Introduction
Pancreatic neuroendocrine tumor (pNET), a group of
endocrine tumors originates from the islets of the Langerhans in
the pancreas, accounts for 1–2% of all pancreatic neoplasms
(1,2). pNET are classified into functional
(including insulinoma, gastrinoma and rare functional pNET) and
non-functional types according to their capacity to stimulate
hormonal hypersecretion, as well as the clinical presentation
itself (1,2). Chromosomal alterations cause pNET
through the loss of tumor suppressor genes, such as MEN1, VHL and
p53, or gain of oncogenes, such as CCND1 (3). Mutations induce precursor lesions, such
as diffuse endocrine cell hyperplasia, dysplasia and microadenoma,
which transform into clinical pNETs accompanied by an accumulation
of additional mutations (2,4). The annual incidence of pNET has
increased in the past few years, from 1.4 to 3.0 per million
between 1973 and 2004, according to an analysis on the pNET cases
the Surveillance, Epidemiology, and End Results database (1), and patients with unresectable or
metastatic tumor tend to present with a poor prognosis (4). Thus, a better understanding of the
molecular mechanisms that trigger and drive cancer progression is
required in order to develop effective strategies for the treatment
of pNET. Matrix metalloproteases (MMPs) have been extensively
studied with regard to migration and invasion activity in cancer
(5). MMP2 and MMP9, known as
gelatinases, modulate degradation of the main component of basal
membranes (collagen type IV) and induce metastasis (6). In pNET, MMP9 is associated with
invasion activity of tumor cells and acts as a potent trigger of
the angiogenic switch in mice models (7,8).
Thrombospondins (THBSs) are a family of
matricellular calcium binding glycoprotein that mediate
cell-to-cell and cell-to-matrix interactions. Among the five
members of the THBSs family, THBS1 and THBS2 are unique due to
their type I repeats and antiangiogenic activities (9). THBS2 reportedly interacted with
multiple cell surface receptors (LRP, CD36, CD47 and various
integrins), growth factors (TGF-β and FGF2), ECM proteins (decorin,
HSPGs and fibronectin) and enzymes (MMPs, elastase and cathepsin G)
(10,11). LRP binds to THBS2 in the pericellular
environment to modulate endocytosis and the lysosomal degradation
of THBS2. Downregulation of THBS2 inhibits the formation of the
THBS2-MMP2 complex, leading to decreased MMP2 recycling via LRP,
which regulates tissue-transglutaminase and VEGF and affects
collagen fibrillogenesis and angiogenesis (12). Through binding to CD36, THBS2
regulates a diverse range of activities including angiogenesis,
apoptosis and metastasis (13,14).
THBS2 is an FGF2 ligand that can block FGF2 interaction with
proangiogenic receptors, such as heparin or FGFR1 (15). THBS2 Hep1 peptide can also bind actin
to alter cytoskeletal reorganization (16). THBS2 participates in the progression
of multiple different types of cancer, but with paradoxical
functions, for example, THBS2 inhibited tumor progression in
cervical cancer, but promoted tumor progression in colorectal
carcinoma, thyroid cancer, gastric cancer and hepatocellular
carcinoma (13). Furthermore, the
detailed function of THBS2 in pNET remains to be elucidated.
THBS trimer has been reported to inhibit the
signaling of two members of the tethered ligand receptor family:
The thrombin receptor and the trypsin receptor. For example,
thrombospondin suppressed protease-activated receptor 2 (PAR2) via
CD36 in human microvascular endothelial cells (17). Simultaneously, PAR2 promoted the DNA
binding activity of CUT-like homeobox 1 (CUX1), a transcription
factor that promoted the aggressiveness of pNET partly through MMP9
(4,18). Thus, the aim of the present study was
to examine whether THBS2 suppressed the transcription activity of
CUX1 via MMP9. In the present study, it was revealed that THBS2 was
significantly downregulated in pNET and modulated the proliferation
and migration activity of pNET tumor cells. The present study also
investigated the effect of THBS2 on the CUX1/MMP9 cascade and
revealed that THBS2 regulated the expression of MMP9 through CUX1.
Furthermore, microRNA(miR)-744-5p activated the Wnt/β-catenin
pathway and functioned as a diagnostic and prognostic biomarker in
pancreatic cancer, according to previous reports (19). The present study also proved that
THBS2 was a target of miR-744-5p in pNET.
Materials and methods
Patients and samples
A total of 10 cases of pNET specimens, which were
histopathologically and clinically diagnosed, and resected between
10th May, 2017 and 10th November, 2017 at the Zhongshan Hospital of
Fudan University (Shanghai, China), were investigated in the
present study. The patients included five men and five women, aged
between 45–84 years. pNET is a well-differentiated neuroendocrine
tumor. They resemble normal endocrine cells, express neuroendocrine
markers (chromogranin A and synaptophysin) and hormones (such as
insulin, gastrin, vasoactive intestinal polypeptide, glucagon),
have mild or moderate nuclear atypical changes, and have low
mitotic numbers [<2/10 high power fields (HPF)]. pNET are
classified into three grades, according to 2010 World Health
Organization classification (20):
Grade 1 where the number of mitotic figures in tumor cells is
<2/10 HPF, and/or Ki-67 positive index <2%; Grade 2 where the
number of mitotic figures in tumor cells is between 2–20/10 HPF
and/or Ki-67 positive index 3–20%; and Grade 3 where the number of
mitotic figure in tumor cells is >20/10 HPF and/or Ki-67
positive index >20%. The mitotic image requires at least 50 HPF,
and the Ki-67 positive index requires that the number of Ki-67
positive cells are calculated on the basis of 500 to 2,000 cells in
the proliferative active region. The resected tissues were frozen
and stored in liquid nitrogen. Paracancerous specimens were at
least 2 cm away from tumor tissue. The present study was approved
by Ethics Committee of the Zhongshan Hospital of Fudan University
(Shanghai, China) and all the specimens were used for scientific
research. All participants provided written informed consent prior
to treatment. The clinical information of the specimens are
summarized in Table I.
 | Table I.Sample descriptions. |
Table I.
Sample descriptions.
| Record number | Specimen
number | Pathological
pattern | Pathological
grading |
|---|
| 80,6194 | A4 | NET | G2 |
| 78,3988 | A5 | NET | G1 |
| 78,2319 | A6 | NET | G1 |
| 83,3221 | B1 | NET | G2 |
| 81,9036 | B2 | NET |
|
| 83,1295 | B3 | NET | G1 |
| 89,1998 | B4 | NET |
|
| 76,3270 | B6 | NET,
Insulinoma | G1 |
| 89,2946 | B9 | NET,
Insulinoma | G2 |
| 91,2247 | B10 | NET,
Insulinoma | G1 |
Cell culture
The pNET cell line BON-1, human kidney epithelial
cell line 293 and human lung fibroblast CCL-153 were cultured in
the DMEM medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (FBS) (HyClone; GE
Healthcare Sciences). QGP-1 cells were maintained in RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% FBS. All cells were supplemented with 2 mM L-glutamine, 100
units/ml penicillin, and 100 µg/ml streptomycin (Invitrogen; Thermo
Fisher Scientific, Inc.) and placed in a humidified chamber with 5%
CO2 at 37°C. All the cells were purchased from Chinese
Academy of Sciences, Shanghai Institutes for Biological Sciences.
In the present study, a normal pancreas cell line corresponding to
pNET cells was not used and therefore, 293 and CCL-153 cell lines
were used instead, according to a previous study (21).
Cell transfection
The human THBS2 gene was cloned into pGV358
lentiviral vector (Shanghai GeneChem Co., Ltd.). The pGV358-THBS2
plasmid (20 µg) was mixed with the Helper 1.0 (15 µg) and Helper
2.0 (10 µg) packaging helper plasmid in 1 ml GeneChem transfection
reagent (Shanghai GeneChem Co., Ltd.) and then added to the 293
cells in 10 cm cell culture dish. Supernatants were collected 48 h
post-transfection and incubated with QGP-1 and BON-1 cells at 37°C
for 24 h. After 14 days of puromycin selection, THBS2 overexpressed
(OE) and negative control (NC) cell lines were established.
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) was used for transfection.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cells/tissues using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). RT was performed using the PrimeScript RT reagent kit
(Takara Bio, Inc.) for the mRNA (at 37°C for 15 min followed by
inactivation at 85°C for 5 sec), and miScript II RT kit (Qiagen
GmbH) for miRNA (at 37°C for 60 min followed by inactivation at
95°C for 5 min). RT-qPCR was performed using a SYBR Premix Ex Taq
kit (Takara Bio, Inc.) and miScript PCR Starter kit (Qiagen GmbH).
GAPDH was selected as the reference gene for proteins and U6 was
used as the internal control for miRNA. The qPCR conditions were
used as follows: Initial denaturation at 95°C for 3 min, followed
by 40 cycles of 95°C for 5 sec and 58°C for 30 sec. Relative gene
expression was analyzed using the 2−ΔΔCq method
(22) and normalized to the controls
(GAPDH or U6).
Primer information: miR-744 forward,
5′-TGCGGGGCTAGGGCTAACAGCA-3′ and the universal reverse primer,
5′-TGTCGTGGAGTCGGC-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-AACGCTTCACGAATTTGCGT-3′; THBS2 forward,
5′-GACACGCTGGATCTCACCTAC-3′ and reverse,
5′-GAAGCTGTCTATGAGGTCGCA-3′; CUX1 forward,
5′-GACGTGTTGCGCACTTAACG-3′ and reverse,
5′-ACGACATAGATTGGGCTTAATGCT-3′; MMP9 forward,
5′-TTGACAGCGACAAGAAGTGG-3′ and reverse, 5′-GCCATTCACGTCGTCCTTAT-3′;
GAPDH forward, 5′-TGCACCACCAACTGCTTAGC-3′ and reverse,
5′-GGCATGGACTGTGGTCATGAG-3′. All primers were purchased from Sangon
Biotech Co., Ltd.).
Western blot analysis
The protein was extracted from pNET cells using RIPA
lysis buffer (Beijing Solarbio Science and Technology Co., Ltd.)
with 1 mM phenylmethylsulfonyl fluoride (Beijing Solarbio Science
and Technology Co., Ltd.) on ice and determined using a BCA protein
assay kit (Bio-Rad Laboratorites, Inc.). A total of 1 mg protein
was separated via SDS-PAGE (12% gel) and transferred to
polyvinylidene difluoride (PVDF) membranes (EMD Millipore). The
membranes were subsequently blocked with 5% skimmed milk powder at
room temperature for 1 h and incubated with antibodies against
THBS2 (cat. no. ab112543; 1:1,000; Abcam) and GAPDH (cat. no.
97166; 1:1,000; Cell Signaling Technology, Inc.) at 4°C overnight.
The membranes were then incubated with a horseradish peroxidase
labeled horse anti-mouse secondary antibodies (cat. no. 7076;
1:1,000; Cell Signaling Technology, Inc.) or a goat anti-rabbit
secondary antibody (cat. no. ab7090; 1:10,000; Abcam) for 1 h at
room temperature and Pierce ECL Substrate were used to visualize
the target proteins.
MTT assay
Cell proliferation was assessed using a MTT Cell
Proliferation kit (Roche Diagnostics). The cells (4×103
cells/well) were seeded into 96-well plates and cultured at 37°C.
Upon reaching about 70% confluence, the cells were transfected and
then incubated for 2, 3, 4 or 6 days. A total of 20 µl MTT reagent
(5 mg/ml) was added to each well for 4 h and then removed, after
which 150 µl DMSO was added to dissolve the formazan crystals.
Finally, the absorbance at 570 nm was measured.
EdU assay
The EdU assay was performed using a Cell-Light EdU
DNA Cell Proliferation kit (Guangzhou RiboBio). The cells
(1×104) were cultured in 24 well plates at 37°C for 24
h. After incubation with 50 µM EdU for 2 h at 37°C, the cells were
fixed with 4% paraformaldehyde for 30 min at 37°C and then
neutralized by Glycine. After the cells were permeabilized by 0.5%
TritonX-100 for 10 min at room temperature, Apollo Dye Solution was
added to react with the EdU at room temperature for 30 min. Nucleic
acid was stained with Hoechst at room temperature for 5 min. Images
were captured with an inverted fluorescence microscope (Olympus) at
×200 magnification and 5 fields of each well were randomly selected
to obtain images.
Transwell assay
A total of 1×104 cells in 100 µl low
serum DMEM medium (0.1% FBS) were plated into the upper chamber, to
ensure cell survival, while 600 µl DMEM supplemented with 20% FBS
was added to the lower chamber as a chemical attractant. The cells
were incubated at 37°C for 24 h, after which the cells on the upper
surface of the filter were removed and the cells on the lower side
of the filter were stained with 0.1% crystal violet at room
temperature for 5 min. The cells in five randomly selected fields
were counted under an inverted light microscope (Olympus) at ×100
magnification.
Bioinformatics analysis
GSE73338 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73338)
and GSE43796 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE43796)
from Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) was analyzed to
identify differentially expressed genes in pNET. The online
databases of TargetScan (http://www.targetscan.org/vert_72/), miRTarBase
(http://mirtarbase.mbc.nctu.edu.tw/php/search.php),
RegRNA2.0 (http://regrna2.mbc.nctu.edu.tw/detection.html) and
RNA22 (https://cm.jefferson.edu/rna22/) were used to predict
the putative binding miRNA and the miRNA response elements (MRE) of
THBS2. The LinkedOmics databases (http://www.linkedomics.org/admin.php) was searched to
identify miRNAs that were negatively correlated with THBS2 in
pancreas-adenocarcinoma-other subtype, which were subsequently
aligned them with the targeted miRNAs that were predicted by
TargetScan, miRTarBase and RegRNA2.0 to obtain the potential
targets.
Luciferase reporter assay
The 293 cells in 96 well plates (4×103
cells/well) were cotransfected with THBS2 or THBS2-MRE mutant
reporter constructs and miR-744 mimics
(5′-TGCGGGGCTAGGGCTAACAGCA-3′) or mi-NC
(5′-UUCUCCGAACGUGUCACGUTT-3′) (Sangon Biotech Co., Ltd.) using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 48 h of incubation, luciferase activity
was measured by dual-luciferase reporter assay system (Promega
Corporation) according to the manufacturer's protocol. The
experiments were performed in triplicate and the relative
luciferase activity was normalized to the Renilla luciferase
activity.
Adhered pNET cells were cotransfected with the
reporter constructs with or without MMP9 promoter (Beijing
Haichuang Keye Biotechnology Co., Ltd.) and the vectors
(pCDH-CMV-MCS-EF1-CopGFP-T2A-puro) with or without CUX1 coding
sequences (Beijing Haichuang Keye Biotechnology Co., Ltd.). The
luciferase activity was determined after 48 h of incubation. Cells
transfected with the MMP9 promoter reporter and the overexpression
vector plasmid (pCDH-CMV-MCS-EF1-CopGFP-T2A-puro) were used as
controls.
Immunohistochemistry (IHC) assay
Fresh samples from patients with pNET were cut into
1.5×1.5×0.2 cm3 sections and fixed with 10% neutral
formalin at room temperature for 24 h. The specimens were
dehydrated by a graded series of ethanol solutions (80, 90, 95 and
100%), the ethanol was gradually removed using 1:1 ethanol and
dimethlbenzene mixture followed by 100% dimethlbenzene, embedded in
paraffin and cut into 4 µm slice by a rotary microtome. The
sections were dewaxed and rehydrated using a graded alcohol series
(100, 95, 90 and 80%) and then applied for antigen retrieval (high
pressure heating at 121°C in repair buffer for 3 min). Afterwards,
the sections were blocked with 10% FBS and 1% BSA (Sigma-Aldrich;
Merck KGaA) TBS solution at room temperature for 2 h and incubated
with THBS2 antibody (cat. no. ab112543; 1:500; Abcam) overnight at
4°C. The sections were incubated with 0.3%
H2O2 at room temperature for 15 min and
subsequently with the goat anti-rabbit secondary antibody (cat. no.
ab7090; 1:300; Abcam) at room temperature for 1 h. After
colouration with 3,3-diaminobenzidin at room temperature without
light for 10 min, all sections were dehydrated, cleared, mounted
and visualized using a light microscope (magnification, ×100;
Phenix Optics Co., Ltd.).
Chromatin immunoprecipitation-qPCR
(ChIP-qPCR)
DNA and proteins of QGP-1 and BON-1 cells were
crosslinked with 1% formaldehyde at room temperature for 20 min,
and then the complexes were sonicated on ice for 2 min (25% power;
4.5 sec shock; 9 sec clearance; 14 times; Sonics and Materials,
Inc.) to achieve 200–500 bp DNA fragments. DNA fragments bound by
specific proteins were immunoprecipitated with immunoglobulin G
(cat. no. sc-2025; 1:80; Santa Cruz Biotechnology, Inc.) or CUX1
antibodies (cat. no. sc-13024; 1:40; Santa Cruz Biotechnology,
Inc.). Eluted DNA fragments were analyzed by semi-qPCR. MMP9
forward primer: 5′-CCAATCACCACCATCCGTTG-3′ and reverse primer:
5′-CCTCGGGCAAATGTCTTACC-3′. The reaction conditions were as
follows: Initial denaturation at 95°C for 3 min, then 40 cycles of
95°C for 5 sec and 58°C for 30 sec. Relative gene expression was
analyzed by 2−ΔΔCq method (22) and normalized to the controls. The
qPCR products were separated by a 2% agarose gel, stained by
10000*SolarRed (Beijing Solarbio Science and Technology Co., Ltd.)
and visualized using a Gel imaging analysis system (WD-9413B;
Beijing Liuyi Biotechnology Co., Ltd.)
Statistical analysis
All statistical analyses were performed using SPSS
software (version 17.0; SPSS, Inc.) and GraphPad Prism software
(version 5.0; GraphPad Software, Inc.). Quantitative data were
presented as mean ± standard deviation. The statistical
significance between two groups was calculated by Student's t-test.
The significance of the difference between more than two groups was
evaluated by one-way analysis of variance followed by Tukey's post
hoc test. Linear correlation analyses were performed in order to
determine correlations between THBS2 and miR-744-5p expression
levels. P<0.05 was considered to indicate a statistically
significant difference.
Results
Altered levels of THBS2 in pancreatic
neuroendocrine tumors (pNET)
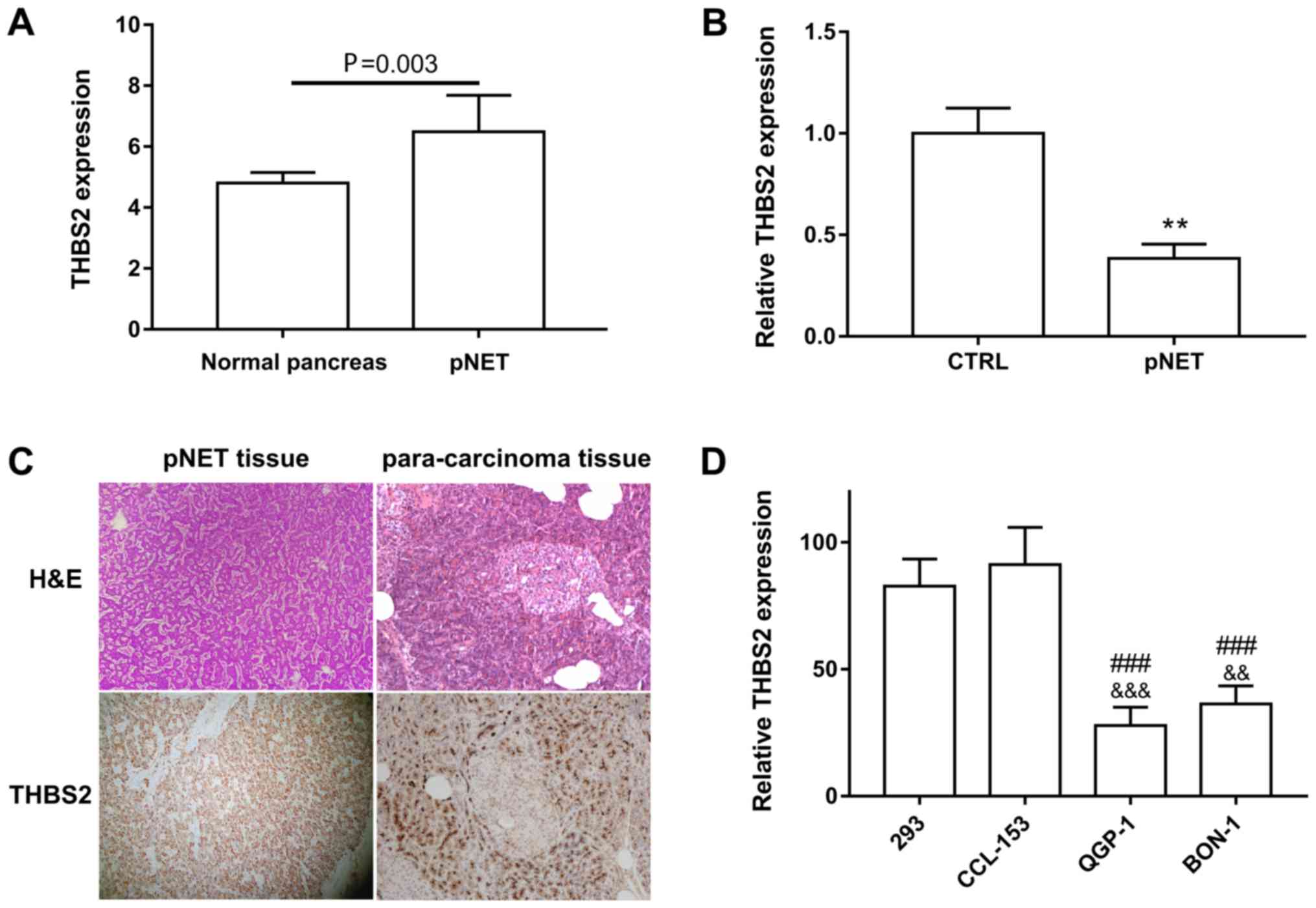
Using publicly available datasets (GSE73338) from
the GEO database, it was revealed that THBS2 was significantly
downregulated in pNET compared with normal tissues (Fig. 1A) (23). In addition, qPCR assay revealed that
THBS2 expression in the 10 pNET samples was 2.6-fold less than that
in para-carcinoma tissues (Fig. 1B).
The expression levels of THBS2 in pNET specimen are presented in
Fig. 1C. Consistently, pNET cell
lines QGP-1 and BON-1 also exhibited decreased levels of THBS2 when
compared with the 293 and human lung fibroblast CCL-153 cell lines
(Fig. 1D).
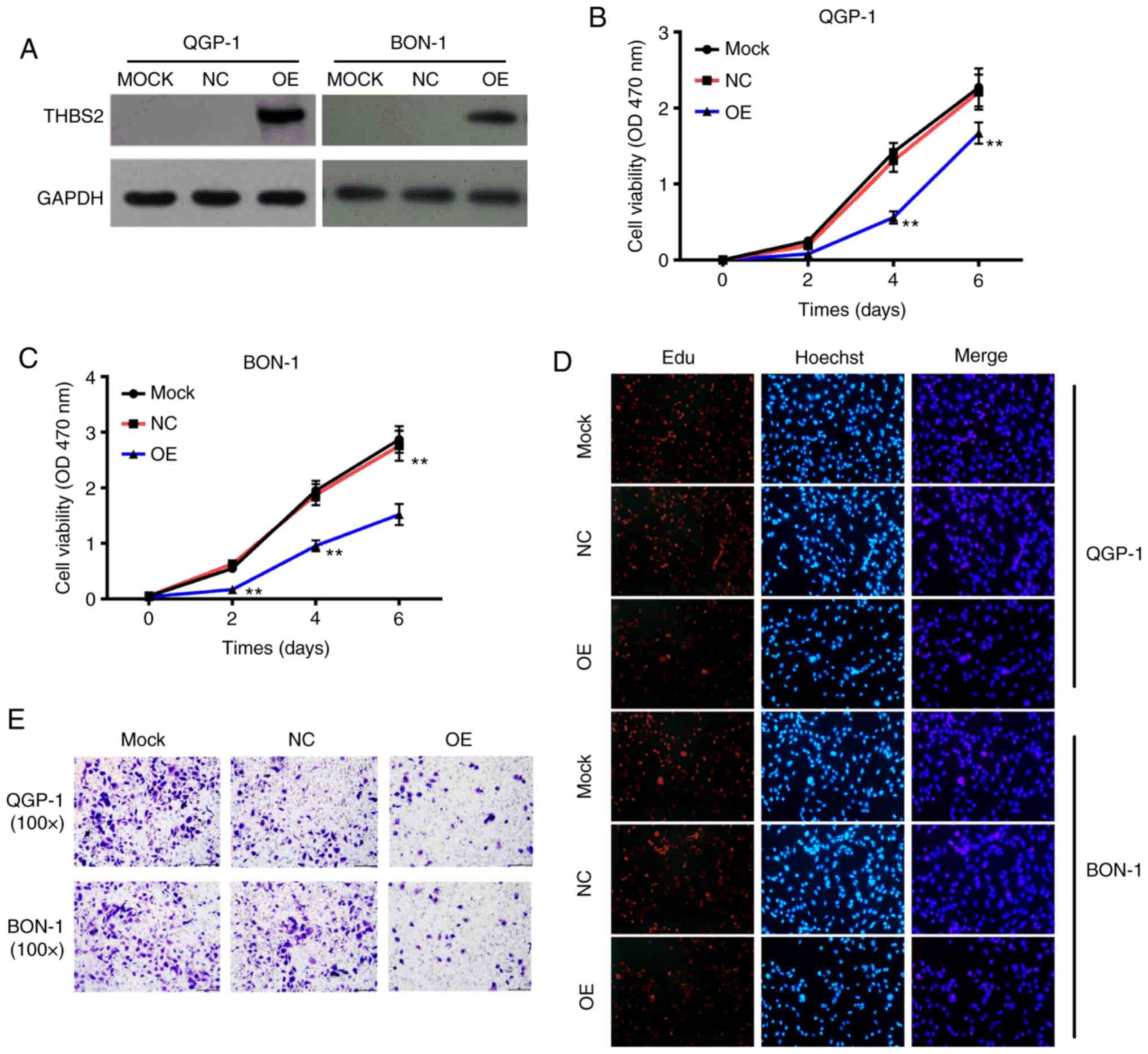
THBS2 overexpression represses the
proliferation and migration of pNET cells in vitro
The present study established THBS2
stable-overexpressed (OE) QGP-1 and BON-1 cells in order to
investigate the effect of THBS2 on pNET development. According to
the qPCR results, OE group expressed >100 times more THBS2
compared with the NC group (data not shown). The western blotting
results further verified the upregulation of THBS2 in THBS2 OE
cells (Fig. 2A). In contrast to the
NC group, THBS2 overexpression impaired the proliferative capacity
of pNET cells, as demonstrated by the MTT and EdU assay (Fig. 2B-D). The migration abilities of
THBS2-overexpressed cells also decreased markedly when compared
with NC (Fig. 2E). These results
indicated that THBS2 significantly inhibited the proliferation and
migration of pNET cells.
THBS2 is regulated by miR-744-5p
Bioinformatics analysis using the TargetScan,
miRTarBase, RegRNA2.0 and RNA22 database predicted that THBS2 mRNA
was the downstream target of miR-744-5p (Fig. 3A). Furthermore, datasets from
LinkedOmics databases demonstrated that the expression level of
miR-744-5p was negatively correlated with THBS2 in
pancreas-adenocarcinoma-other subtype (Fig. 3B). Therefore, the cross-regulation
between THBS2 and miR-744-5p was assayed. First, it was revealed
that miR-744-5p was upregulated in pNET (NCBI/GEO/GSE43796)
(24) and pancreatic cancer samples.
miR-744-5p was also upregulated in BON-1 and QGP-1 cells when
compared with 293 cells (Fig. 3C)
(21). The luciferase reporter assay
was then performed. miR-744-5p cotransfection induced a significant
decrease in the signal of the luciferase reporter containing THBS2
MRE, but not the mutant THBS2 MRE reporter, demonstrating that
THBS2 was a direct target of miR-744-5p (Fig. 3D). In addition, miR-744-5p
overexpression or interference inhibited or promoted THBS2
expression both at transcription and translation level in BON1 and
QGP-1 cells, respectively (Fig. 3E and
F). Furthermore, Pearson's correlation analysis revealed a
negative correlation between miR-744-5p and THBS2 in 30 tumor
tissues (Fig. 3G). These results
indicated that upregulation of miR-744-5p led to the suppression of
THBS2 in pNET.
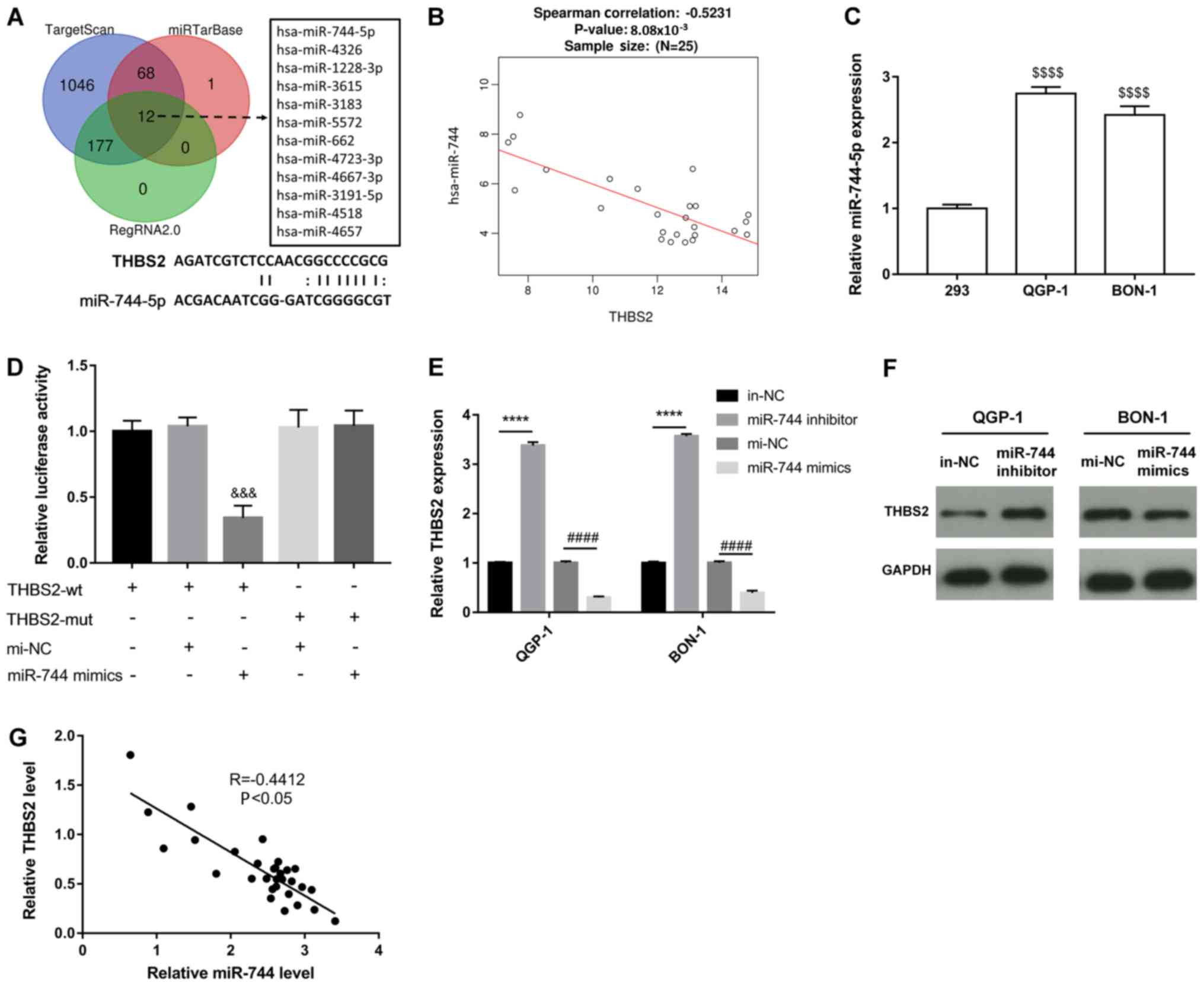 | Figure 3.THBS2 was a direct target of
miR-744-5p. (A) Venn diagram demonstrating that 12 genes are
putative miR-744 targets predicted by TargetScan, miRTarBase and
RegRNA2.0 database. (B) The correlation between miR-744-5p and
THBS2 in pancreas-adenocarcinoma-other subtype from the LinkedOmics
databases. (C) qPCR analysis of the expression of miR-744-5p in
BON-1, QGP-1 and 293 cells. (D) Luciferase activity of wt THBS2
reporter and mutant THBS2 reporter constructs in 293 cells
following transfection of miR-744-5p mimics. (E) qPCR and (F)
western blot analysis the expression of THBS2 following
transfection of miR-744-5p mimics or inhibitor. (G) The expression
of THBS2 was negatively correlated with miR-744-5p in pNET tissues.
$$$$P<0.0001 vs. 293,
&&&P<0.001 vs. THBS2 wt+mi-NC,
****P<0.0001 vs. ni-NC, ####P<0.0001 vs. mi-NC.
THBS2, thrombospondin 2; miR, microRNA; qPCR, quantitative PCR; wt,
wild-type; Mut, mutation; NC, negative control. |
THBS2 inhibits the CUX1/MMP9 axis
Thrombospondin/CD36 cascade regulated PAR2 signaling
in microvascular endothelial cells (17). CUX1 was reported to be the downstream
effector of PAR2 (18). Upregulated
CUX1 also stimulated the expression of MMP9 in pNET. The present
study thus predicted that THBS2 regulated MMP9 through CUX1 in
pNET. First, the present study verified that CUX1 was a
transactivator for MMP9 transcription using ChIP and luciferase
reporter assays (Fig. 4A and B). The
effect of THBS2 on CUX1 was then investigated. CUX1 levels did not
change in either mRNA or protein levels when the expression of
THBS2 was artificially changed in QGP-1 and BON-1 cells (Fig. 4C and D). However, in the ChIP assay,
the amount of CUX1 proteins bound to MMP9 fragments in THBS2 OE
cells was lower compared with that in NC cells, implying that THBS2
may impair the interaction between CUX1 and the MMP9 promoter
(Fig. 4E and F). Consistently, the
transcription stimulation of CUX1 on MMP9 promoter was also
decreased in THBS2 OE cells when compared with NC cells, validated
by luciferase reporter assay (Fig. 4G
and H). Furthermore, THBS2 inhibited the expression of MMP9 at
transcriptional level, whereas CUX1 overexpression alleviated this
effect in QGP-1 cells (Fig. 4I and
K). On the other hand, THBS2 interference stimulated the
production of MMP9, which was counteracted by CUX1 knockdown in
BON-1 cells, indicating that THBS2 regulated the production of MMP9
is CUX1-dependent (Fig. 4J and
K).
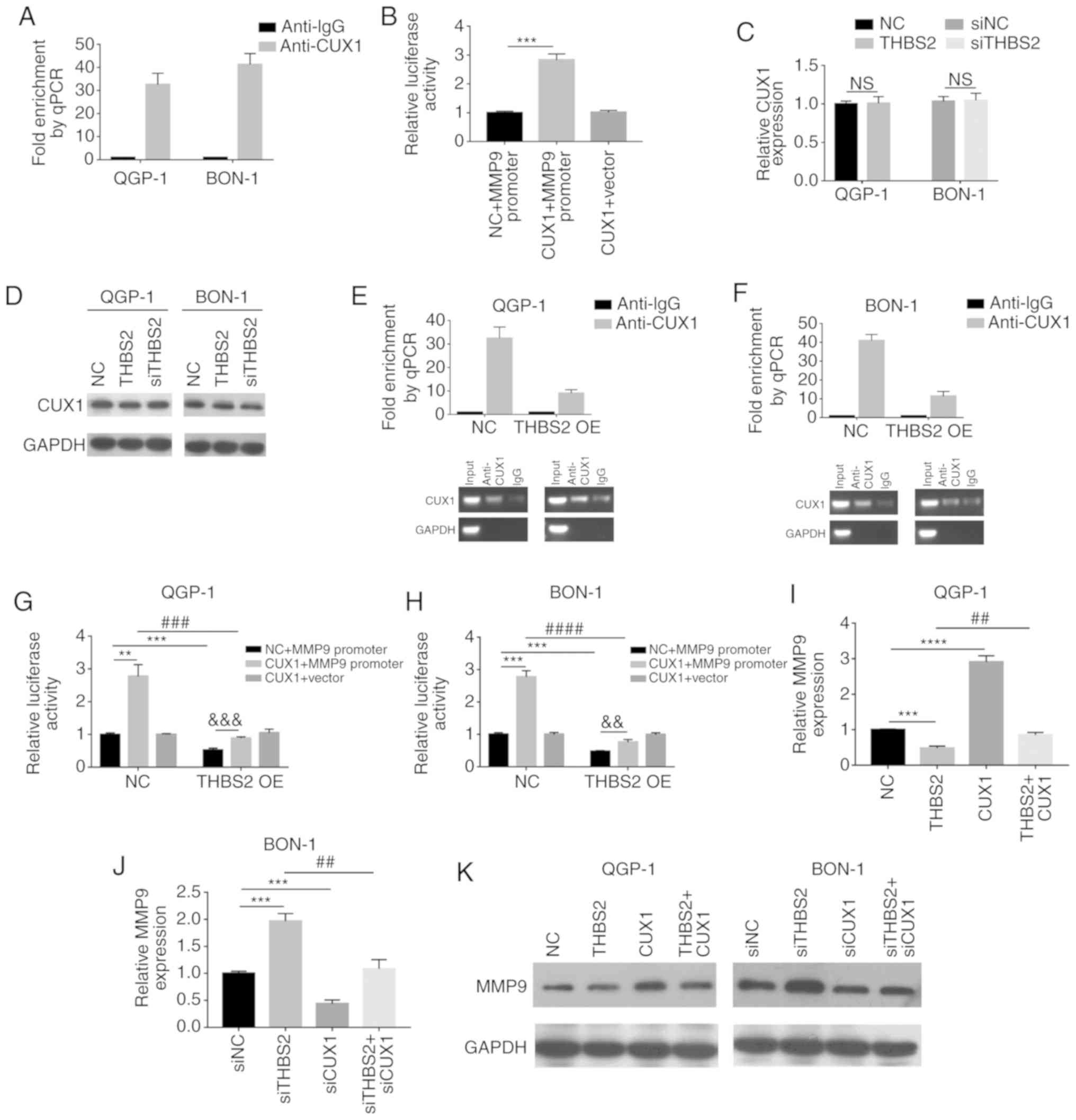 | Figure 4.THBS2 regulates the CUX1/MMP9
cascade. (A) CUX1 directly interacted with the MMP9 promoter. qPCR
analysis following ChIP revealed enrichment of the MMP9 promoter
sequence in the anti-CUX1-precipitated DNA compared with the IgG
group. (B) Luciferase reporter analysis after 293 cells were
co-transfected with plasmid, with or without CUX1 and reporter,
with or without MMP9 promoter for 48 h. (C) qPCR and (D) western
blot analysis demonstrating the expression of CUX1 after THBS2 was
overexpressed or knocked-down. (E and F) qPCR analysis of the MMP9
sequences bound with CUX1 after ChIP analysis in NC or (E) THBS2 OE
QGP-1 and (F) BON-1 cells. (G and H) The relative luciferase
activity was detected after NC or (G) THBS2 OE QGP-1 and (H) BON-1
cells were co-transfected with plasmid with or without CUX1 and
reporter with or without MMP9 promoter for 48 h. (I-K) MMP9
expression was detected by qPCR (I and J) and western blot (K)
after THBS2 or CUX1 was overexpressed or knockdown. **P<0.01,
***P<0.001 or ****P<0.0001 vs. NC or siNC;
##P<0.01 vs. THBS2 or siTHBS2;
###P<0.001 or ####P<0.0001 vs.
CUX1+MMP9 promoter of NC cells; &&P<0.01 or
&&&P<0.001 vs. NC of THBS2 OE cells.
THBS2, thrombospondin 2; CUX1, CUT-like homeobox 1; MMP, matrix
metalloproteinase; qPCR, quantitative PCR; OE, THBS2 lentivirus;
NC, negative control; NS, not significant; si, small
interfering. |
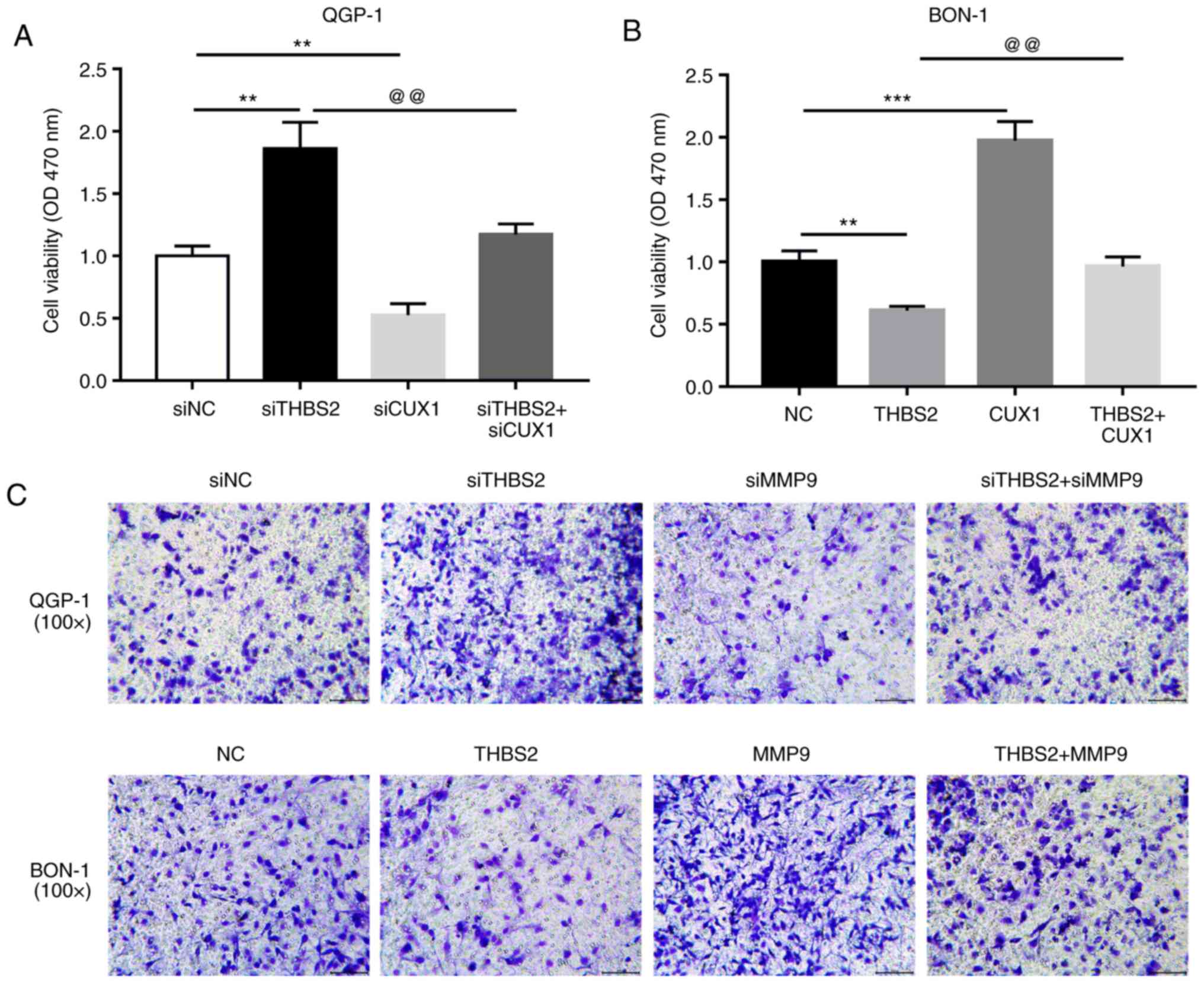
THBS2/CUX1/MMP9 axis is critical for
pNET cell proliferation and migration
The present study assessed whether THBS2 repression
promoted pNET cell proliferation and migration through activating
CUX1 and MMP9. As presented in the MTT and Transwell assay, CUX1
and MMP9 knockdown reversed downregulated THBS2-induced cell
proliferation and migration in QGP-1 cells (Fig. 5A and C), respectively. On the
contrary, CUX1 and MMP9 exogenous expression prevented the
proliferation and migration repression caused by THBS2 upregulation
in BON-1 cells (Fig. 5B and C),
respectively. In summary, these data suggested that THBS2
downregulation stimulated the activity and expression of CUX1 and
MMP9, respectively, which was associated with pNET development.
Discussion
In the present study, it was revealed that THBS2 was
downregulated in pNET tissue and cells. Furthermore, THBS2
overexpression inhibited the proliferation and migration of pNET
cells. Upregulation of miR-744-5p resulted in THBS2 repression.
THBS2 could regulate MMP9 through affecting the transcriptional
activity of CUX1. The THBS2/CUX1/MMP9 cascade modulated
proliferation and migration in pNET.
THBS2 contains the type II repeats similar to
epidermal growth factor repeats, the type III repeats for calcium
binding, the thrombospondin common C-terminal domain for cell
binding, as well as the THBS1 and THBS2 unique type I repeats used
for interacting with TGFβ, MMP9 and CD36 (13). Deregulation of THBS2 induces
adhesion, apoptosis, migration and cytoskeletal organization of
cancer cells (25). Chen et
al (26) reported that THBS2
promoted prostate cancer bone metastasis by inducing
miR-376c-mediated MMP2 upregulation. Cancer-associated fibroblasts
also suppressed prostate cancer invasion via modulation of the
ERα/THBS2/MMP3 axis (27). THBS2
silencing inhibited gastric cancer progression through the PI3K/AKT
signaling pathway (28). THBS2
potentiated Notch3/Jagged1 signaling, which decreased cancer cell
proliferation (29). THBS2
repression receded the degradation of MMP9 and thus promoted VEGF
production (13). In addition, THBS2
modulated a series of processes comprising chondrogenesis collagen
fibrillogenesis, wound healing, angiogenesis, tumor growth and cell
apoptosis (10,30). The present study proved that
downregulation of THBS2 induced proliferation of pNET cells through
CUX1 and promoted migration through MMP9.
PI3K/AKT/NOS, Rac/ROS, CYP1B1/ROS, NF-κB,
adrenocorticotrophic hormone-receptor and estrogen receptor α (ERα)
signaling, as well as DNA methylation and miRNA deregulation, have
all been reported as involved in the regulation of THBS2 expression
(27,30–32).
However, to the best of our knowledge, the regulators of THBS2 in
pNET have not yet been investigated. The present study revealed
that miR-744-5p targeted THBS2 transcripts directly, and that
upregulation of miR-744-5p may induce THBS2 inhibition. Aberrant
expression of miR-744-5p has been identified in a number of
different types of cancer, which affected cancer progression by
targeting different proteins, such as Bcl-2, cMyc, TGF-β1, Notch1,
PTP1B, PAX2, RING1, MAFG, NFIX and HNRNPC (33–38).
miR-744-5p targeted SFRP1, GSK3β and TLE3 to modulate Wnt/β-catenin
signaling, which was associated with lymph node metastasis,
recurrences, prognosis and chemoresistance in pancreatic cancer
(39,40). Furthermore, miR-744-3p stimulated
MMP9 production via different ways in laryngeal squamous cell
carcinoma (19), suggesting that
miR-744 clustering may be a potent regulator of MMP9 as well as
metastasis. Furthermore, transcription factor c-Jun, TLR4/NF-κB
signaling, DNA hypermethylated and T-cell intracellular antigen
(TIA) were revealed to regulate the expression of miR-744-5p
(33,34,41).
However, the factors that stimulate the upregulation of miR-744-5p
in pNET remain unknown.
The function of CUX1 consists of tumor suppression
(via promoting base excision repair and transcriptionally
inhibiting the PI3K/AKT signaling pathway), as well as tumor
promoting (via promoting cell cycle progression and cell
proliferation, stimulating cell migration and invasion, inducing
apoptosis resistance, modulating the tumor microenvironment,
reinforcing spindle assembly checkpoints to promote bipolar
mitosis, and accelerating oxidative DNA damage repair) (42,43).
Upregulation of CUX1 stimulated proliferation, tumor growth,
resistance to apoptosis and angiogenesis in Pnet (8). In the present study, CUX1 functioned as
a transactivator for MMP9 transcription and induced the
proliferation of pNET cells (potentially through modulating the
transcription of certain effectors, for example p21, FGF1, VAV2),
which was consistent with previous studies (44–46).
In the present study, it was speculated that THBS2
inhibited CUX1 through PAR2, as calcium mobilization of PAR2 can be
repressed by thrombospondin/CD36 signaling, and transcription
activity of CUX1 can be stimulated by PAR2 by enhancing its DNA
binding ability (17,18). As demonstrated in the present study,
THBS2 could not regulate the production of CUX1 transcripts or
proteins. However, CUX1 bound much less MMP9 and indicated weaker
transcriptional activity for MMP9 in THBS2 OE cells when compared
with NE cells. These results indicated that THBS2 inhibited the
transcriptional activity of CUX1 for MMP9, which was in accordance
with previous studies. However, whether this effect was indeed
mediated by PAR2 requires further investigation. In addition, CUX1
prevented the affect of THBS2 change on proliferation, which
suggested that CUX1 may be a crucial effector of THBS2.
MMP2/9 forms complexes with THBS2 to interact with
LRP1 and gets degraded; however, THBS2 could also regulate MMPs
indirectly (26). The results from
the present study demonstrated that CUX1 mediated the effect of
THBS2 on MMP9. Whether THBS2 can regulate MMP9 directly in pNET
cells remains uncertain. According to the results of the present
study, THBS2 should not regulate exogenous MMP9 expression through
CUX1, as the MMP9 plasmid lacks CUX1 binding sequences. However,
Fig. 5C demonstrates that THBS2
upregulation inhibited MMP9 overexpression-induced migration,
implying that THBS2 also regulates MMP9 expression at
post-transcriptional level. In addition, MMP9 may be the major
prometastatic effector of THBS2 as MMP9 knockdown or overexpression
almost completely prevented the affect of THBS2 up- or
downregulation on migration, respectively. Thus, the results of the
present study suggested that inhibition of CUX1 and MMP9 may be an
effective method to prevent THBS2 repression-caused pNET
development.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural
Science Foundation of China (grant no. 81773068).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Gene Expression Omnibus,
(https://www.ncbi.nlm.nih.gov/geo/),
the miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/search.php),
the RegRNA2.0 (http://regrna2.mbc.nctu.edu.tw/detection.html), and
the LinkedOmics (http://www.linkedomics.org/admin.php)
repositories.
Authors' contributions
HJ and WL designed the study, and wrote and revised
the manuscript. LZ, JZ and SY performed the study and analyzed the
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Zhongshan Hospital of Fudan University (Shanghai,
China). All patients provided written informed consent prior to the
study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Franko J, Feng W, Yip L, Genovese E and
Moser AJ: Non-functional neuroendocrine carcinoma of the pancreas:
Incidence, tumor biology, and outcomes in 2,158 patients. J
Gastrointest Surg. 14:541–548. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Batukbhai BDO and De Jesus-Acosta A: The
molecular and clinical landscape of pancreatic neuroendocrine
tumors. Pancreas. 48:9–21. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Batcher E, Madaj P and Gianoukakis AG:
Pancreatic neuroendocrine tumors. Endocr Res. 36:35–43. 2017.
View Article : Google Scholar
|
|
4
|
Stevenson M, Lines KE and Thakker RV:
Molecular genetic studies of pancreatic neuroendocrine tumors: New
therapeutic approaches. Endocrinol Metab Clin North Am. 47:525–548.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kessenbrock K, Plaks V and Werb Z: Matrix
metalloproteinases: Regulators of the tumor microenvironment. Cell.
141:52–67. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bauvois B: New facets of matrix
metalloproteinases MMP-2 and MMP-9 as cell surface transducers:
Outside-in signaling and relationship to tumor progression. Biochim
Biophys Acta. 1825:29–36. 2012.PubMed/NCBI
|
|
7
|
Shchors K, Nozawa H, Xu J, Rostker F,
Swigart-Brown L, Evan G and Hanahan D: Increased invasiveness of
MMP-9-deficient tumors in two mouse models of neuroendocrine
tumorigenesis. Oncogene. 32:502–513. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Krug S, Kühnemuth B, Griesmann H, Neesse
A, Mühlberg L, Boch M, Kortenhaus J, Fendrich V, Wiese D, Sipos B,
et al: CUX1: A modulator of tumour aggressiveness in pancreatic
neuroendocrine neoplasms. Endocr Relat Cancer. 21:879–890. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lawler J: The functions of
thrombospondin-1 and −2. Curr Opin Cell Biol. 12:634–640. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Calabro NE, Kristofik NJ and Kyriakides
TR: Thrombospondin-2 and extracellular matrix assembly. Biochim
Biophys Acta. 1840:2396–2402. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mir FA, Contreras-Ruiz L and Masli S:
Thrombospondin-1-dependent immune regulation by transforming growth
factor-β2-exposed antigen-presenting cells. Immunology.
146:547–556. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang Z, Strickland DK and Bornstein P:
Extracellular matrix metalloproteinase 2 levels are regulated by
the low density lipoprotein-related scavenger receptor and
thrombospondin 2. J Biol Chem. 276:8403–8408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lawler J and Detmar M: Tumor progression:
The effects of thrombospondin-1 and −2. Int J Biochem Cell Biol.
36:1038–1045. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Koch M, Hussein F, Woeste A, Gründker C,
Frontzek K, Emons G and Hawighorst T: CD36-mediated activation of
endothelial cell apoptosis by an N-terminal recombinant fragment of
thrombospondin-2 inhibits breast cancer growth and metastasis in
vivo. Breast Cancer Res Treat. 128:337–346. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Farberov S and Meidan R: Functions and
transcriptional regulation of thrombospondins and their
interrelationship with fibroblast growth factor-2 in bovine luteal
cells. Biol Reprod. 91:582014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lopes N, Gregg D, Vasudevan S, Hassanain
H, Goldschmidt-Clermont P and Kovacic H: Thrombospondin 2 regulates
cell proliferation induced by Rac1 redox-dependent signaling. Mol
Cell Biol. 23:5401–5408. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Enenstein J, Gupta K, Vercellotti GM and
Hebbel RP: Thrombin-stimulated calcium mobilization is inhibited by
thrombospondin via CD36. Exp Cell Res. 238:465–471. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wilson BJ, Harada R, LeDuy L, Hollenberg
MD and Nepveu A: CUX1 transcription factor is a downstream effector
of the proteinase-activated receptor 2 (PAR2). J Biol Chem.
284:36–45. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou W, Li Y, Gou S, Xiong J, Wu H, Wang
C, Yan H and Liu T: MiR-744 increases tumorigenicity of pancreatic
cancer by activating Wnt/β-catenin pathway. Oncotarget.
6:37557–37569. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fléjou JF: WHO Classification of digestive
tumors: The fourth edition. Ann Pathol. 31 (5 Suppl):S27–S31.
2011.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang YY and Feng HM: MEG3 suppresses
human pancreatic neuroendocrine tumor cells growth and metastasis
by down-regulation of Mir-183. Cell Physiol Biochem. 44:345–356.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sadanandam A, Wullschleger S, Lyssiotis
CA, Grötzinger C, Barbi S, Bersani S, Körner J, Wafy I, Mafficini
A, Lawlor RT, et al: A cross-species analysis in pancreatic
neuroendocrine tumors reveals molecular subtypes with distinctive
clinical, metastatic, developmental, and metabolic characteristics.
Cancer Discov. 5:1296–1313. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Park M, Kim M, Hwang D, Park M, Kim WK,
Kim SK, Shin J, Park ES, Kang CM, Paik YK and Kim H:
Characterization of gene expression and activated signaling
pathways in solid-pseudopapillary neoplasm of pancreas. Mod Pathol.
27:580–593. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nezu Y, Hagiwara K, Yamamoto Y, Fujiwara
T, Matsuo K, Yoshida A, Kawai A, Saito T and Ochiya T: miR-135b, a
key regulator of malignancy, is linked to poor prognosis in human
myxoid liposarcoma. Oncogene. 35:6177–6188. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen PC, Tang CH, Lin LW, Tsai CH, Chu CY,
Lin TH and Huang YL: Thrombospondin-2 promotes prostate cancer bone
metastasis by the up-regulation of matrix metalloproteinase-2
through down-regulating miR-376c expression. J Hematol Oncol.
10:332017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Slavin S, Yeh CR, Da J, Yu S, Miyamoto H,
Messing EM, Guancial E and Yeh S: Estrogen receptor alpha in
cancer-associated fibroblasts suppresses prostate cancer invasion
via modulation of thrombospondin 2 and matrix metalloproteinase 3.
Carcinogenesis. 35:1301–1309. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ao R, Guan L, Wang Y and Wang JN:
Silencing of COL1A2, COL6A3, and THBS2 inhibits gastric cancer cell
proliferation, migration, and invasion while promoting apoptosis
through the PI3k-Akt signaling pathway. J Cell Biochem.
119:4420–4434. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Meng H, Zhang X, Hankenson KD and Wang MM:
Thrombospondin 2 potentiates notch3/jagged1 signaling. J Biol Chem.
284:7866–7874. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou Q, Dong J, Luo R, Zhou X, Wang J and
Chen F: MicroRNA-20a regulates cell proliferation, apoptosis and
autophagy by targeting thrombospondin 2 in cervical cancer. Eur J
Pharmacol. 844:102–109. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
MacLauchlan S, Yu J, Parrish M, Asoulin
TA, Schleicher M, Krady MM, Zeng J, Huang PL, Sessa WC and
Kyriakides TR: Endothelial nitric oxide synthase controls the
expression of the angiogenesis inhibitor thrombospondin 2. Proc
Natl Acad Sci USA. 108:E1137–E1145. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
De Stefano D, Nicolaus G, Maiuri MC,
Cipolletta D, Galluzzi L, Cinelli MP, Tajana G, Iuvone T and
Carnuccio R: NF-kappaB blockade upregulates Bax, TSP-1, and TSP-2
expression in rat granulation tissue. J Mol Med (Berl). 87:481–492.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hatakeyama H, Nishizawa M, Nakagawa A,
Nakano S, Kigoshi T, Miyamori I and Uchida K: Thrombospondin
expression in aldosterone-producing adenomas. Hypertens Res.
25:523–527. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sha Z, Zhu X, Li N, Li Y and Li D:
Proto-oncogenic miR-744 is upregulated by transcription factor
c-Jun via a promoter activation mechanism. Oncotarget.
7:64977–64986. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sánchez-Jiménez C, Carrascoso I, Barrero J
and Izquierdo JM: Identification of a set of miRNAs differentially
expressed in transiently TIA-depleted HeLa cells by genome-wide
profiling. BMC Mol Biol. 14:42013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen S, Shi F, Zhang W, Zhou Y and Huang
J: miR-744-5p inhibits non-small cell lung cancer proliferation and
invasion by directly targeting PAX2. Technol Cancer Res Treat.
18:15330338198769132019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sui Y, Lin G, Zheng Y and Huang W: LncRNA
MAFG-AS1 boosts the proliferation of lung adenocarcinoma cells via
regulating miR-744-5p/MAFG axis. Eur J Pharmacol. 859:1724652019.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kleemann M, Schneider H, Unger K, Sander
P, Schneider EM, Fischer-Posovszky P, Handrick R and Otte K:
MiR-744-5p inducing cell death by directly targeting HNRNPC and
NFIX in ovarian cancer cells. Sci Rep. 8:90202018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ching T, Song MA, Tiirikainen M, Molnar J,
Berry M, Towner D and Garmire L: Genome-wide hypermethylation
coupled with promoter hypomethylation in the chorioamniotic
membranes of early onset pre-eclampsia. Mol Hum Reprod. 20:885–904.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Miyamae M, Komatsu S, Ichikawa D,
Kawaguchi T, Hirajima S, Okajima W, Ohashi T, Imamura T, Konishi H,
Shiozaki A, et al: Plasma microRNA profiles: Identification of
miR-744 as a novel diagnostic and prognostic biomarker in
pancreatic cancer. Br J Cancer. 113:1467–1476. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang JC, Wu SC, Rau CS, Chen YC, Lu TH, Wu
YC, Tzeng SL, Wu CJ and Hsieh CH: TLR4/NF-κB-responsive microRNAs
and their potential target genes: A mouse model of skeletal muscle
ischemia-reperfusion injury. Biomed Res Int.
2015:4107212015.PubMed/NCBI
|
|
42
|
Ramdzan ZM and Nepveu A: CUX1: A
haploinsufficient tumour suppressor gene overexpressed in advanced
cancers. Nat Rev Cancer. 14:673–682. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hulea L and Nepveu A: CUX1 transcription
factors: From biochemical activities and cell-based assays to mouse
models and human diseases. Gene. 497:18–26. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Paul BM, Vassmer D, Taylor A, Magenheimer
L, Carlton CG, Piontek KB, Germino GG and Vanden Heuvel GB: Ectopic
expression of Cux1 is associated with reduced p27 expression and
increased apoptosis during late stage cyst progression upon
inactivation of Pkd1 in collecting ducts. Dev Dyn. 240:1493–1501.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Latreille R, Servant R, Darsigny M,
Marcoux S, Jones C, Perreault N and Boudreau F: Transcription
factor CUX1 is required for intestinal epithelial wound healing and
targets the VAV2-RAC1 signalling complex. Biochim Biophys Acta Mol
Cell Res. 1864:2347–2355. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen J, Zhou Z, Yao Y, Dai J, Zhou D, Wang
L and Zhang QQ: Dipalmitoylphosphatidic acid inhibits breast cancer
growth by suppressing angiogenesis via inhibition of the
CUX1/FGF1/HGF signalling pathway. J Cell Mol Med. 22:4760–4770.
2018. View Article : Google Scholar : PubMed/NCBI
|