsatellite instability in colon tumors. J Natl Cancer Inst.
92:1831–1836. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chia VM, Newcomb PA, Bigler J, Morimoto
LM, Thibodeau SN and Potter JD: Risk of microsatellite-unstable
colorectal cancer is associated jointly with smoking and
nonsteroidal anti-inflammatory drug use. Cancer Res. 66:6877–6883.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wood LD, Parsons DW, Jones S, Lin J,
Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al: The
genomic landscapes of human breast and colorectal cancers. Science.
318:1108–1113. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fodde R: The APC gene in colorectal
cancer. Eur J Cancer. 38:867–871. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Iacopetta B: TP53 mutation in colorectal
cancer. Hum Mutat. 21:271–276. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ficari F, Cama A, Valanzano R, Curia MC,
Palmirotta R, Aceto G, Esposito DL, Crognale S, Lombardi A,
Messerini L, et al: APC gene mutations and colorectal adenomatosis
in familial adenomatous polyposis. Br J Cancer. 82:348–353. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu J and Attisano L: Mutations in the
tumor suppressors Smad2 and Smad4 inactivate transforming growth
factor beta signaling by targeting Smads to the
ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 97:4820–4825.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li X, Liu B, Xiao J, Yuan Y, Ma J and
Zhang Y: Roles of VEGF-C and Smad4 in the lymphangiogenesis,
lymphatic metastasis, and prognosis in colon cancer. J Gastrointest
Surg. 15:2001–2010. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang B, Zhang B, Chen X, Bae S, Singh K,
Washington MK and Datta PK: Loss of Smad4 in colorectal cancer
induces resistance to 5-fluorouracil through activating Akt
pathway. Br J Cancer. 110:946–957. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mehrvarz Sarshekeh A, Advani S, Overman
MJ, Manyam G, Kee BK, Fogelman DR, Dasari A, Raghav K, Vilar E,
Manuel S, et al: Association of SMAD4 mutation with patient
demographics, tumor characteristics, and clinical outcomes in
colorectal cancer. PLoS One. 12:e01733452017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Therkildsen C, Bergmann TK,
Henrichsen-Schnack T, Ladelund S and Nilbert M: The predictive
value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment
in metastatic colorectal cancer: A systematic review and
meta-analysis. Acta Oncol. 53:852–864. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mao C, Yang ZY, Hu XF, Chen Q and Tang JL:
PIK3CA exon 20 mutations as a potential biomarker for resistance to
anti-EGFR monoclonal antibodies in KRAS wild-type metastatic
colorectal cancer: A systematic review and meta-analysis. Ann
Oncol. 23:1518–1525. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sartore-Bianchi A, Martini M, Molinari F,
Veronese S, Nichelatti M, Artale S, Di Nicolantonio F, Saletti P,
De Dosso S, Mazzucchelli L, et al: PIK3CA mutations in colorectal
cancer are associated with clinical resistance to EGFR-targeted
monoclonal antibodies. Cancer Res. 69:1851–1857. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
De Roock W, Claes B, Bernasconi D, De
Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V,
Papamichael D, Laurent-Puig P, et al: Effects of KRAS, BRAF, NRAS,
and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy
in chemotherapy-refractory metastatic colorectal cancer: A
retrospective consortium analysis. Lancet Oncol. 11:753–762. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Italiano A, Hostein I, Soubeyran I, Fabas
T, Benchimol D, Evrard S, Gugenheim J, Becouarn Y, Brunet R, Fonck
M, et al: KRAS and BRAF mutational status in primary colorectal
tumors and related metastatic sites: Biological and clinical
implications. Ann Surg Oncol. 17:1429–1434. 2010. View Article : Google Scholar : PubMed/NCBI
|