Introduction
Adamantinomatous craniopharyngioma (ACP) is a rare
epithelial tumor of the sellar region (1). As the tumor is adjacent to the
hypothalamus-pituitary axis, craniopharyngioma is typically
intractable (2). Therefore, gaining
a better understanding of the molecular pathology of
craniopharyngioma is of major importance for the development of
targeted therapies to improve patient outcomes (3,4).
With the extensive application of high-throughput
omics in craniopharyngioma research, the understanding of the
molecular pathology of the tumor has been further extended. Almost
all patients with ACP have been reported to have mutations in exon
3 of the catenin β1 (CTNNB1) gene (5). As a result, β-catenin cannot be
degraded in the cytoplasm and is translocated to the nucleus,
continuously activating the Wnt/β-catenin pathway (6–8). Using
the mRNA microarray gene expression analysis method, several
pharmaceutical targets have been identified, which have been
reported to be significantly and consistently upregulated in ACP
compared with that in pituitary tumors and normal pituitary tissue
(9). Previous studies have also
indicated that other signaling pathways, including the epidermal
growth factor (EGF), NOTCH, bone morphogenetic protein
(BMP)/fibroblast growth factor (FGF) and Sonic hedgehog (Shh)
pathways are involved in ACP tumorigenesis (8,9).
In order to better understand this disease, several
models of ACP have been established (5–8), ranging
from primary cell cultures to transgenic mouse models. Two murine
models (Sox2CreERT2/+; Ctnnb1lox(ex3)/+ and
Hesx1Cre/+; Ctnnb1lox(ex3)/+) resembling
human ACP in the anterior pituitary were established (10,11). On
the other hand, craniopharyngioma primary cell (CPC) lines have the
advantage of being derived from human tumors and can be used to
screen pharmacological agents relatively easily; however, the
tissue disruption required for cell culture means that the complex
architecture of the ACP is lost (12). Furthermore, culture conditions cannot
replicate microenvironmental signals to the tumor from the
surrounding tissue. To the best of our knowledge, it has yet to be
examined whether CPC culture can be used as appropriate model of
tumor tissues.
It is unclear how much difference in transcriptome
between cultured primary cells and tumor tissues of ACP. Therefore,
the aim of this study was to investigate whether cultured primary
cells in vitro can make a good simulation of tumors in
vivo, and whether the effect of simulation is weakened with the
prolongation of culture time.
Materials and methods
Tumor samples
A total of three ACP tumor samples were included in
this study. Samples were obtained from three patients who underwent
primary tumor resection at the Neurosurgery Department of Nanfang
Hospital between May and July 2017. Three patients were all female,
and their ages were 42, 56 and 59 years old. Tumor samples were
collected during surgery and stored at 4°C until culture procedures
were performed. All ACP tumor samples were pathologically confirmed
by hematoxylin and eosin staining. The staining was performed at
25°C for 5 min with hematoxylin and 2 min with eosin. The diagnosis
was made by two pathologists who were blinded to the conditions of
the study. Written informed consent was provided by all subjects in
accordance with the Declaration of Helsinki. The present study was
approved by the local Ethics Committee of Nanfang Hospital,
Southern Medical University.
Cell culture
ACP specimens were cultured as previously reported
(13–15). Tumor tissues were washed in
triplicate with PBS and then cut into fragments ~5 µm. Tissues were
digested with 0.25% trypsin for 40 min, and DNase (20 µg/ml;
Sigma-Aldrich; Merck KGaA) was added for 5 min at 37°C. Tumor cells
were centrifuged at 300 × g for 5 min in room temperature,
resuspended and cultured at 37°C with 5% CO2. For the
first 3 days, the culture medium was DMEM-F12 (Gibco; Thermo Fisher
Scientific, Inc.) with 20% FBS, after which FBS concentration was
decreased by 10% at each passage until it was zero. Finally, cells
were cultured in serum-free medium with additional 5 µg/ml insulin
(Sigma-Aldrich; Merck KGaA) and 10 µg/ml EGF (R&D Systems,
Inc.) as previously described (13–15). The
anchorage velocity-dependent separation method was used to exclude
mesenchymal cells (14).
Immunofluorescence
Cells were cultured at 37°C with 5% CO2
in cover glass-bottomed dishes. At 24 h, cells were washed with PBS
and fixed in 4% paraformaldehyde for 30 min at 4°C. Cells were then
ruptured and blocked in 0.1% Triton X-100 and 5% BSA at 25°C for 30
min. Cells were incubated at 4°C overnight with primary antibodies,
including pan-cytokeratin (pan-CK; dilution, 1:100; cat. no.,
ab7753; Abcam) and epithelial cell adhesion molecule (EpCAM;
dilution 1:100, cat. no., ab32392; Abcam), followed by incubation
at 25°C for 1 h with Alexa Fluor 488 anti-mouse or 594 anti-rabbit
secondary antibody (dilution, 1:1,000; cat. nos. A-11001 and
A-11012; Invitrogen; Thermo Fisher Scientific, Inc.). Nuclei were
counterstained with DAPI (Sigma-Aldrich; Merck KGaA). Images were
captured on an inverted LSM880 confocal system (magnification,
×400; Zeiss AG).
Western blot analysis
Cells were lysed in RIPA Buffer (50 Mm Tris-HCl pH
8.0, 1 mM EDTA pH 8.0, 5 mM DTT, 2% SDS), and the protein
concentration was determined using a BCA assay (Beyotime Institute
of Biotechnology). Total protein (30 µg) was resolved using 10%
SDS-PAGE gel, electro-transferred to polyvinylidene fluoride
membranes (Invitrogen; Thermo Fisher Scientific, Inc.) and blocked
with 5% non-fat dry milk at 25°C for 1 h in Tris-buffered saline
(pH 7.5). Membranes were immunoblotted overnight at 4°C with rabbit
monoclonal antibodies at a dilution of 1:1,000, including keratin 5
(cat. no., 25807), E-cadherin (cat. no., 3195), vimentin (cat. no.,
5741) and GAPDH (cat. no., 2118), all purchased from CST Biological
Reagents Co., Ltd. A HRP-conjugated anti-rabbit IgG antibody was
used as the secondary antibody (dilution, 1:2,000; cat. no., 7074;
CST Biological Reagents Co., Ltd.). The incubation was performed at
25°C for 1 h. Signals were detected using enhanced
chemiluminescence reagents (EMD Millipore). Signal intensities were
obtained using ImageJ software (v1.51, National Institutes of
Health).
RNA-Seq library preparation and
sequencing
Total RNA was extracted from all three surgical
specimens and the corresponding primary cells. The RNA
concentration was measured using a Qubit® RNA Assay kit
in a Qubit® 2.0 Fluorometer (both from Thermo Fisher
Scientific, Inc.), and RNA integrity was assessed using the RNA
Nano 6000 Assay kit with the Bioanalyzer 2100 system (both from
Agilent Technologies, Inc.) according to the manufacturer's
protocol. A total of 3 µg RNA per sample was used as the input
material. Ribosomal RNA (rRNA) was removed using an Epicentre
Ribo-zero™ rRNA Removal kit (Epicentre; Illumina, Inc.), according
to the manufacturer's protocol. Subsequently, sequencing libraries
were generated using rRNA-depleted RNA with the NEBNext®
Ultra™ Directional RNA Library Prep kit for Illumina®
(New England BioLabs, Inc.) according to the manufacturer's
protocol. PCR was performed with Phusion High-Fidelity DNA
polymerase, Universal PCR primers and Index (X) Primer. Finally,
the products were purified (AMPure XP system), and library quality
was assessed on the Agilent Bioanalyzer 2100 system. Clustering of
the index-coded samples was performed using a cBot Cluster
Generation System with a TruSeq PE Cluster kit v3-cBot-HS
(Illumina, Inc.). Following cluster generation, the libraries were
sequenced on an Illumina Hiseq X-ten platform and 150-bp paired-end
reads were generated (sequenced by Novogene Co., Ltd.).
Data analysis
Raw data of fastq format were firstly processed
through in-house perl scripts. Clean data were obtained by removing
reads containing adapter or poly-N and low-quality reads from the
raw data. All downstream analyses were performed using the clean
high-quality data. The reference genome index was built using
Bowtie2 v2.2.8 software (16) and
paired-end clean reads were aligned to the reference genome using
HISAT2 software (v2.0.4) (17,18).
StringTie software (v1.3.1) (19)
was used to calculate the fragments per kilobase of exon per
million fragments mapped (FPKMs) of coding genes in each sample
(20). Gene FPKMs were computed by
adding the FPKMs of the transcripts in each gene group. Ballgown R
package (21) was used to compare
all transcripts across conditions and produces tables and plots of
differentially expressed genes and transcripts. Transcripts with
q-value <0.05 were designated as differentially expressed. Genes
were ranked using the pre-ranked tool in Gene Set Enrichment
Analysis (GSEA) (v3.0, Broad Institute). Gene sets were downloaded
from the hallmark molecular signatures database (v6.1; Broad
Institute) (22,23).
Statistical analysis
Statistical analyses were performed using GraphPad
Prism (v7.0 GraphPad Software, Inc.). The Pearson's correlation
coefficient was used to assess the association between two groups.
The results are presented as the mean ± standard deviation. When
calculating the expression value of signature genes, a two-tailed
Student's t-test was performed to compare the means of two groups.
ImageJ software (v1.51, National Institutes of Health) was used to
process semi-quantitative analysis of the densitometry obtained
from western blot experiments. All western blot and
immunofluorescence experiments were performed in triplicate.
One-way ANOVA was performed to compare the group means followed by
Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference. No samples were excluded from
the analyses.
Results
Morphological evolution of primary
cells
The initial CPCs exhibited a paving stone-like
appearance and spontaneously formed keratin pearls (Fig. 1A and B). With increased culture time
and passages (10, 20 and 30 days), cell morphology gradually
changed from the initial epithelial irregular polygon shape to a
spindle shape, and further stabilized in the latter form (Fig. 1C-F).
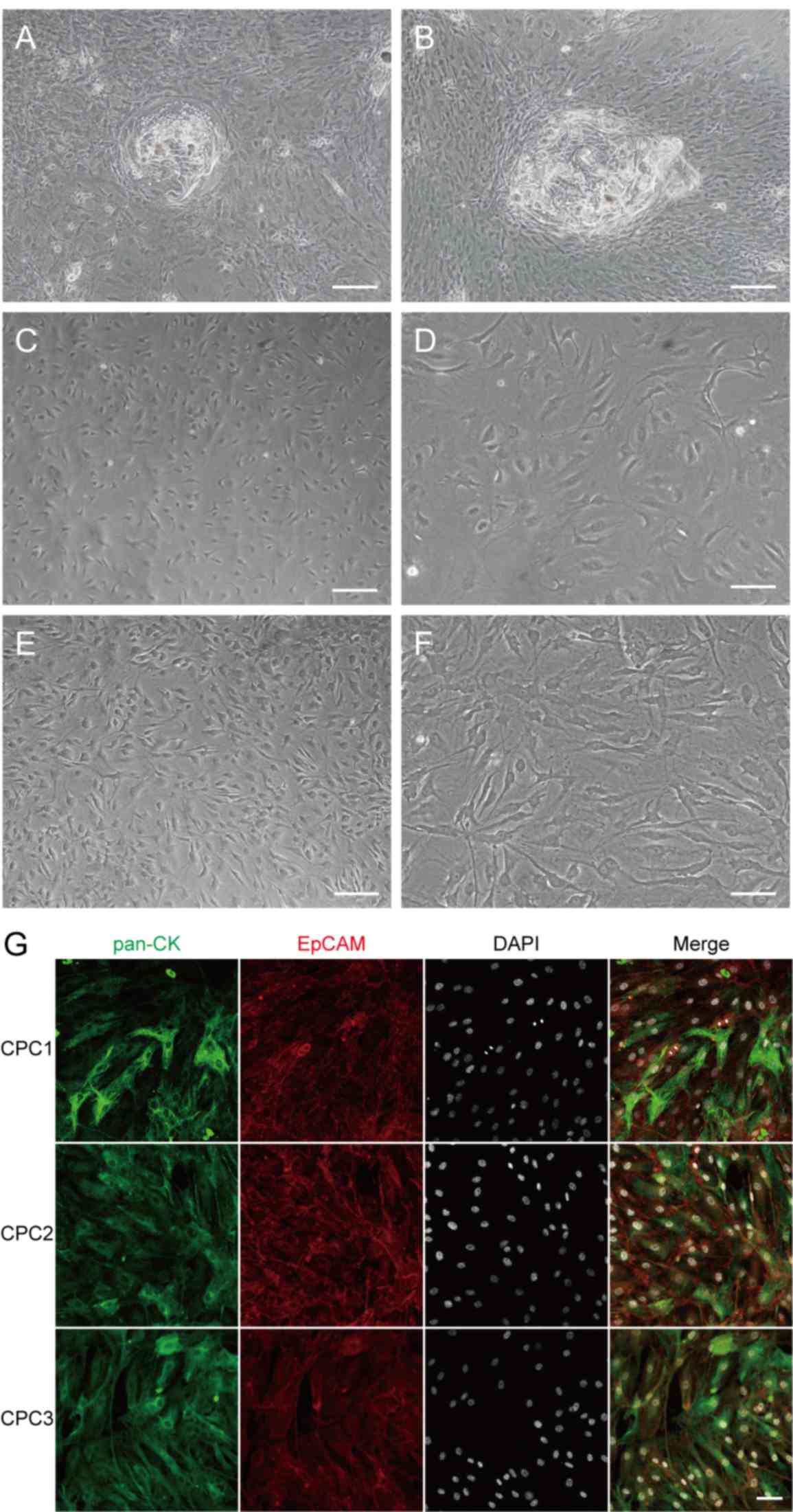 | Figure 1.Morphology and lineage identification
of primary cells. At the start of culture, primary cells exhibited
paving stone-like appearance and spontaneously formed keratin
pearls on (A) the second and (B) the third day of CPC culture. With
the prolongation of culture time and passages, the morphology of
cells gradually changed from the initial epithelial irregular
polygon shape to spindle shape. (C) Cells in low density;
magnification, ×200. (D) Cells in low density; magnification, ×400.
(E) cells in high density; magnification, ×200. (F) Cells in high
density; magnification, ×400. (G) Double immunostaining of
long-term cultured CPCs (CPC1, CPC2 and CPC3) demonstrated that
pan-CK and EpCAM remained positive with prolonged culture time,
although cell morphology had changed. Scale bars: A, B, C and E,
100 µm; D, F and G, 50 µm. CPCs, craniopharyngioma primary cells;
CPC1, 10 days culture; CPC2, 20 days culture; CPC3, 30 days of
culture; pan-CK, pan-cytokeratin; EpCAM, epithelial cell adhesion
molecule. |
CPCs maintain the properties of
epithelial cells
Pan-CK and EpCAM are classical epithelial markers
(24). The expression of pan-CK and
EpCAM was measured in cells cultured for 10 days (CPC1), 20 days
(CPC2) and 30 days (CPC3) using immunofluorescence. The results
demonstrated that pan-CK and EpCAM remained positively expressed as
culture time increased (Fig. 1G).
The epithelial properties of CPCs did not change even with
prolonged culture time and increased passages.
Transcriptome differences between ACP
tissues and CPCs
To analyze the characteristics of CPCs and identify
whether they can be used as a stable research model, three pairs of
CPCs and their parental ACP tissues were analyzed using RNA-Seq.
Pearson's correlation coefficient was used to compare the
consistency of the transcriptome between CPCs and ACP tissues. With
increased cell culture time, the correlation coefficient decreased
from 0.657 (CPC1) to 0.610 (CPC2) and then to 0.547 (CPC3)
(Fig. 2A-C). This indicated that the
difference of transcriptome between CPCs and ACP tissues increased
with prolonged culture time.
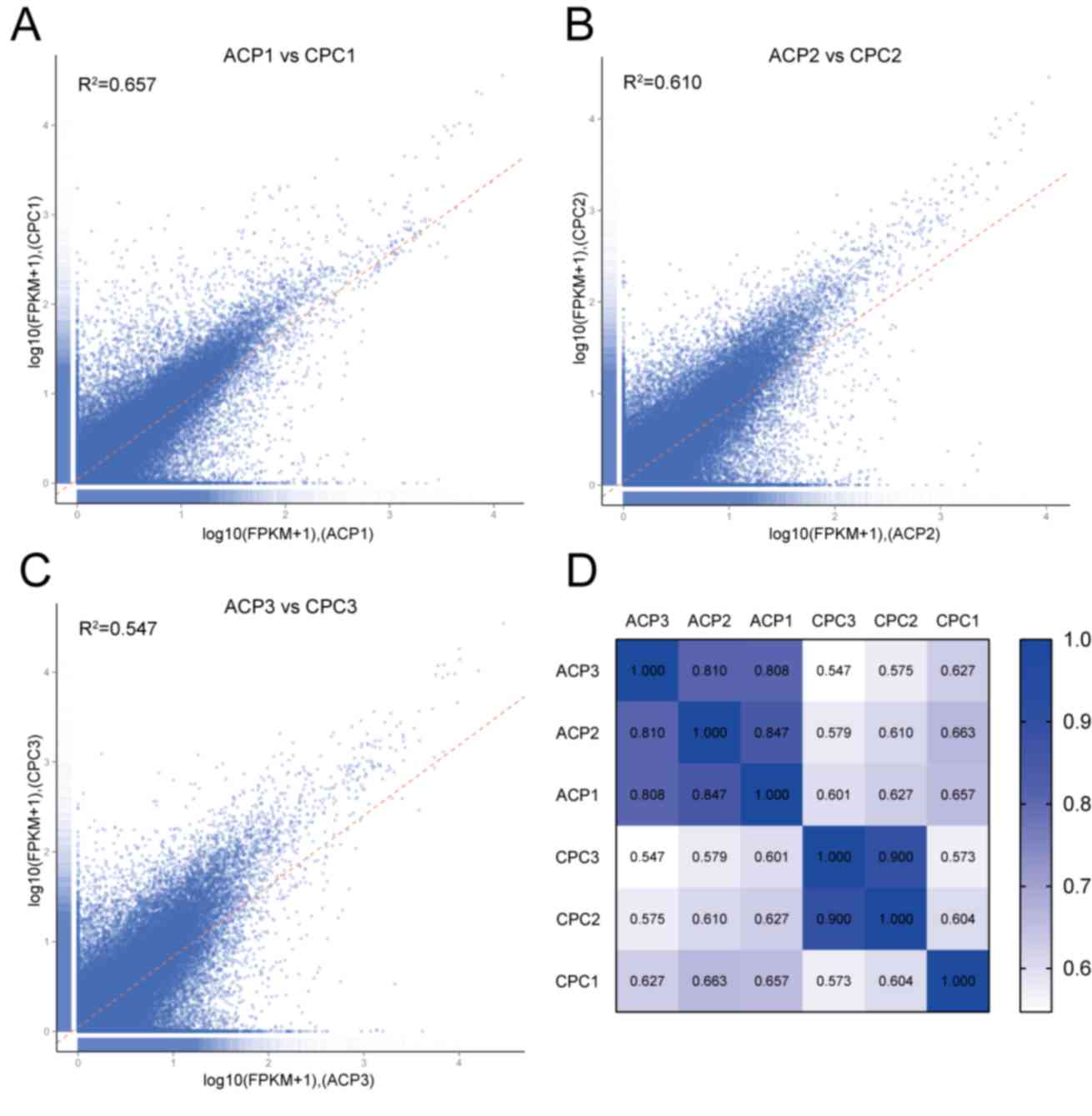 | Figure 2.Consistency of transcriptome between
ACP tissues and CPCs. With increasing culture time, the correlation
coefficient of transcriptome between ACP tumors and CPCs gradually
decreased. (A-C) Transcriptome correlation between tissues and
cells at (A) 10, (B) 20 and (C) 30 days of culture. (D) The heatmap
of the correlation coefficient was at a high level (>0.8) among
the three ACP tissues, whereas with increased culturing time, the
correlation coefficient of transcriptome between ACP tissues and
CPCs gradually decreased. Deeper blue represents stronger
correlation. ACP1, 2 and 3 respectively represent tumor tissues
used to culture CPC1, 2 and 3. ACP, adamantinomatous
craniopharyngioma; CPCs, craniopharyngioma primary cells; CPC1, 10
days culture; CPC2, 20 days culture; CPC3, 30 days of culture;
FPKMs, fragments per kilobase of exon per million fragments
mapped. |
The heat map revealed that the correlation
coefficient was high (>0.8) for the three ACP tissues; however,
as culture time increased, the transcriptome correlation
coefficient between ACP tissues and CPCs decreased gradually. The
correlation coefficient among CPC groups also decreased gradually
with increased culture time (CPC3 vs. CPC2, 0.900; CPC3 vs. CPC1,
0.573; CPC2 vs. CPC1, 0.604) (Fig.
2D).
Differential gene expression between
ACP tissues and CPCs
Differentially expressed genes and samples were
hierarchically clustered. The results suggested that ACP tumor
tissues and CPCs were grouped into two categories (Fig. 3A), and they were represented by black
and white. This indicated that the intra-group similarity between
tumor tissues and primary cells was greater compared with the
similarity between groups. To further analyze the differential gene
expression between CPCs and ACP tissues, differential genes were
screened using the standard criteria, including FPKM >1 and
q-value <0.05. The results indicated that the number of
differentially expressed genes was increased between ACP tissues
and CPCs in a culture time-dependent manner; the number of
differentially expressed genes (DEGs) was increased from 1,247
(CPC1) to 1,643 (CPC2) and further to 1,949 (CPC3). Compared with
the ACP tissues, the number of upregulated genes in CPCs was 901
(CPC1), 1,359 (CPC2) and 1,453 (CPC3), whereas the number of
downregulated genes was 346 (CPC1), 284 (CPC2) and 496 (CPC3)
(Fig. 3B).
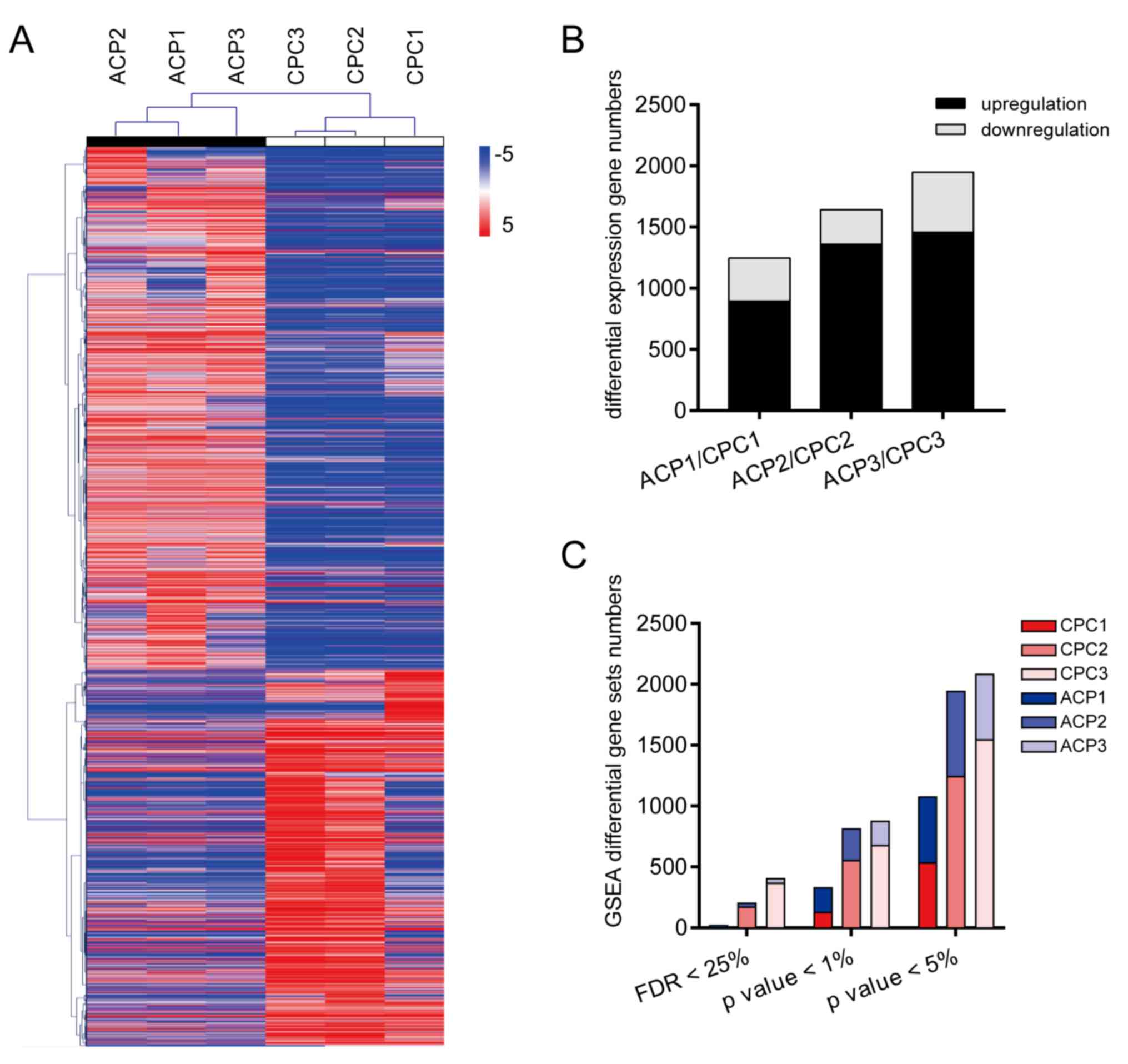 | Figure 3.Description of differentially
expressed genes and gene sets between tumor samples and primary
cells. (A) Hierarchical clustering analysis of differential genes
between ACP tissues and CPCs demonstrated that samples were divided
into two categories. ACP tissues and CPCs are represented by black
and white. (B) Comparison of differentially expressed genes between
ACP tissues and CPCs. With the increase of culture duration, the
number of differential genes increased significantly. (C) GSEA
revealed that with prolonged incubation time, the diversity of gene
sets between ACP tissues and CPCs also increased. ACP1, 2 and 3
respectively represent tumor tissues used to culture CPC1, 2 and 3.
ACP, adamantinomatous craniopharyngioma; CPCs, craniopharyngioma
primary cells; CPC1, 10 days culture; CPC2, 20 days culture; CPC3,
30 days of culture; FPKMs, fragments per kilobase of exon per
million fragments mapped; FDR, false discovery rate; GSEA, Gene Set
Enrichment Analysis. |
The transcriptome information for GSEA analysis was
subsequently used to identify DEG sets between ACP tissues and CPC.
The results showed that the diversity of gene sets between ACP
tissues and CPCs increased with culture duration (FDR <0.25;
P<0.01 and P<0.05). DEG sets increased from 19 to 203 and
further to 405 under the criteria of FDR <0.25; DEG sets
increased from 330 to 814 and then to 877 under P<0.01, whereas
DEG sets increased from 1,075 to 1,943 and then to 2,083 when
P<0.05 (Fig. 3C).
Similarities and differences between
characteristic genes and gene sets in ACP tissues and CPCs
In order to illustrate the simulation potential of
CPC models for ACP tissues, 20 ACP-characteristic genes were
selected based on previous reports (9) and the differences in these genes
between ACP tissues and CPCs were analyzed. The results
demonstrated that the majority of the genes were not significantly
differentially expressed in the tumors and cultured cells. No
significant difference was observed in the expression of the
following genes (tumors vs. primary cells): Amphiregulin
[AREG; fold-change (FC)=0.588; P=0.734], a member of the
epidermal growth factor family; epidermal growth factor receptor
(FC=0.919; P=0.705); EPH receptor A2 (FC=0.638; P=0.599), which
belongs to the ephrin receptor subfamily of the protein-tyrosine
kinase family, and MUC1 (FC=0.512; P=0.479), encoded a
membrane-bound protein for forming protective mucous barriers on
epithelial surfaces. However, a number of significant gene changes
were identified, including TNF superfamily member 11 (FC=21.99;
P=0.003), SRC proto-oncogene (FC=1.352; P=0.013), interleukin 2
receptor subunit β (FC=23.636; P=0.009) and interleukin 6 receptor
(FC=3.496; P=0.035), which were involved in cell proliferation,
differentiation and immune responses (Fig. 4A).
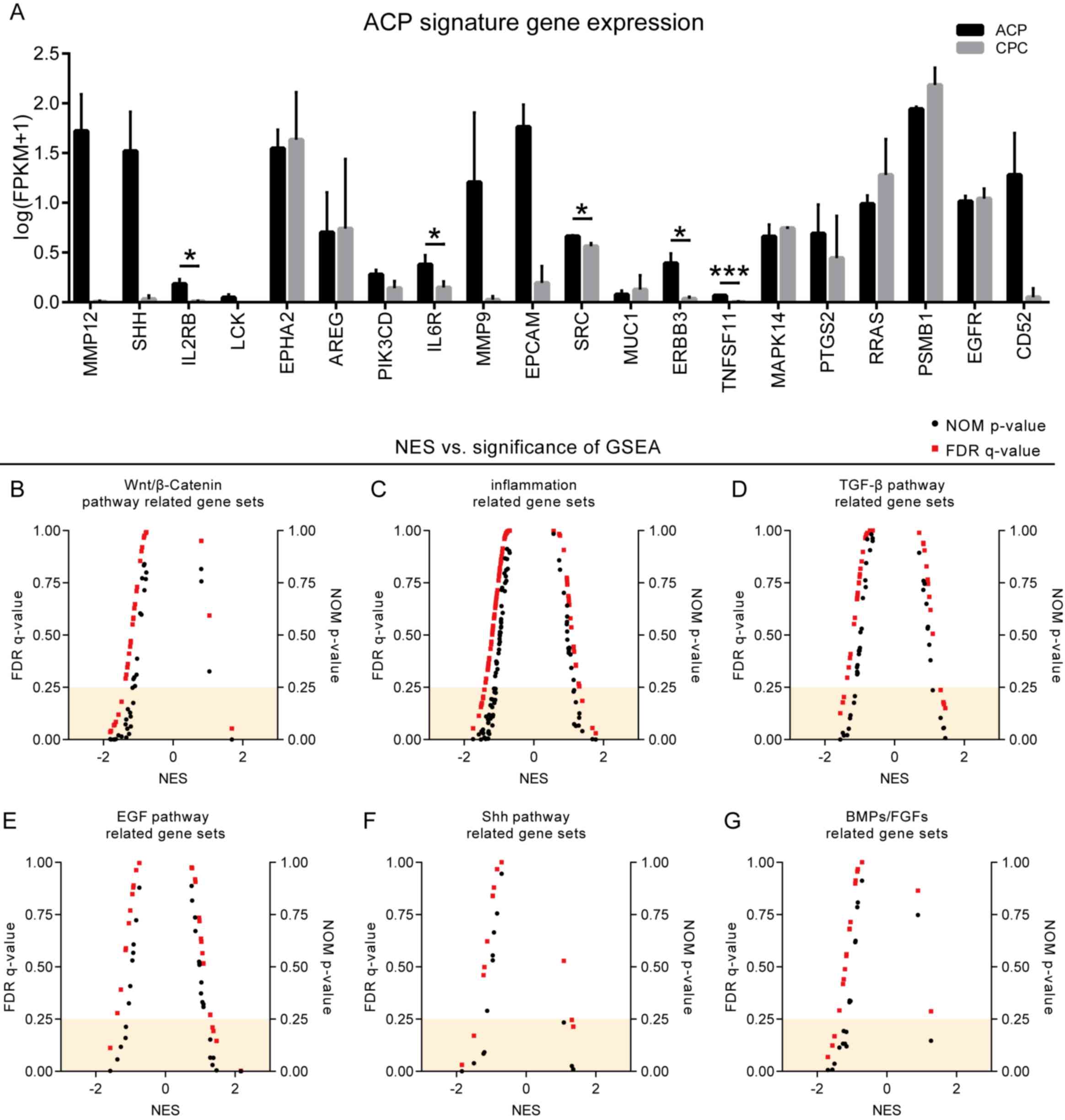 | Figure 4.Description of differential
characteristic genes and gene sets between ACP tissues and CPCs.
(A) The majority of characteristic genes and possible therapeutic
targets between ACP tissues and CPCs were not significantly
different. (B-G) GSEA revealed that characteristic gene sets,
including (B) ‘Wnt/β-catenin pathway’, (C) ‘Inflammation’, (D)
‘TGF-β pathway’, (E) ‘EGF pathway’, (F) ‘Shh pathway’ and (G)
‘BMPs/FGFs’ were not significantly different between ACP tissues
and CPCs. *P<0.05 and ***P<0.001. ACP, adamantinomatous
craniopharyngioma; CPC, craniopharyngioma primary cells; FPKMs,
fragments per kilobase of exon per million fragments mapped; NOM,
nominal; FDR, false discovery rate; GSEA, Gene Set Enrichment
Analysis; NES, normalized enrichment score; TGF-β, transforming
growth factor-β; EGF, epidermal growth factor; Shh, Sonic hedgehog;
BMPs, bone morphogenetic proteins; FGFs, fibroblast growth
factors. |
GSEA was performed for all samples and selected
characteristic gene sets (‘Wnt/β-catenin pathway’, ‘Inflammation’,
‘TGF-β pathway’, ‘EGF pathway’, ‘Shh pathway’ and ‘BMPs/FGFs’) were
selected for further analysis. No significant differences were
observed between ACP tissues and CPCs in the majority of the
characteristic gene sets (Fig.
4B-G). However, in CPC, the ‘keratinization’ [normalized
enrichment score (NES)=−2.02, false discovery rate (FDR)=0.0038]
and ‘epithelial cell proliferation’ (NES=−1.82, FDR=0.032)
phenotypes were significantly weakened. At the same time, cell
proliferation- (NES=1.78, FDR=0.028) and mitosis- (NES=1.93,
FDR=0.012) associated phenotypes were significantly enhanced
(Fig. 5A-H).
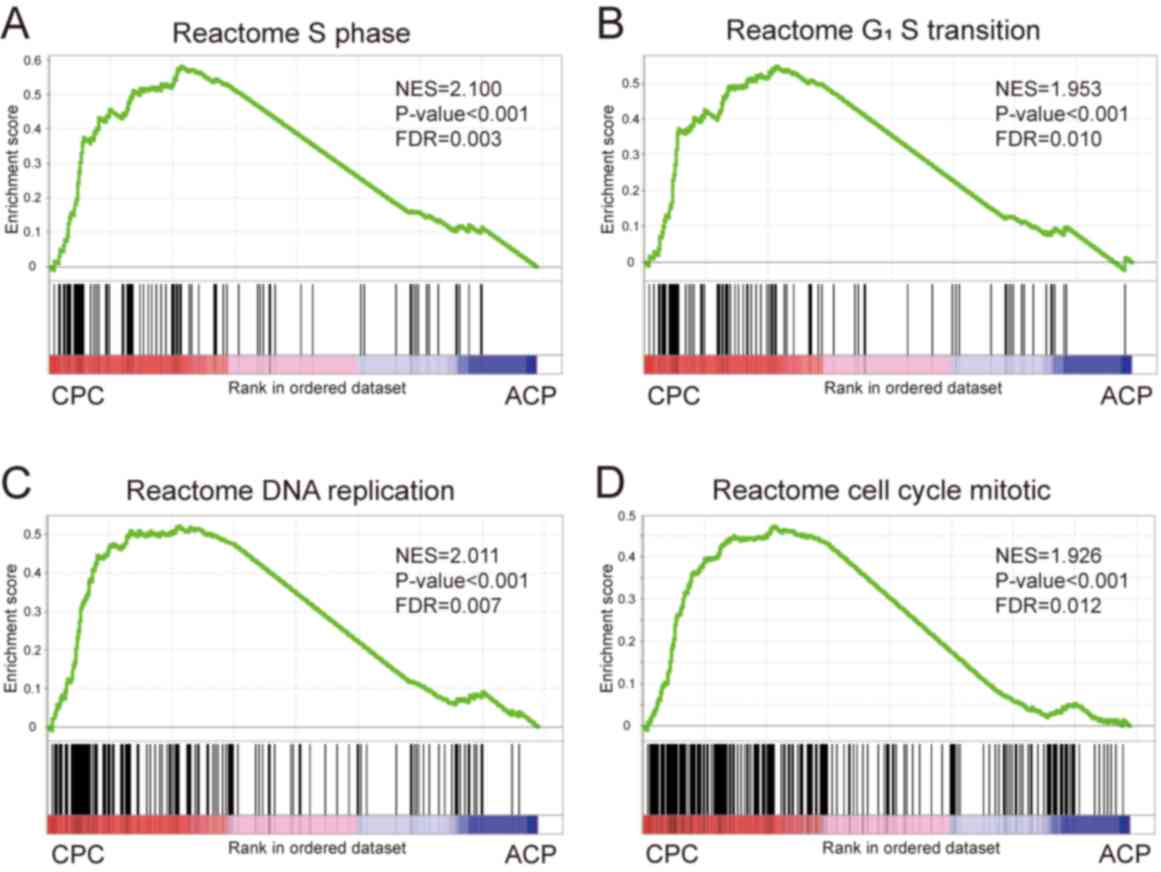 | Figure 5.Significantly different gene sets
between ACP tissues and CPCs. (A-D) GSEA results revealed that the
gene sets associated with cell proliferation (A) S phase, (B)
G1/S transition, (C) DNA replication and (D) cell cycle
were significantly enriched in primary cells compared with ACP
tissues. (E-H) The epithelial phenotype-associated gene sets (E)
keratinization, (F) regulation of keratinocyte proliferation, (G)
epithelial proliferation and (H) morphogenesis of an epithelium
decreased significantly in CPCs compared with ACP tissues. (I)
Western blot analysis results suggested that with prolonged culture
time, the marker of keratinization phenotype keratin 5 and the
marker of epithelial phenotype E-cadherin were significantly
downregulated, whereas the marker of mesenchymal phenotypes
vimentin was significantly upregulated in CPCs compared with APC
tissues. *P<0.05 and ***P<0.001. GSEA, Gene Set Enrichment
Analysis; GO, gene ontology; ACP, adamantinomatous
craniopharyngioma; CPC, craniopharyngioma primary cells; CPC1, 10
days culture; CPC2, 20 days culture; CPC3, 30 days of culture. |
Western blot analysis of tumor tissues and primary
cells after different culture times was used to detect the
attenuation of primary cell epithelialization and keratinization
phenotypes. Keratin 5, a marker of the keratinization phenotype,
and E-cadherin, a marker of the epithelial phenotype, were
significantly downregulated, whereas vimentin, a marker of the
mesenchymal phenotype, was significantly upregulated in CPCs
compared with APC tissues (Fig.
5I).
Discussion
Craniopharyngioma is a rare type of intracranial
tumor (1,4). The use of transgenic mouse models has
resulted in breakthroughs in craniopharyngioma research (10,25).
Craniopharyngioma is a tumor with a low frequency of mutation,
therefore transgenic mice modified to express the known
CTNNB1 gene mutation can be used to simulate human
craniopharyngioma (5,26–28).
However, the animal model cannot fully simulate human
craniopharyngioma considering species differences and complex
clinical manifestations in patients, especially the multiple
manifestations of hormonal abnormalities. Basic research into human
craniopharyngioma has been progressing slowly in recent years,
which may be due to the lack of a stable and accessible research
platform for human ACP.
Craniopharyngioma is a benign tumor, and therefore
it is challenging to produce a stable cell line; when the cells are
not immortalized, the primary cells exhibit growth stagnation and
trait changes in the late stage of in vitro culture. Primary
cells are one of the most important research platforms at this
stage (12). Whether primary cells
can effectively simulate the tumor microenvironment remains
unknown. The transcriptome represents the entire manifestation of
RNA transcripts in cells or tissues and DEGs at various life
stages, in different tissue types, physiological states and
environmental conditions. Transcriptome analysis provides a
comprehensive understanding of gene expression and its regulation
(29). RNA-Seq is a powerful tool
used for the comprehensive characterization of the whole
transcriptome at both the gene and exon levels (30). RNA-Seq is based on next generation
sequencing technology. Compared with traditional technology, which
can only be used detect known genes, RNA-Seq is a cost-efficient
technology that can detect almost all genes expressed in samples,
including novel genes and dynamic changes in gene expression
(31–34). RNA-Seq technology was used in this
study to detect genetic differences between tumor samples and
primary cells and to demonstrate whether primary cells can be used
as a model for ACP research at the gene level.
Changes in the morphology and properties of CPCs
were first compared following culture for different durations. The
results revealed that the morphology of primary cells notably
changed with increased culture time. The morphology of primary
cells changed from an epithelial to a stromal state, but maintained
original epithelial lineage markers, including pan-CK and
EpCAM.
ACP tissues and CPCs were further analyzed using
RNA-Seq analysis. To the best of our knowledge, this was the first
time that transcriptome differences between ACP tissues and CPCs
were analyzed using RNA-Seq. The results demonstrated that the
correlation between tumor tissues and primary cell transcriptome
gradually decreased with increased culture duration, whereas DEGs
and GSEA-assembled differential gene sets gradually increased.
However, the results suggested that the CPCs resembled and
maintained the overall genomic signatures of ACP tissues from which
they were derived.
A number of studies have reported morphological and
phenotypic changes in long-term cultured cells (35–37).
This is consistent with the results of the present study. It was
concluded that such changes were mainly associated with
alternations in the tumor microenvironment. Hypoxia is likely to
occur after long-term cell culture, resulting in hypoxia-inducible
factor 1α upregulation and the subsequent activation of a series of
pathways, including the AKT/PI3K and the transforming growth
factor-β1/SMAD pathways. This activation may contribute to the
occurrence of epithelial-mesenchymal transition. These pathways may
also lead to enhanced invasive and proliferative capacities with
reduced apoptosis in long-term cultured cells.
In summary, ACP primary cells may be used as a
suitable research platform; however, the relevant experiments
should be concluded as early as possible to maintain consistency
between primary cells and tumor tissues. The recommended in
vitro culture time should be <30 days to ensure that the
primary cell model is representative of tumor tissues. However, the
present study is not without limitations. The sample size was small
and thus the number of cases included in this experiment is
insufficient. In future studies, larger sample sizes should be used
and in vivo experiments should be performed to confirm the
present results of the present study.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from Science and
Technology Program of Guangdong (grant nos. 2016A020213006,
2017A020215048 and 2017A020215191); Natural Science Foundation of
Guangdong (grant no. 2016A030310377); Science and Technology
Program of Guangzhou (grant no. 201707010149); President Foundation
of Nanfang Hospital, Southern Medical University (grant nos.
2015C018, 2016L002 and 2017Z009).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
SQ contributed to experimental design. PZ and CW
performed the experiments. PZ and YL performed the bioinformatics
analysis on RNA sequencing data. CW carried out the immunostaining
analysis. YL, JF, JXP and JP contributed to the pathological
analysis of human samples. YL and JP wrote the manuscript.
Ethics approval and consent to
participate
Written informed consent was provided by all
subjects in accordance with the Declaration of Helsinki. The
present study was approved by the local Ethics Committee of Nanfang
Hospital, Southern Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Erfurth EM, Holmer H and Fjalldal SB:
Mortality and morbidity in adult craniopharyngioma. Pituitary.
16:46–55. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Olsson DS, Andersson E, Bryngelsson IL,
Nilsson AG and Johannsson G: Excess mortality and morbidity in
patients with craniopharyngioma, especially in patients with
childhood onset: A population-based study in Sweden. J Clin
Endocrinol Metab. 100:467–474. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tomlinson JW, Holden N, Hills RK, Wheatley
K, Clayton RN, Bates AS, Sheppard MC and Stewart PM: Association
between premature mortality and hypopituitarism. West midlands
prospective hypopituitary study group. Lancet. 357:425–431. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Muller HL, Merchant TE, Puget S and
Martinez-Barbera JP: New outlook on the diagnosis, treatment and
follow-up of childhood-onset craniopharyngioma. Nat Rev Endocrinol.
13:299–312. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brastianos PK, Taylor-Weiner A, Manley PE,
Jones RT, Dias-Santagata D, Thorner AR, Lawrence MS, Rodriguez FJ,
Bernardo LA, Schubert L, et al: Exome sequencing identifies BRAF
mutations in papillary craniopharyngiomas. Nat Genet. 46:161–165.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kato K, Nakatani Y, Kanno H, Inayama Y,
Ijiri R, Nagahara N, Miyake T, Tanaka M, Ito Y, Aida N, et al:
Possible linkage between specific histological structures and
aberrant reactivation of the Wnt pathway in adamantinomatous
craniopharyngioma. J Pathol. 203:814–821. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sekine S, Shibata T, Kokubu A, Morishita
Y, Noguchi M, Nakanishi Y, Sakamoto M and Hirohashi S:
Craniopharyngiomas of adamantinomatous type harbor beta-catenin
gene mutations. Am J Pathol. 161:1997–2001. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hölsken A, Sill M, Merkle J, Schweizer L,
Buchfelder M, Flitsch J, Fahlbusch R, Metzler M, Kool M, Pfister
SM, et al: Adamantinomatous and papillary craniopharyngiomas are
characterized by distinct epigenomic as well as mutational and
transcriptomic profiles. Acta Neuropathol Commun. 4:202016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gump JM, Donson AM, Birks DK, Amani VM,
Rao KK, Griesinger AM, Kleinschmidt-DeMasters BK, Johnston JM,
Anderson RC, Rosenfeld A, et al: Identification of targets for
rational pharmacological therapy in childhood craniopharyngioma.
Acta Neuropathol Commun. 3:302015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Andoniadou CL, Matsushima D, Mousavy
Gharavy SN, Signore M, Mackintosh AI, Schaeffer M, Gaston-Massuet
C, Mollard P, Jacques TS, Le Tissier P, et al: Sox2(+)
stem/progenitor cells in the adult mouse pituitary support organ
homeostasis and have tumor-inducing potential. Cell Stem Cell.
13:433–445. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gaston-Massuet C, Andoniadou CL, Signore
M, Jayakody SA, Charolidi N, Kyeyune R, Vernay B, Jacques TS,
Taketo MM, Le Tissier P, et al: Increased Wingless (Wnt) signaling
in pituitary progenitor/stem cells gives rise to pituitary tumors
in mice and humans. Proc Natl Acad Sci USA. 108:11482–11487. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hölsken A and Buslei R: Models of human
adamantinomatous craniopharyngioma tissue: Steps toward an
effective adjuvant treatment. Brain Pathol. 27:358–363. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hölsken A, Buchfelder M, Fahlbusch R,
Blümcke I and Buslei R: Tumour cell migration in adamantinomatous
craniopharyngiomas is promoted by activated Wnt-signalling. Acta
Neuropathol. 119:631–639. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen M, Zheng SH, Liu Y, Shi J and Qi ST:
Periostin activates pathways involved in epithelial-mesenchymal
transition in adamantinomatous craniopharyngioma. J Neurol Sci.
360:49–54. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu Y, Wang CH, Li DL, Zhang SC, Peng YP,
Peng JX, Song Y, Qi ST and Pan J: TREM-1 expression in
craniopharyngioma and Rathke's cleft cyst: Its possible implication
for controversial pathology. Oncotarget. 7:50564–50574.
2016.PubMed/NCBI
|
|
16
|
Langmead B and Salzberg SL: Fast
gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pertea M, Kim D, Pertea GM, Leek JT and
Salzberg SL: Transcript-level expression analysis of RNA-seq
experiments with HISAT, StringTie and Ballgown. Nat Protoc.
11:1650–1667. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pertea M, Pertea GM, Antonescu CM, Chang
TC, Mendell JT and Salzberg SL: StringTie enables improved
reconstruction of a transcriptome from RNA-seq reads. Nat
Biotechnol. 33:290–295. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Trapnell C, Williams BA, Pertea G,
Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ and Pachter
L: Transcript assembly and quantification by RNA-Seq reveals
unannotated transcripts and isoform switching during cell
differentiation. Nat Biotechnol. 28:511–515. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Frazee AC, Sabunciyan S, Hansen KD,
Irizarry RA and Leek JT: Differential expression analysis of
RNA-seq data at single-base resolution. Biostatistics. 15:413–426.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mootha VK, Lindgren CM, Eriksson KF,
Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E,
Ridderstråle M, Laurila E, et al: PGC-1alpha-responsive genes
involved in oxidative phosphorylation are coordinately
downregulated in human diabetes. Nat Genet. 34:267–273. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Thimsen V, Hölsken A, Buchfelder M,
Flitsch J, Fahlbusch R, Stefanits H, Losa M, Jones DT and Buslei R:
EpCAM (CD326) is differentially expressed in craniopharyngioma
subtypes and Rathke's cleft cysts. Sci Rep. 6:297312016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gonzalez-Meljem JM, Haston S, Carreno G,
Apps JR, Pozzi S, Stache C, Kaushal G, Virasami A, Panousopoulos L,
Mousavy-Gharavy SN, et al: Stem cell senescence drives
age-attenuated induction of pituitary tumours in mouse models of
paediatric craniopharyngioma. Nat Commun. 8:18192017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Campanini ML, Colli LM, Paixao BM, Cabral
TP, Amaral FC, Machado HR, Neder LS, Saggioro F, Moreira AC,
Antonini SR and de Castro M: CTNNB1 gene mutations, pituitary
transcription factors, and MicroRNA expression involvement in the
pathogenesis of adamantinomatous craniopharyngiomas. Horm Cancer.
1:187–196. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sekine S, Sato S, Takata T, Fukuda Y,
Ishida T, Kishino M, Shibata T, Kanai Y and Hirohashi S:
Beta-catenin mutations are frequent in calcifying odontogenic
cysts, but rare in ameloblastomas. Am J Pathol. 163:1707–1712.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hölsken A, Kreutzer J, Hofmann BM, Hans V,
Oppel F, Buchfelder M, Fahlbusch R, Blümcke I and Buslei R: Target
gene activation of the Wnt signaling pathway in nuclear
beta-catenin accumulating cells of adamantinomatous
craniopharyngiomas. Brain Pathol. 19:357–364. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
van Dijk EL, Auger H, Jaszczyszyn Y and
Thermes C: Ten years of next-generation sequencing technology.
Trends Genet. 30:418–426. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Foo JN, Liu JJ and Tan EK: Whole-genome
and whole-exome sequencing in neurological diseases. Nat Rev
Neurol. 8:508–517. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pickrell JK, Marioni JC, Pai AA, Degner
JF, Engelhardt BE, Nkadori E, Veyrieras JB, Stephens M, Gilad Y and
Pritchard JK: Understanding mechanisms underlying human gene
expression variation with RNA sequencing. Nature. 464:768–772.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang Z, Gerstein M and Snyder M: RNA-Seq:
A revolutionary tool for transcriptomics. Nat Rev Genet. 10:57–63.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Byron SA, Van Keuren-Jensen KR,
Engelthaler DM, Carpten JD and Craig DW: Translating RNA sequencing
into clinical diagnostics: Opportunities and challenges. Nat Rev
Genet. 17:257–271. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jeppesen M, Hagel G, Glenthoj A, Vainer B,
Ibsen P, Harling H, Thastrup O, Jørgensen LN and Thastrup J:
Short-term spheroid culture of primary colorectal cancer cells as
an in vitro model for personalizing cancer medicine. PLoS One.
12:e01830742017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mahajan AS, Sugita BM, Duttargi AN, Saenz
F, Krawczyk E, McCutcheon JN, Fonseca AS, Kallakury B, Pohlmann P,
Gusev Y and Cavalli LR: Genomic comparison of early-passage
conditionally reprogrammed breast cancer cells to their
corresponding primary tumors. PLoS One. 12:e01861902017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Broutier L, Mastrogiovanni G, Verstegen
MM, Francies HE, Gavarró LM, Bradshaw CR, Allen GE, Arnes-Benito R,
Sidorova O, Gaspersz MP, et al: Human primary liver cancer-derived
organoid cultures for disease modeling and drug screening. Nat Med.
23:1424–1435. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mortazavi A, Williams BA, McCue K,
Schaeffer L and Wold B: Mapping and quantifying mammalian
transcriptomes by RNA-Seq. Nat Methods. 5:621–628. 2008. View Article : Google Scholar : PubMed/NCBI
|



















