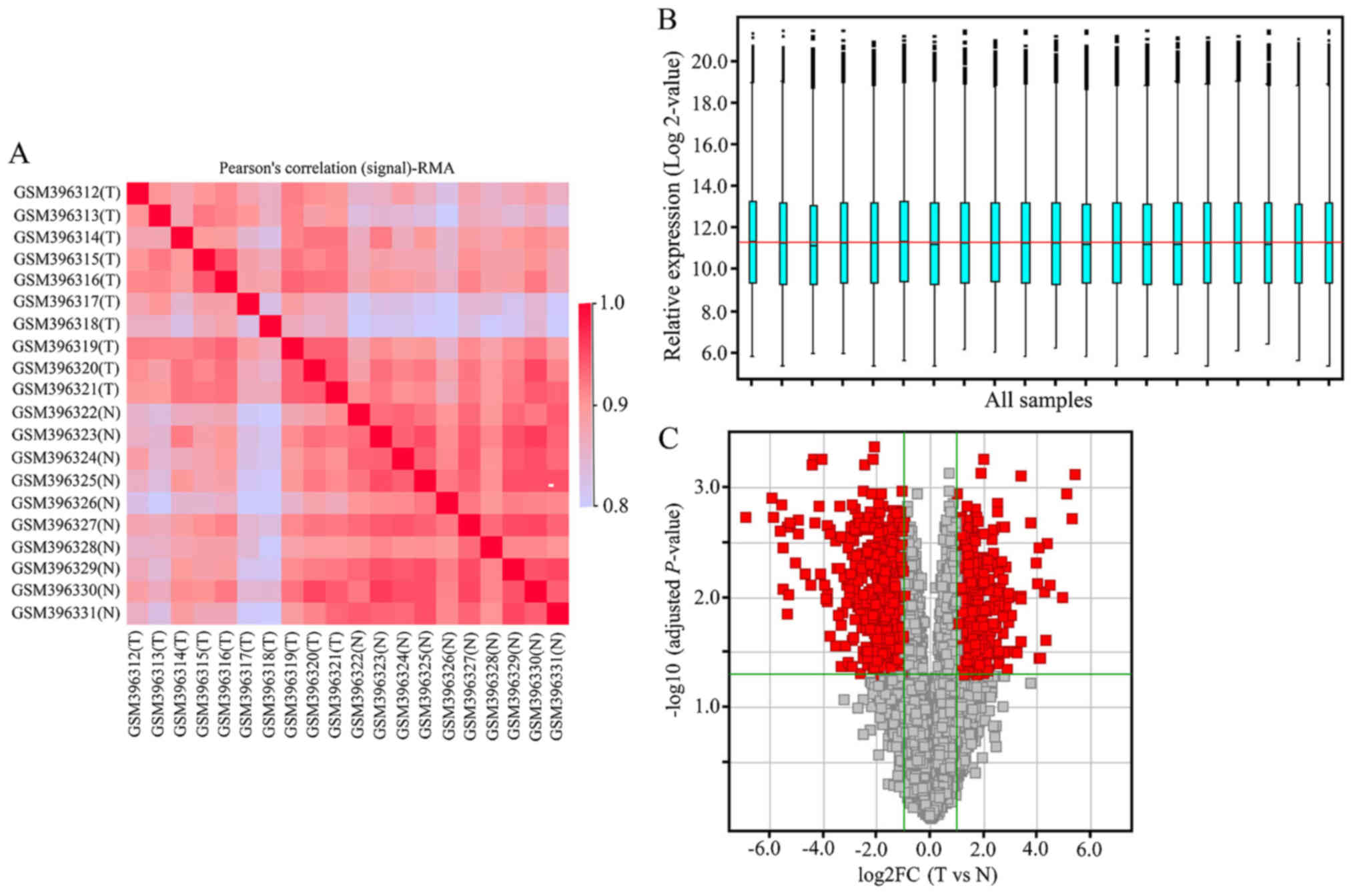
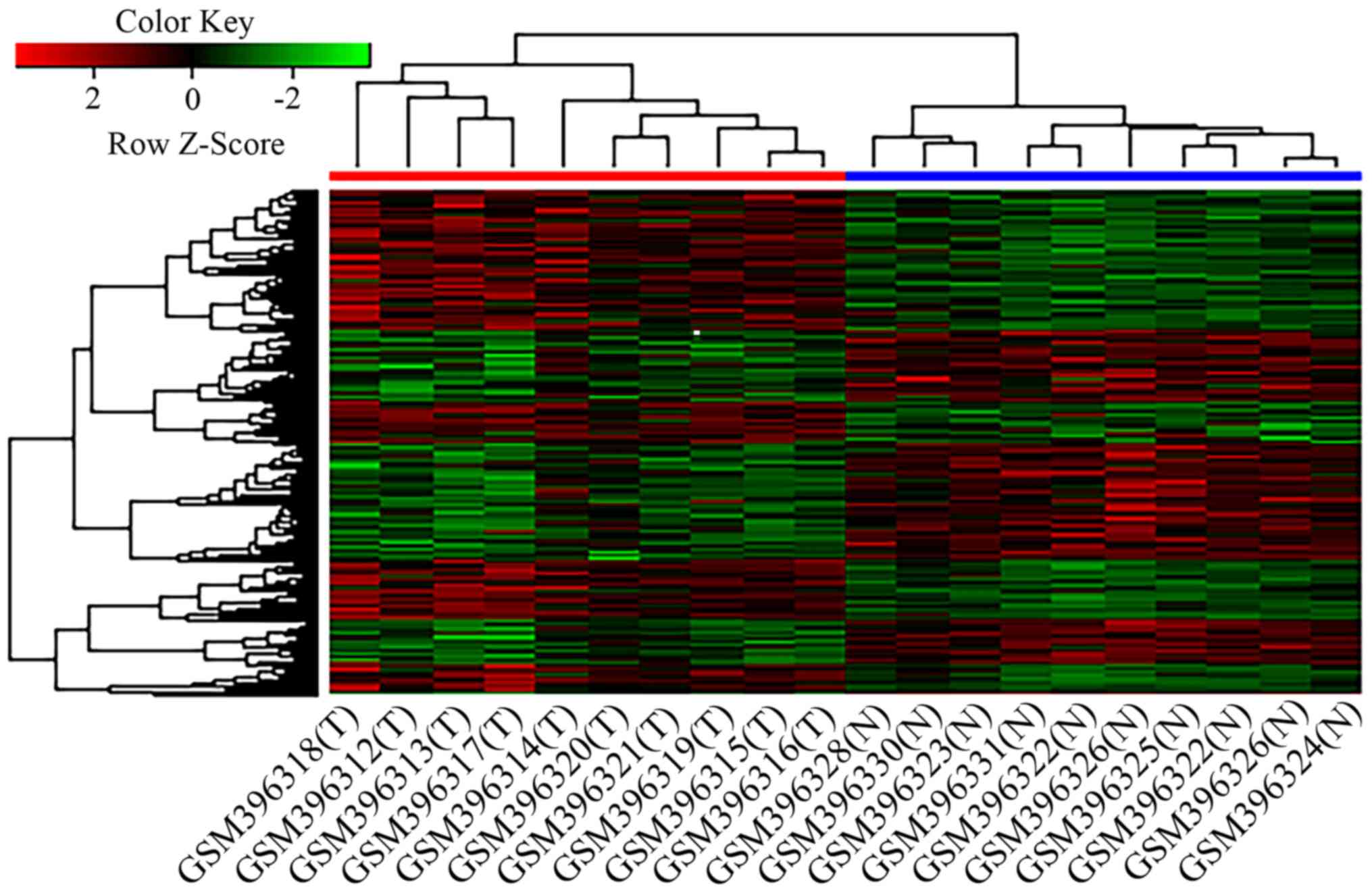
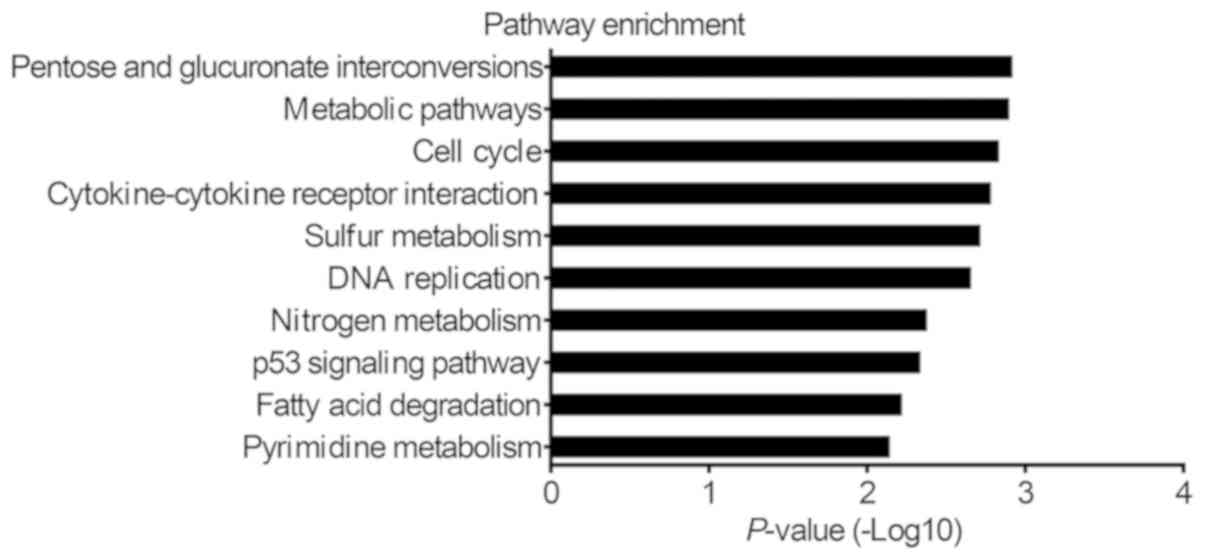
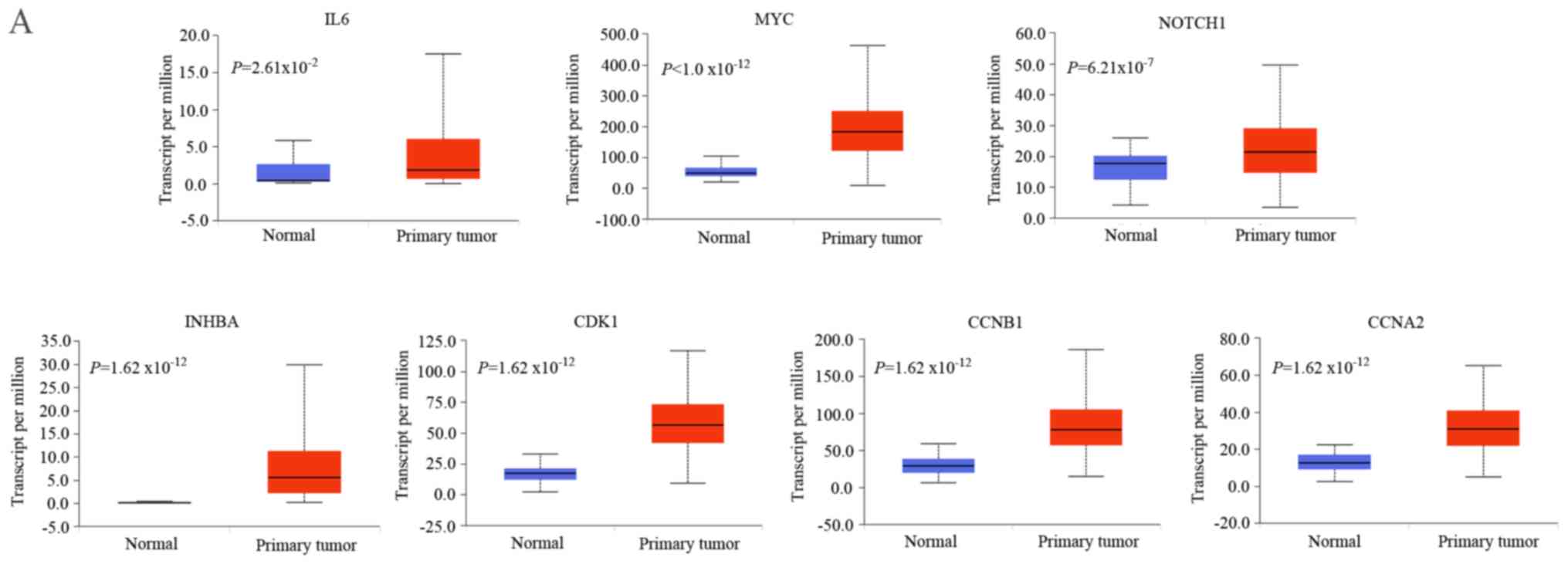
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Al Bandar MH and Kim NK: Current status
and future perspectives on treatment of liver metastasis in
colorectal cancer (Review). Oncol Rep. 37:2553–2564. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim SC, Shin YK, Kim YA, Jang SG and Ku
JL: Identification of genes inducing resistance to ionizing
radiation in human rectal cancer cell lines: Re-sensitization of
radio-resistant rectal cancer cells through down regulating NDRG1.
BMC Cancer. 18:5942018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Andersen SE, Andersen IB, Jensen BV,
Pfeiffer P, Ota T and Larsen JS: A systematic review of
observational studies of trifluridine/tipiracil (TAS-102) for
metastatic colorectal cancer. Acta Oncol. 58:1149–1157. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jin S, Mu Y, Wang X, Liu Z, Wan L, Xiong
Y, Zhang Y, Zhou L and Li L: Overexpressed RACK1 is positively
correlated with malignant degree of human colorectal carcinoma. Mol
Biol Rep. 41:3393–3399. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li XY, Hu Y, Li NS, Wan JH, Zhu Y and Lu
NH: RACK1 acts as a potential tumor promoter in colorectal cancer.
Gastroenterol Res Pract. 2019:56250262019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ouyang S, Zhou X, Chen Z, Wang M, Zheng X
and Xie M: LncRNA BCAR4, targeting to miR-665/STAT3 signaling,
maintains cancer stem cells stemness and promotes tumorigenicity in
colorectal cancer. Cancer Cell Int. 19:722019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shangguan H, Tan SY and Zhang JR:
Bioinformatics analysis of gene expression profiles in
hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 19:2054–2061.
2015.PubMed/NCBI
|
|
10
|
Kosti A, Harry Chen HI, Mohan S, Liang S,
Chen Y and Habib SL: Microarray profile of human kidney from
diabetes, renal cell carcinoma and renal cell carcinoma with
diabetes. Genes Cancer. 6:62–70. 2015.PubMed/NCBI
|
|
11
|
Christgen M, Geffers R, Kreipe H and
Lehmann U: IPH-926 lobular breast cancer cells are triple-negative
but their microarray profile uncovers a luminal subtype. Cancer
Sci. 104:1726–1730. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu J, Li X, Zhong C, Li D, Zhai X, Hu W,
Guo C, Yuan Y and Zheng S: High-throughput proteomics integrated
with gene microarray for discovery of colorectal cancer potential
biomarkers. Oncotarget. 7:75279–75292. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shen X, Yue M, Meng F, Zhu J, Zhu X and
Jiang Y: Microarray analysis of differentially-expressed genes and
linker genes associated with the molecular mechanism of colorectal
cancer. Oncol Lett. 12:3250–3258. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Snipstad K, Fenton CG, Kjaeve J, Cui G,
Anderssen E and Paulssen RH: New specific molecular targets for
radio-chemotherapy of rectal cancer. Mol Oncol. 4:52–64. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hardcastle TJ: Generalised empirical
Bayesian methods for discovery of differential data in
high-throughput biology. Bioinformatics. 32:195–202.
2016.PubMed/NCBI
|
|
17
|
Huang DW, Sherman BT, Tan Q, Kir J, Liu D,
Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC and Lempicki RA:
DAVID Bioinformatics Resources: Expanded annotation database and
novel algorithms to better extract biology from large gene lists.
Nucleic Acids Res. 35:W169–W175. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tang Y, Li M, Wang J, Pan Y and Wu FX:
CytoNCA: A cytoscape plugin for centrality analysis and evaluation
of protein interaction networks. Biosystems. 127:67–72. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gao X, Wang X and Zhang S: Bioinformatics
identification of crucial genes and pathways associated with
hepatocellular carcinoma. Biosci Rep. 38:BSR201814412018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hassanain M, Al-Alem F, Simoneau E, Traiki
TA, Alsaif F, Alsharabi A, Al-Faris H and Al-Saleh K: Colorectal
cancer liver metastasis trends in the kingdom of Saudi Arabia.
Saudi J Gastroenterol. 22:370–374. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Katsidzira L, Ocvirk S, Wilson A, Li J,
Mahachi CB, Soni D, DeLany J, Nicholson JK, Zoetendal EG and
O'Keefe SJD: Differences in fecal gut microbiota, short-chain fatty
acids and bile acids link colorectal cancer risk to dietary changes
associated with urbanization among zimbabweans. Nutr Cancer.
71:1313–1324. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Park Y, Park SJ, Cheon JH, Kim WH and Kim
TI: Association of family history with cancer recurrence, survival,
and the incidence of colorectal adenoma in patients with colorectal
cancer. J Cancer Prev. 24:1–10. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cordier-Bussat M, Thibert C, Sujobert P,
Genestier L, Fontaine É and Billaud M: Even the Warburg effect can
be oxidized: Metabolic cooperation and tumor development. Med Sci
(Paris). 34:701–708. 2018.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Subramanian C and Cohen MS: Over
expression of DNA damage and cell cycle dependent proteins are
associated with poor survival in patients with adrenocortical
carcinoma. Surgery. 165:202–210. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Suehiro Y, Takemoto Y, Nishimoto A, Ueno
K, Shirasawa B, Tanaka T, Kugimiya N, Suga A, Harada E and Hamano
K: Dclk1 Inhibition cancels 5-FU-induced cell-cycle arrest and
decreases cell survival in colorectal cancer. Anticancer Res.
38:6225–6230. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Thorenoor N, Faltejskova-Vychytilova P,
Hombach S, Mlcochova J, Kretz M, Svoboda M and Slaby O: Long
non-coding RNA ZFAS1 interacts with CDK1 and is involved in
p53-dependent cell cycle control and apoptosis in colorectal
cancer. Oncotarget. 7:622–637. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fang Y, Yu H, Liang X, Xu J and Cai X:
Chk1-induced CCNB1 overexpression promotes cell proliferation and
tumor growth in human colorectal cancer. Cancer Biol Ther.
15:1268–1279. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gan Y, Li Y, Li T, Shu G and Yin G: CCNA2
acts as a novel biomarker in regulating the growth and apoptosis of
colorectal cancer. Cancer Manag Res. 10:5113–5124. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Patel SA and Gooderham NJ: IL6 mediates
immune and colorectal cancer cell cross-talk via miR-21 and
miR-29b. Mol Cancer Res. 13:1502–1508. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Miteva LD, Stanilov NS, Cirovski GМ and
Stanilova SA: Upregulation of Treg-related genes in addition with
IL6 showed the significant role for the distant metastasis in
colorectal cancer. Cancer Microenviron. 10:69–76. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lv Z, Wei J, You W, Wang R, Shang J, Xiong
Y, Yang H, Yang X and Fu Z: Disruption of the
c-Myc/miR-200b-3p/PRDX2 regulatory loop enhances tumor metastasis
and chemotherapeutic resistance in colorectal cancer. J Transl Med.
15:2572017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Okano M, Yamamoto H, Ohkuma H, Kano Y, Kim
H, Nishikawa S, Konno M, Kawamoto K, Haraguchi N, Takemasa I, et
al: Significance of INHBA expression in human colorectal cancer.
Oncol Rep. 30:2903–2908. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang H, Jiang H, Chen L, Liu J, Hu X and
Zhang H: Inhibition of Notch1/Hes1 signaling pathway improves
radiosensitivity of colorectal cancer cells. Eur J Pharmacol.
818:364–370. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sajadimajd S and Yazdanparast R:
Differential behaviors of trastuzumab-sensitive and -resistant
SKBR3 cells treated with menadione reveal the involvement of
Notch1/Akt/FOXO1 signaling elements. Mol Cell Biochem. 408:89–102.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Prabakaran DS, Muthusami S, Sivaraman T,
Yu JR and Park WY: Silencing of FTS increases radiosensitivity by
blocking radiation-induced Notch1 activation and spheroid formation
in cervical cancer cells. Int J Biol Macromol. 126:1318–1325. 2019.
View Article : Google Scholar : PubMed/NCBI
|