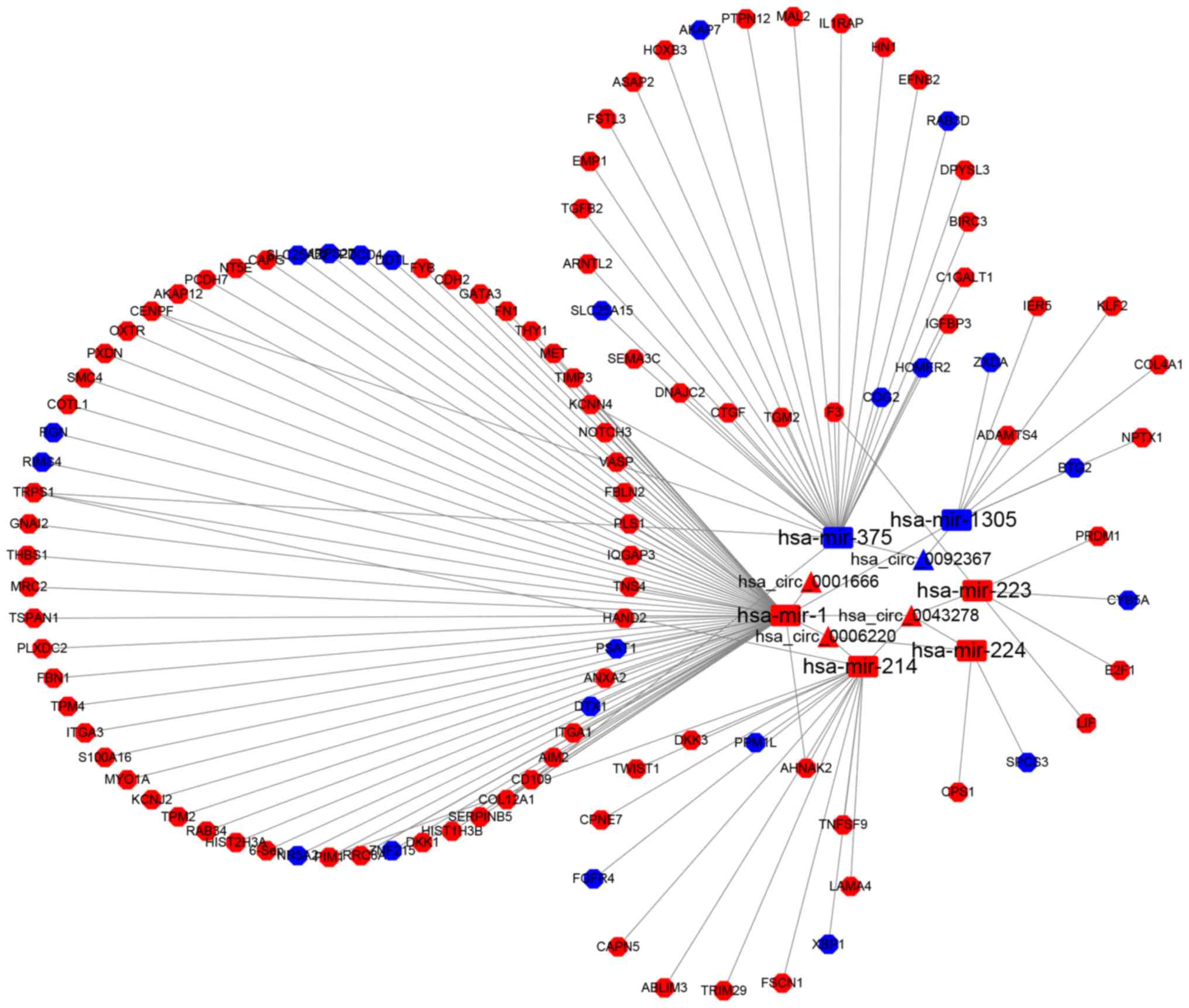
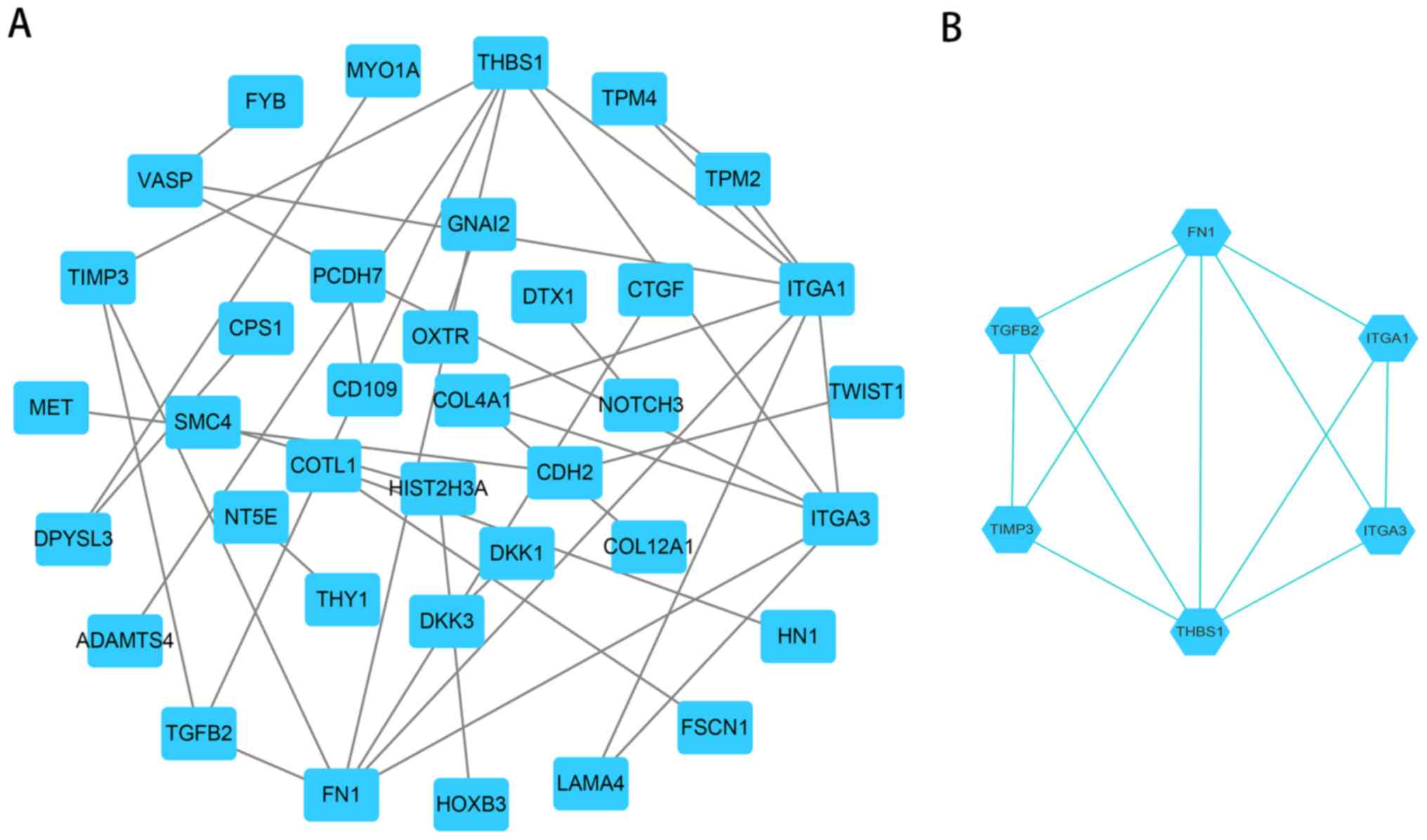
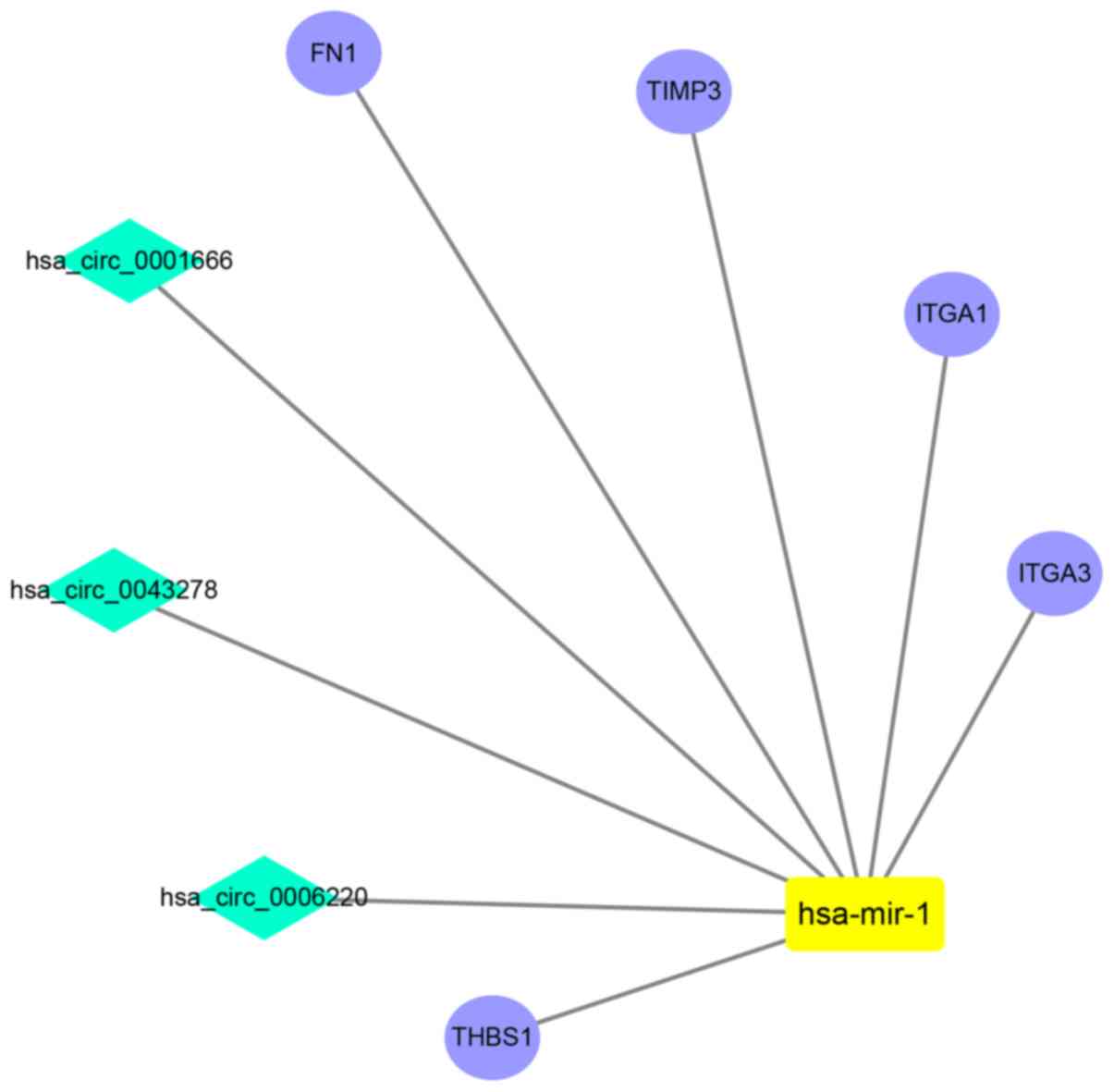
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
DeSantis CE, Lin CC, Mariotto AB, Siegel
RL, Stein KD, Kramer JL, Alteri R, Robbins AS and Jemal A: Cancer
treatment and survivorship statistics, 2014. CA Cancer J Clin.
64:252–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hidalgo M: Pancreatic cancer. N Engl J
Med. 362:1605–1617. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chrystoja CC, Diamandis EP, Brand R,
Ruckert F, Haun R and Molina R: Pancreatic cancer. Clin Chem.
59:41–46. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Du WW, Zhang C, Yang W, Yong T, Awan FM
and Yang BB: Identifying and characterizing circRNA-protein
interaction. Theranostics. 7:4183–4191. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu J, Liu T, Wang X and He A: Circles
reshaping the RNA world: From waste to treasure. Mol Cancer.
16:582017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li J, Yang J, Zhou P, Le Y, Zhou C, Wang
S, Xu D, Lin HK and Gong Z: Circular RNAs in cancer: Novel insights
into origins, properties, functions and implications. Am J Cancer
Res. 5:472–480. 2015.PubMed/NCBI
|
|
9
|
Lv C, Sun L, Guo Z, Li H, Kong D, Xu B,
Lin L, Liu T, Guo D, Zhou J and Li Y: Circular RNA regulatory
network reveals cell-cell crosstalk in acute myeloid leukemia
extramedullary infiltration. J Transl Med. 16:3612018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu
K, Fan Q, Li J, Ning T, Tian F, et al: Exosome circRNA secreted
from adipocytes promotes the growth of hepatocellular carcinoma by
targeting deubiquitination-related USP7. Oncogene. 38:2844–2859.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang
S, Song W, Li X, Li L, Du Z, et al: A novel FLI1 exonic circular
RNA promotes metastasis in breast cancer by coordinately regulating
TET1 and DNMT1. Genome Biol. 19:2182018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong
F, Ren D, Ye X, Li C, Wang Y, et al: Circular RNAs function as
ceRNAs to regulate and control human cancer progression. Mol
Cancer. 17:792018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Retraction: Circular RNA-0007874
(circMTO1) reverses chemoresistance to temozolomide by acting as a
sponge of microRNA-630 in glioblastoma by Jiang Rao, Xinxin Cheng,
Huimin Zhu, Lifeng Wang, Li Liu. Cell Biol Int. Dec 11–2018.(Epub
ahead of print).
|
|
14
|
Wei H, Pan L, Tao D and Li R: Circular RNA
circZFR contributes to papillary thyroid cancer cell proliferation
and invasion by sponging miR-1261 and facilitating C8orf4
expression. Biochem Biophys Res Commun. 503:56–61. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li H, Hao X, Wang H, Liu Z, He Y, Pu M,
Zhang H, Yu H, Duan J and Qu S: Circular RNA expression profile of
pancreatic ductal adenocarcinoma revealed by microarray. Cell
Physiol Biochem. 40:1334–1344. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guo S, Xu X, Ouyang Y, Wang Y, Yang J, Yin
L, Ge J and Wang H: Microarray expression profile analysis of
circular RNAs in pancreatic cancer. Mol Med Rep. 17:7661–7671.
2018.PubMed/NCBI
|
|
17
|
Sandhu V, Bowitz Lothe IM, Labori KJ,
Skrede ML, Hamfjord J, Dalsgaard AM, Buanes T, Dube G, Kale MM,
Sawant S, et al: Differential expression of miRNAs in
pancreatobiliary type of periampullary adenocarcinoma and its
associated stroma. Mol Oncol. 10:303–316. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kolde R, Laur S, Adler P and Vilo J:
Robust rank aggregation for gene list integration and
meta-analysis. Bioinformatics. 28:573–580. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chou CH, Shrestha S, Yang CD, Chang NW,
Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al: miRTarBase
update 2018: A resource for experimentally validated
microRNA-target interactions. Nucleic Acids Res. 46(D1): D296–D302.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
eLife. 4:e050052015. View Article : Google Scholar
|
|
22
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bandettini WP, Kellman P, Mancini C,
Booker OJ, Vasu S, Leung SW, Wilson JR, Shanbhag SM, Chen MY and
Arai AE: MultiContrast delayed enhancement (MCODE) improves
detection of subendocardial myocardial infarction by late
gadolinium enhancement cardiovascular magnetic resonance: A
clinical validation study. J Cardiovasc Magn Reson. 14:832012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rybak-Wolf A, Stottmeister C, Glažar P,
Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss
R, et al: Circular RNAs in the mammalian brain are highly abundant,
conserved, and dynamically expressed. Mol Cell. 58:870–885. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P
and Wu M: CircRNA: Functions and properties of a novel potential
biomarker for cancer. Mol Cancer. 16:942017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jiang XM, Li ZL, Li JL, Xu Y, Leng KM, Cui
YF and Sun DJ: A novel prognostic biomarker for cholangiocarcinoma:
circRNA Cdr1as. Eur Rev Med Pharmacol Sci. 22:365–371.
2018.PubMed/NCBI
|
|
30
|
Chen X, Chen RX, Wei WS, Li YH, Feng ZH,
Tan L, Chen JW, Yuan GJ, Chen SL, Guo SJ, et al: PRMT5 circular RNA
promotes metastasis of urothelial carcinoma of the bladder through
sponging miR-30c to induce epithelial-mesenchymal transition. Clin
Cancer Res. 24:6319–6330. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu X, Abraham JM, Cheng Y, Wang Z, Wang
Z, Zhang G, Ashktorab H, Smoot DT, Cole RN, Boronina TN, et al:
Synthetic Circular RNA functions as a miR-21 sponge to suppress
gastric carcinoma cell proliferation. Mol Ther Nucleic Acids.
13:312–321. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cao S, Chen G, Yan L, Li L and Huang X:
Contribution of dysregulated circRNA_100876 to proliferation and
metastasis of esophageal squamous cell carcinoma. Onco Targets
Ther. 11:7385–7394. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang F, Liu DY, Guo JT, Ge N, Zhu P, Liu
X, Wang S, Wang GX and Sun SY: Circular RNA circ-LDLRAD3 as a
biomarker in diagnosis of pancreatic cancer. World J Gastroenterol.
23:8345–8354. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang WJ, Wang Y, Liu S, Yang J, Guo SX,
Wang L, Wang H and Fan YF: Silencing circular RNA hsa_circ_0000977
suppresses pancreatic ductal adenocarcinoma progression by
stimulating miR-874-3p and inhibiting PLK1 expression. Cancer Lett.
422:70–80. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu Y, Yao Y, Gao P and Cui Y: Upregulated
circular RNA circ_0030235 predicts unfavorable prognosis in
pancreatic ductal adenocarcinoma and facilitates cell progression
by sponging miR-1253 and miR-1294. Biochem Biophys Res Commun.
509:138–142. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu C, Liu R, Zhang D, Deng Q, Liu B, Chao
HP, Rycaj K, Takata Y, Lin K, Lu Y, et al: MicroRNA-141 suppresses
prostate cancer stem cells and metastasis by targeting a cohort of
pro-metastasis genes. Nat Commun. 8:142702017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
An Y, Cai H, Zhang Y, Liu S, Duan Y, Sun
D, Chen X and He X: circZMYM2 competed endogenously with miR-335-5p
to regulate JMJD2C in pancreatic cancer. Cell Physiol Biochem.
51:2224–2236. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hao L, Rong W, Bai L, Cui H, Zhang S, Li
Y, Chen D and Meng X: Upregulated circular RNA circ_0007534
indicates an unfavorable prognosis in pancreatic ductal
adenocarcinoma and regulates cell proliferation, apoptosis, and
invasion by sponging miR-625 and miR-892b. J Cell Biochem.
120:3780–3789. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cheng Q, Han LH, Zhao HJ, Li H and Li JB:
Abnormal alterations of miR-1 and miR-214 are associated with
clinicopathological features and prognosis of patients with PDAC.
Oncol Lett. 14:4605–4612. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cao TH, Ling X, Chen C, Tang W, Hu DM and
Yin GJ: Role of miR-214-5p in the migration and invasion of
pancreatic cancer cells. Eur Rev Med Pharmacol Sci. 22:7214–7221.
2018.PubMed/NCBI
|
|
42
|
Zhu G, Zhou L, Liu H, Shan Y and Zhang X:
MicroRNA-224 promotes pancreatic cancer cell proliferation and
migration by targeting the TXNIP-mediated HIF1α pathway. Cell
Physiol Biochem. 48:1735–1746. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
He D, Huang C, Zhou Q, Liu D, Xiong L,
Xiang H, Ma G and Zhang Z: HnRNPK/miR-223/FBXW7 feedback cascade
promotes pancreatic cancer cell growth and invasion. Oncotarget.
8:20165–20178. 2017.PubMed/NCBI
|
|
44
|
Yang D, Yan R and Zhang X, Zhu Z, Wang C,
Liang C and Zhang X: Deregulation of MicroRNA-375 inhibits cancer
proliferation migration and chemosensitivity in pancreatic cancer
through the association of HOXB3. Am J Transl Res. 8:1551–1559.
2016.PubMed/NCBI
|
|
45
|
Begum A, Ewachiw T, Jung C, Huang A,
Norberg KJ, Marchionni L, McMillan R, Penchev V, Rajeshkumar NV,
Maitra A, et al: The extracellular matrix and focal adhesion kinase
signaling regulate cancer stem cell function in pancreatic ductal
adenocarcinoma. PLoS One. 12:e01801812017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang J, Dumartin L, Mafficini A, Ulug P,
Sangaralingam A, Alamiry NA, Radon TP, Salvia R, Lawlor RT, Lemoine
NR, et al: Splice variants as novel targets in pancreatic ductal
adenocarcinoma. Sci Rep. 7:29802017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dhayat SA, Traeger MM, Rehkaemper J,
Stroese AJ, Steinestel K, Wardelmann E, Kabar I and Senninger N:
Clinical impact of epithelial-to-mesenchymal transition regulating
MicroRNAs in pancreatic ductal adenocarcinoma. Cancers (Basel).
10(pii): E3282018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kumar S, Inigo JR, Kumar R, Chaudhary AK,
O'Malley J, Balachandar S, Wang J, Attwood K, Yadav N, Hochwald S,
et al: Nimbolide reduces CD44 positive cell population and induces
mitochondrial apoptosis in pancreatic cancer cells. Cancer Lett.
413:82–93. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Asama H, Suzuki R, Hikichi T, Takagi T,
Masamune A and Ohira H: MicroRNA let-7d targets thrombospondin-1
and inhibits the activation of human pancreatic stellate cells.
Pancreatology. 19:196–203. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
D'Cruz OJ, Qazi S, Hwang L, Ng K and Trieu
V: Impact of targeting transforming growth factor β-2 with
antisense OT-101 on the cytokine and chemokine profile in patients
with advanced pancreatic cancer. Onco Targets Ther. 11:2779–2796.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gharibi A, La Kim S, Molnar J, Brambilla
D, Adamian Y, Hoover M, Hong J, Lin J, Wolfenden L and Kelber JA:
ITGA1 is a pre-malignant biomarker that promotes therapy resistance
and metastatic potential in pancreatic cancer. Sci Rep.
7:100602017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Idichi T, Seki N, Kurahara H, Fukuhisa H,
Toda H, Shimonosono M, Yamada Y, Arai T, Kita Y, Kijima Y, et al:
Involvement of anti-tumor miR-124-3p and its targets in the
pathogenesis of pancreatic ductal adenocarcinoma: Direct regulation
of ITGA3 and ITGB1 by miR-124-3p. Oncotarget. 9:28849–28865. 2018.
View Article : Google Scholar : PubMed/NCBI
|