Introduction
Gastric cancer (GC), or stomach adenocarcinoma, was
the most common malignancy of the gastrointestinal system,
according to statistics in 2017 (1).
It was the second leading cause of cancer-associated mortalities
worldwide in 2018 and is a regularly diagnosed form of cancer
(2). The GC cases in China in 2017
represented approximately 42% of all cases worldwide (3). This may be due to the highly documented
incidence of Helicobacter pylori infection among the Chinese
population (1,3). Throughout the years, several attempts
have been made to identify methods for ameliorating GC-associated
mortality. Efforts have been directed towards the discovery of
sensitive diagnostic biomarkers and specific prognostic markers and
therapeutic targets. Nevertheless, minimal progress has been made,
as GC remains a major global health burden with a median survival
of only 3–5 months (4). Thus,
enhanced comprehension of the molecular and genetic changes that
accompany gastric tumorigenesis is essential. Also important is the
identification of oncogenes that promote gastric tumor growth.
These oncogenes may serve as biomarkers for GC progression and as
molecular therapeutic targets.
Genomic studies have revealed that approximately 90%
of the human genome is actively transcribed (5). Only ~2% is transcribed into
protein-coding genes, whereas the remainder is transcribed into
non-protein-coding genes (6).
Non-coding RNAs consist of microRNAs (miRNAs/miRs), small
interfering RNAs (siRNAs) and long non-coding RNAs (lncRNAs).
lncRNAs are RNA transcripts over 200 nucleotides in length. Their
expression is highly cell- and tissue-specific (7). They affect various processes of
protein, DNA and RNA interactions, thereby regulating their
expression and functions. Consequently, lncRNAs are capable of
potently altering the proliferative, invasive and metastatic
potential of malignant cells (8).
They have been implicated in various types of cancer, such as
colorectal, non-small cell lung, non-muscle invasive bladder,
breast and prostate cancers (9–13),
although their functional mechanisms are largely underexplored.
Recently, certain studies have focused on the analyses of lncRNA
expression patterns and their associations with clinical and
demographic characteristics of patients. Of note, several lncRNAs
have been associated with tumor growth, metastasis and patient
clinical outcomes (1,14,15).
lncRNAs drive several oncogenic processes (16,17).
Findings have demonstrated that the expression of lncRNAs is
dysregulated between tumor and adjacent normal tissues (18,19), and
between cancer patients and healthy subjects (20,21).
lncRNAs are increasingly gaining attention as potential biomarkers
and therapeutic targets for various kinds of malignancies (22). Specific GC biomarkers are currently
limited. Although advancements have been made and a number of
treatment options are available, the survival rates of patients
with GC have not been markedly improved. For instance, trastuzumab,
a human epidermal growth factor receptor 2 (HER2) antibody, has
been recommended as the first-line regimen for advanced GC
treatment (16). However, only
HER2+ patients, which correspond to 12–20% of GC cases,
may benefit from this regimen (22).
Further understanding of the molecular mechanisms and exploration
of corresponding GC biomarkers could contribute to the early
detection and optimal prediction of prognosis. Identification of
GC-associated lncRNAs could lead to the development of novel
markers and effective therapeutic targets for patients with GC.
The lncRNA PTPRG antisense RNA 1 (PTPRG-AS1) is the
antisense of protein tyrosine phosphatase receptor type G (PTPRG),
a tumor suppressor gene. High expression of PTPRG-AS1 has been
linked to poor clinical outcomes for lung cancer patients (23). In breast cancer, high expression of
this lncRNA has been suggested to downregulate the expression of
PTPRG (24), thereby enhancing
disease progression.
Forkhead box P4 antisense RNA 1 (FOXP4-AS1) is an
lncRNA associated with colorectal cancer (25). It is significantly upregulated in
colorectal cancer tissues, promotes cell proliferation and inhibits
apoptosis both in vitro and in vivo (25).
The lncRNA LINC00958, also termed bladder
cancer-associated transcript 2 (BLACAT2), has been found to be
markedly upregulated in lymph node-metastatic bladder cancer and
was associated with lymph node metastasis (LNM) (26). It is implicated in bladder
cancer-associated lymphangiogenesis and lymphatic metastasis.
Patients with bladder cancer and high BLACAT2 expression have been
shown to have shorter overall and metastasis-free survival
(26).
lncRNA ZXF2 is highly expressed in lung
adenocarcinoma (27). It is believed
that ZXF2 enhances lung cancer progression via the c-myc/e-cadherin
pathway. Suppression of ZXF2 inhibits c-myc, a potent
proto-oncogene, and enhances the expression of e-cadherin, a tumor
suppressor gene. In addition, ZXF2 is located adjacent to c-myc in
chromosome locus 8q24.2, which is a hotspot for multiple types of
human cancer. Interaction of ZXF2 and c-myc results in cell cycle
progression, proliferation, migration and invasion of tumor cells.
Upregulation of ZXF2 was also associated with lymph node metastasis
and poor prognosis (27).
Huang et al (15) demonstrated that the long intergenic
non-coding RNA ENST00000602992 (CTD-2227E11.1), an oncogenic lncRNA
also known as upregulated in colorectal cancer (UCC), promotes the
progression of colorectal cancer by upregulating KRAS and acting as
a sponge for miR-143. Furthermore, it was shown that suppression of
UCC in colorectal cancer cells inhibited proliferation and
invasion, and induced apoptosis. A positive association was also
observed between UCC expression, LNM and overall survival (OS) of
patients with colorectal cancer.
Thus far, limited data exist on the expression
profiles, functions and prognostic efficacy of PTPRG-AS1,
FOXP4-AS1, BLACAT2, ZXF2 and UCC in GC. The present study aimed to
examine their expression profiles in GC tissues and cell lines and
to determine their functions and clinical significance in GC. It
was found that lncRNAs PTPRG-AS1, ZXF2, FOXP4-AS1, BLACAT2 and UCC
are upregulated in GC tissues and cell lines. Also reported in the
present study are the functions, clinical significance and
potential for application of the aforementioned lncRNAs as
diagnostic, prognostic and therapeutic markers in GC.
Materials and methods
Study design
Based on previous studies, nine lncRNAs aberrantly
expressed in other adenocarcinomas, namely UCC, FOXP4-AS1, SCAL1,
ANRASSF1, PTPRG-AS1, ZXF2, SChLAP1, BLACAT2 and SBF2-AS1 (5,13,15,23,25–29),
were selected as potential candidates in the present study. This
selection was based on the fact that many types of cancer, though
substantially different, share common gene expression signatures.
In the screening stage, the expression levels of the nine lncRNAs
were examined in GC tissues and five upregulated lncRNAs became the
focus of subsequent studies.
Patients and specimens
The current study was approved through the Clinical
Research Ethics Committee of the Second Hospital of Shandong
University. Written informed consent was obtained from all
participants. In total, 61 pairs of GC tissues and adjacent normal
tissues (ANTs) collected at the Second Hospital of Shandong
University were used in the present study. All patients were
confirmed to have GC via histopathological evaluation. None of the
included patients had received pre-operative chemotherapy or
radiotherapy. The obtained samples were immediately snap-frozen in
liquid nitrogen and then stored at −80°C until further use for
total RNA extraction.
Cell culture
The non-tumorigenic human gastric epithelial cell
line GES-1 and four GC cell lines, namely MKN-45 (indicated not to
have a TP53 mutation), NCI-N87, AGS and HGC-27, which were
purchased from the American Type Culture Collection and maintained
in liquid nitrogen or at −80°C in the laboratory, were used in the
present study. All the cells were cultured in RPMI-1640 medium
(Invitrogen; Thermo Fisher Scientific, Inc.). All culture media
were supplemented with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin. The cells
were plated and incubated in 5% CO2 at 37°C and 95%
humidity.
Total RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the tissue samples
using TRIzol reagent (Thermo Fisher Scientific, Inc.) and from the
cell lines using the miRNeasy Mini Kit (Qiagen N.V.), according to
the manufacturers' protocol. The quantity and quality of extracted
RNA were determined by a NanoDrop spectrophotometer (Thermo Fisher
Scientific, Inc.). For RT-qPCR, 1 µg of RNA was reverse transcribed
to cDNA using a Takara reverse transcription kit (Takara
Biotechnology Co., Ltd.). RT-qPCR analyses were conducted on a
CFX-96 real-time PCR System (Bio-Rad Laboratories, Inc.) using SYBR
Premix Ex TaqTM (Takara Biotechnology Co., Ltd.). The temperature
protocol was as follows: 37°C for 15 min, followed by 85°C for 5
sec. The thermocycling conditions were as follows: 95°C for 30 sec,
followed by 40 cycles of 95°C for 5 sec, 60°C for 30 sec and 72°C
for 30 sec, followed by a final step of 72°C for 2 min. The primer
sequences used for target amplification are listed in Table SI. Data were normalized to GAPDH
expression. Relative expression levels were determined using the
comparative 2−ΔΔCq method (30).
UCC siRNA transfection
Three siRNA sequences specifically targeted against
UCC and si-NC, a scramble siRNA used as negative control, were
synthesized from Shanghai GenePharma Co., Ltd. The AGS cells were
transfected with the siRNA oligonucleotides at a concentration of
20 µM using Lipofectamine® 2000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturers'
protocol. The nucleotide sequences of the siRNAs are listed in
Table SI. The cells were harvested
for further study 24 h after transfection. The efficiency of
siRNA-mediated knockdown was determined using RT-qPCR by comparing
the expression of UCC in siRNA-treated cells with that in
si-NC-treated cells.
Cell proliferation assay
To determine the effect of UCC on cell
proliferation, a cell proliferation assay was performed using Cell
Counting kit-8 (CCK8) according to the manufacturer's protocol. AGS
cells transfected with si-UCC were seeded into 96-well plates at a
density of 3×103 cells/well and incubated for 24, 48 and
72 h, respectively. si-NC treatment was used as the negative
control. The effect of UCC on cell proliferation was determined
using CCK-8 (Wuhan Boster Biological Technology, Ltd.) as follows:
At the end of each 24, 48 or 72 h incubation, 10 µl of CCK-8
solution was added to each well. After 4 h, the number of viable
cells was determined by measuring the absorbance of water-soluble
formazan at 465 nm using a FlexStation 3 plate reader (Molecular
Devices, LLC). The experiments were performed in six
replicates.
Colony formation assay
Transfected AGS cells were seeded in 6-well plates
at a density of 1×103/well and incubated at 37°C and 5%
CO2 for 14 days. Subsequently, colonies were stained
with 10% Giemsa, visualized under a light microscope (Olympus
Corporation) at ×100 magnification and then physically quantified.
The experiments were performed in duplicate.
Survival analysis
Survival analyses for FOXP4-AS1, BLACAT2 and UCC
were conducted using Gene Expression Profiling Interactive Analysis
(GEPIA; http://gepia.cancer-pku.cn), an
online tool (31). A dataset from an
independent cohort (Affymetrix ID: 232242_at) was utilized for the
evaluation of PTPRG-AS1 expression as a prognostic factor of GC.
The results from the survival analyses of 631 GC tissue samples
from The Cancer Genome Atlas (http://cancer.gov/tcga) are available at the Kaplan
Meier Plot database (http://kmplot.com) (32–34).
Gene alteration frequencies of PTPRG-AS1 and BLACAT2 mRNA in GC
were assessed using cBioPortal for Cancer Genomics (http://www.cbioportal.org) (35).
Statistical analysis
Statistical analyses were performed using SPSS
software (version 22.0; IBM Corp.) and GraphPad Prism (GraphPad
Software, Inc.). Data were presented as the mean ± standard
deviation. Differences between experimental groups were assessed by
the Student's t-test. Associations between lncRNA expression and
clinical characteristics were analyzed using the simple Chi-square
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
PTPRG-AS1, FOXP4-AS1, BLACAT2, ZXF2
and UCC are upregulated in GC tissues and cell lines
To determine the relative expression of the lncRNAs
in GC, their expression levels were examined in GC cell lines and
GC tissues in comparison with ANTs. Fig.
1 shows the relative expression and mean-fold changes of
PTPRG-AS1 (P=0.001; Fig. 1A),
FOXP4-AS1 (P<0.001; Fig. 1B),
BLACAT2 (P<0.001; Fig. 1C), ZXF2
(P=0.102, Fig. 1D) and UCC (P=0.012;
Fig. 1E) in 61 pairs of GC tissues
and ANTs. All five lncRNAs were upregulated in GC tissues compared
with their corresponding ANTs. Apart from ZXF2, relative expression
levels of the lncRNAs in GC tissues were significant compared with
those observed in ANTs. The expression levels of the five lncRNAs
were verified in four GC cell lines, specifically MKN-45, NCI-N87,
AGS and HGC-27, relative to the non-tumorigenic human gastric
epithelial cell line, GES-1. All five lncRNAs were highly expressed
in GC cell lines. Elevated PTPRG-AS1 expression was observed in two
cell lines, MKN-45 and NCI-N87. FOXP4-AS1 was highly expressed in
three cell lines, AGS, NCI-N87 and MKN-45. BLACAT2 and ZXF2 were
upregulated in all four cell lines and UCC was upregulated in AGS,
NCI-N87 and HGC-27 compared with the GES-1 cell line (Fig. 2A-E). These results demonstrate that
PTPRG-AS1, FOXP4-AS1, BLACAT2, ZXF2 and UCC are upregulated in GC
tissues and cell lines.
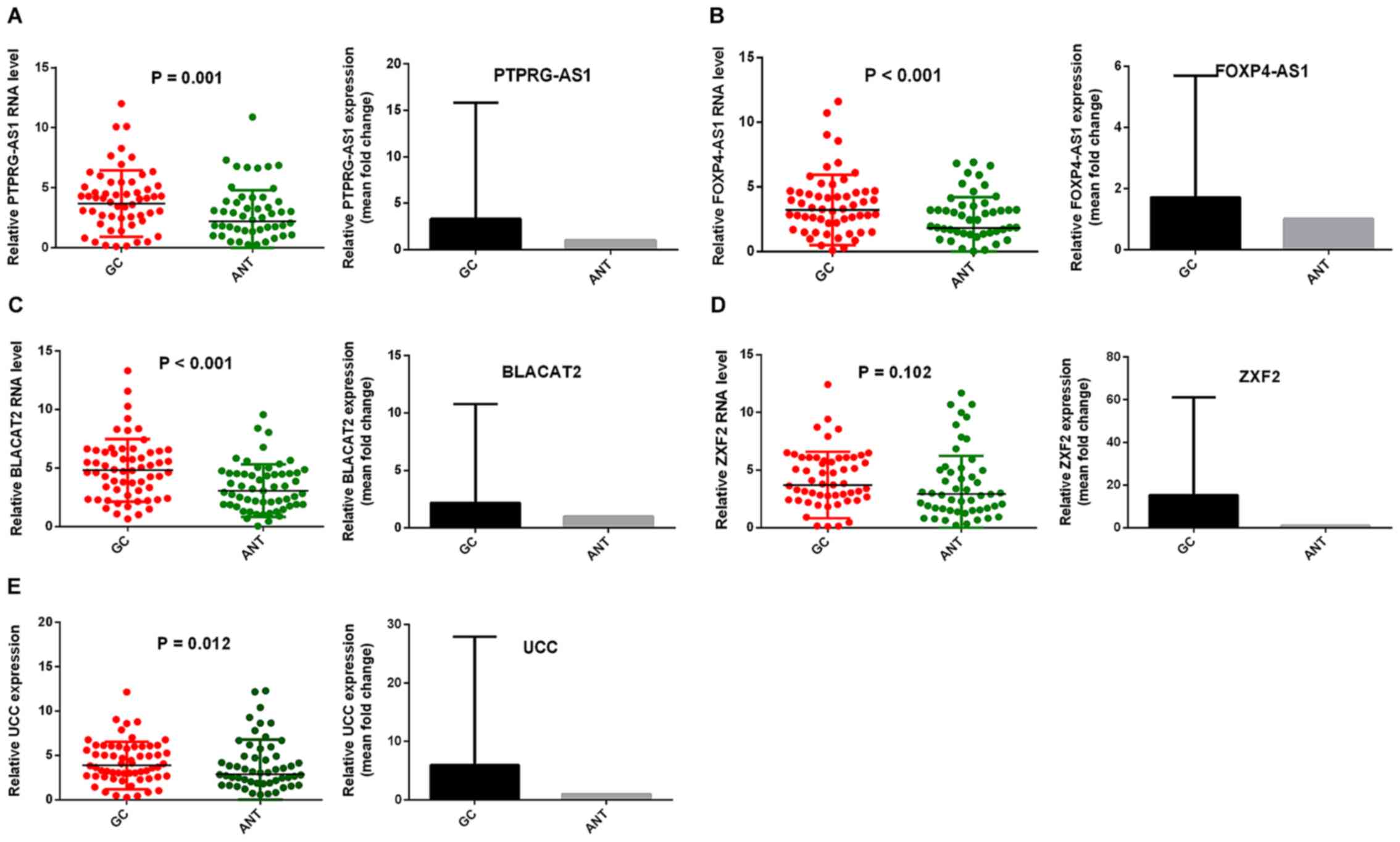 | Figure 1.PTPRG-AS1, FOXP4-AS1, BLACAT2, ZXF2
and UCC were upregulated in GC tissues. Reverse
transcription-quantitative PCR results demonstrated that the levels
of (A) PTPRG-AS1, (B) FOXP4-AS1, (C) BLACAT2, (D) ZXF2, and (E) UCC
in 61 pairs of GC tissues were higher than those in ANTs. Error
bars represent the mean ± SD. GC, gastric cancer, ANT, adjacent
normal tissues; PTPRG-AS1, PTPRG antisense RNA 1; FOXP4-AS1,
forkhead box P4 antisense RNA 1; BLACAT2, bladder cancer-associated
transcript 2; UCC, upregulated in colorectal cancer. |
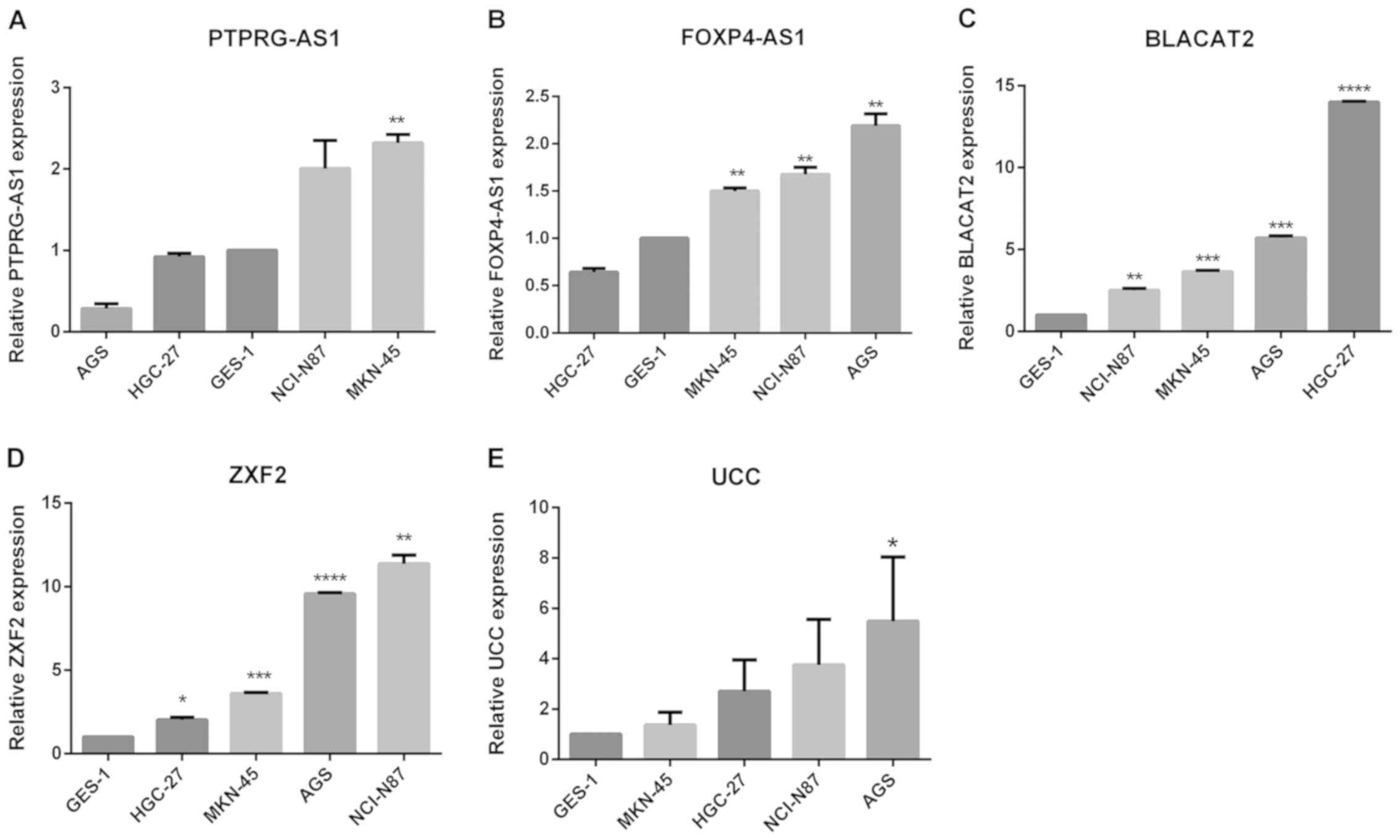 | Figure 2.Expression of lncRNAs in GC cell
lines. Reverse transcription-quantitative PCR results revealed the
baseline mRNA levels of (A) PTPRG-AS1, (B) FOXP4-AS1, (C) BLACAT2,
(D) ZXF2, and (E) UCC in the non-tumorigenic gastric epithelial
cell line GES-1 and four gastric cancer cell lines. The expression
levels of lncRNAs were normalized to that of GES-1. Error bars
represent the mean ± SD of triplicate experiments. *P<0.05,
**P<0.01, ***P<0.001 and ****P<0.0001. lncRNAs, long
non-coding RNAs; PTPRG-AS1, PTPRG antisense RNA 1; FOXP4-AS1,
forkhead box P4 antisense RNA 1; BLACAT2, bladder cancer-associated
transcript 2; UCC, upregulated in colorectal cancer. |
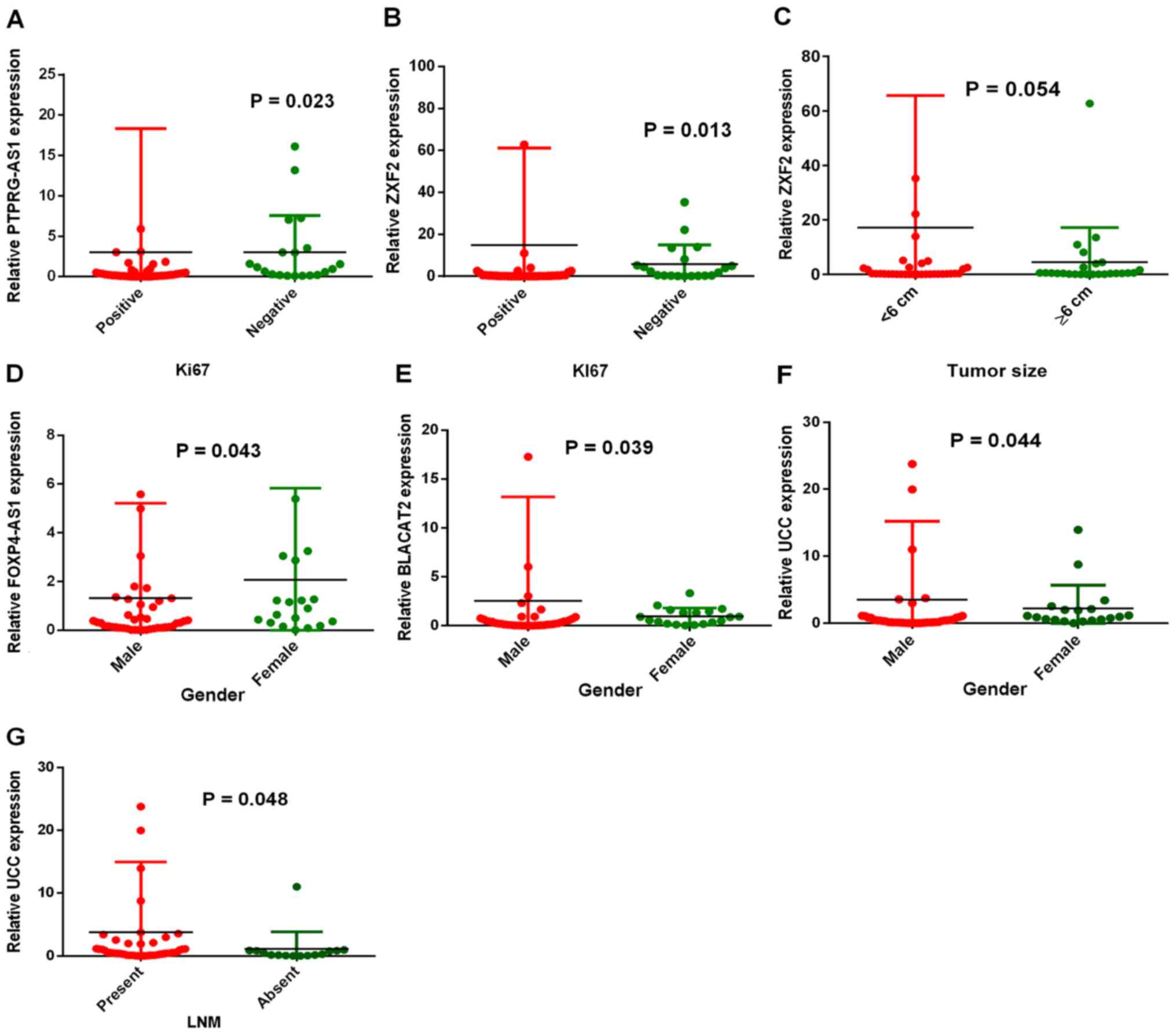
PTPRG-AS1, FOXP4-AS1, BLACAT2, ZXF2
and UCC are associated with the clinical characteristics of
patients with GC
In order to study the roles of these lncRNAs in GC,
the associations between overexpression and the clinical
characteristics of patients were evaluated. Table I summarizes the associations between
lncRNA expression levels and the clinical characteristics of
patients. The characteristics that were studied include: i) Sex;
ii) age; iii) lymph node metastasis status; iv) tumor grade; v)
tumor size; vi) Ki67 expression status; and vii) Ki67 percentage of
expression. PTPRG-AS1 expression was associated with the cell
proliferation marker Ki67 (P=0.023; Fig.
3A). Similarly, patients in the Ki67-positive group (n=41) were
shown to have significantly higher ZXF2 expression than patients in
the Ki67-negative group (n=20) (P=0.013; Fig. 3B). Both PTPRG-AS1 and ZXF2 expression
levels increased with increase in the percentage of Ki67
expression. Patients with Ki67 expression levels ≥50% (n=24) had
higher PTPRG-AS1 and ZXF2 expression than those with Ki67
expression levels <50% (n=37). A relationship between ZXF2
expression and tumor size was also found. Patients with tumors
<6 cm (n=36) had increased ZXF2 expression compared with
patients with tumors ≥6 cm (n=25). However, this was not
statistically significant (P=0.054; Fig.
3C). In addition, differential expression levels of FOXP4-AS1,
BLACAT2 and UCC were observed between males and females. FOXP4-AS1
was significantly increased in females (n=9) compared with males
(n=42) (P=0.043; Fig. 3D). BLACAT2
expression was significantly higher in males (n=42) than in females
(n=19) (P=0.037; Fig. 3E).
Similarly, UCC expression was higher in males (n=42) than females
(n=19) (P=0.044; Fig. 3F). Patients
with lymph node metastasis (n=46) had higher UCC expression than
patients with no lymph node metastasis (n=15) (P=0.048; Fig. 3G), implicating UCC overexpression in
GC metastasis. UCC expression was also closely associated with the
expression status of cell proliferation marker Ki67, although this
was not statistically significant (P=0.074). Collectively, these
results indicate the associations of PTPRG-AS1, ZXF2, BLACAT2,
FOXP4-AS1 and UCC expression levels with clinical and demographic
characteristics of patients with GC.
 | Table I.PTPRG-AS1, ZXF2, BLACAT2, FOXP4-AS1
and UCC expression levels and their associations with clinical
characteristics of patients. |
Table I.
PTPRG-AS1, ZXF2, BLACAT2, FOXP4-AS1
and UCC expression levels and their associations with clinical
characteristics of patients.
|
|
| PTPRG-AS1 | ZXF2 | BLACAT2 | FOXP4-AS1 | UCC |
|---|
|
|
|
|
|
|
|
|
|---|
|
Characteristics | n (%) | High | Low | P-value | High | Low | P-value | High | Low | P-value | High | Low | P-value | High | Low | P-value |
|---|
| Sex | 61 | 27 | 34 | 0.743 | 35 | 26 | 0.241 | 33 | 28 | 0.039 | 30 | 31 | 0.043 | 21 | 40 | 0.044 |
|
Male | 42 (68.9) | 18 | 24 |
| 22 | 20 |
| 19 | 23 |
| 17 | 25 |
| 11 | 31 |
|
|
Female | 19 (31.1) | 9 | 10 |
| 13 | 6 |
| 14 | 5 |
| 13 | 6 |
| 10 | 9 |
|
| Age (years) |
|
<60 | 25 (41.0) | 11 | 14 | 0.973 | 16 | 9 | 0.383 | 15 | 10 | 0.441 | 12 | 13 | 0.878 | 6 | 19 | 0.153 |
|
≥60 | 36 (59.0) | 16 | 20 |
| 19 | 7 |
| 18 | 18 |
| 18 | 18 |
| 15 | 21 |
|
| LNM |
|
Present | 46 (75.4) | 20 | 26 | 0.829 | 29 | 17 | 0.117 | 24 | 22 | 0.597 | 25 | 21 | 0.157 | 19 | 27 | 0.048 |
|
Absent | 15 (24.6) | 7 | 8 |
| 6 | 9 |
| 9 | 6 |
| 5 | 10 |
| 2 | 13 |
|
| Tumor grade |
|
1/2 | 10 (16.4) | 6 | 4 | 0.273 | 7 | 3 | 0.494 | 7 | 3 | 0.319 | 7 | 3 | 0.182 | 5 | 5 | 0.291 |
| 3 | 51 (83.6) | 21 | 30 |
| 28 | 23 |
| 26 | 25 |
| 23 | 28 |
| 16 | 35 |
|
| Tumor size |
| <6
cm | 36 (59.0) | 16 | 20 | 0.973 | 17 | 19 | 0.054 | 19 | 17 | 0.804 | 19 | 17 | 0.500 | 12 | 24 | 0.829 |
| ≥6
cm | 25 (41.0) | 11 | 14 |
| 18 | 7 |
| 14 | 11 |
| 11 | 14 |
| 9 | 16 |
|
| Ki67 status |
|
Positive | 41 (67.2) | 14 | 27 | 0.023 | 19 | 22 | 0.013 | 21 | 20 | 0.518 | 20 | 21 | 0.929 | 11 | 30 | 0.074 |
|
Negative | 20 (32.8) | 13 | 7 |
| 16 | 4 |
| 12 | 8 |
| 10 | 10 |
| 10 | 10 |
|
| Ki67 expression
(%) |
|
<50 | 37 (60.7) | 18 | 19 | 0.392 | 23 | 14 | 0.348 | 20 | 17 | 0.993 | 18 | 19 | 0.918 | 12 | 25 | 0.684 |
|
≥50 | 24 (39.3) | 9 | 15 |
| 12 | 12 |
| 13 | 11 |
| 12 | 12 |
| 9 | 15 |
|
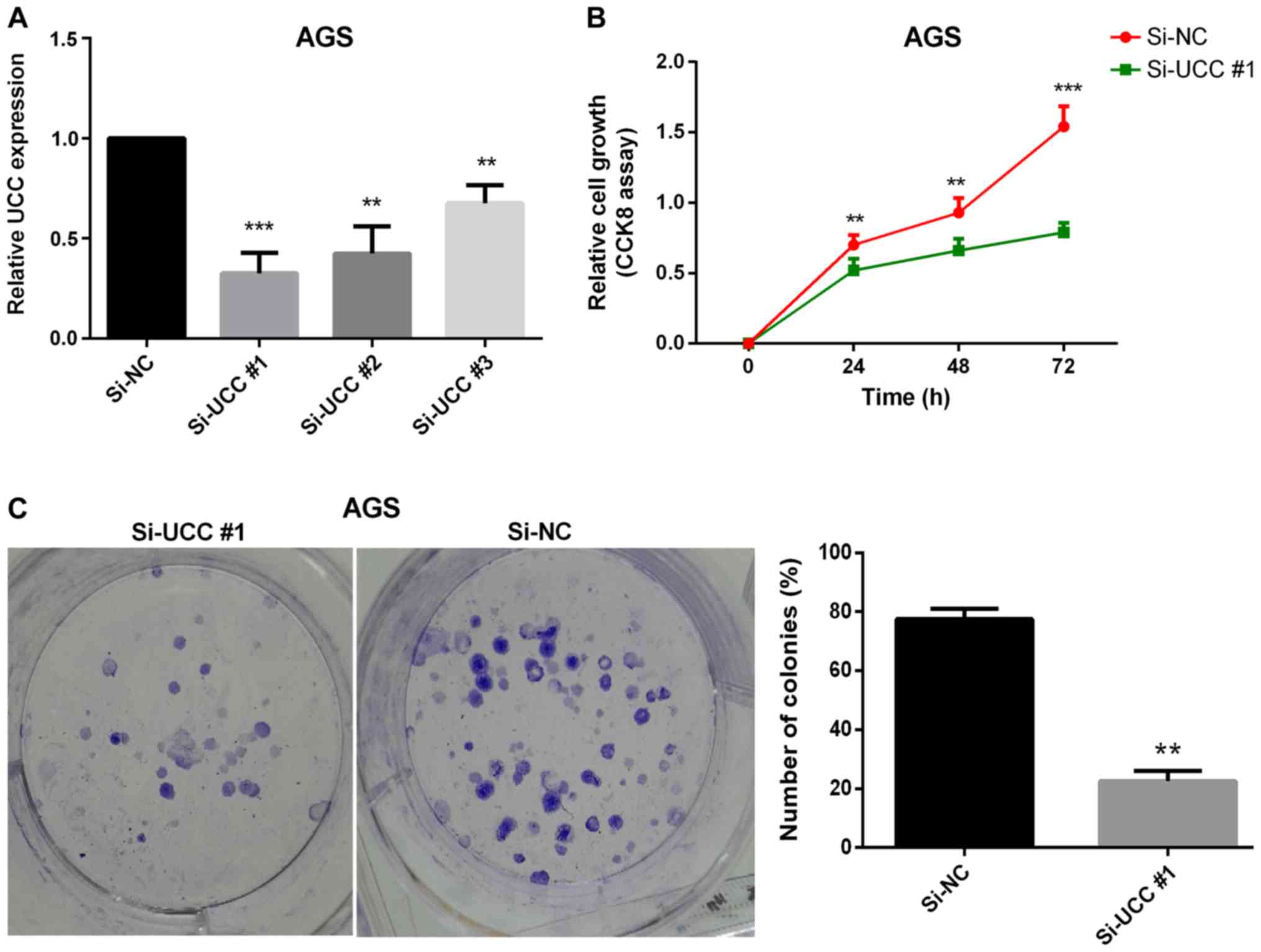
UCC promotes GC cell proliferation in
vitro
Uncontrolled cell proliferation has been proven to
be one of the hallmarks of cancer (36). Thus, the effect of one of the
upregulated lncRNAs (UCC) on GC cell proliferation was
investigated. Loss-of-function experiments were performed on UCC
using siRNAs to ascertain its effect on GC cell proliferation. The
GC cell line AGS, in which UCC expression was found to be
significantly upregulated (Fig. 2E),
was transfected with three siRNA oligonucleotides in order to
achieve siRNA-mediated UCC knockdown. Twenty-four hours after
transfection, the efficiency of transfections was verified by
RT-qPCR analysis (Fig. 4A). The
CCK-8 cell proliferation assay results revealed that silencing UCC
markedly reduced the rate of cell proliferation (Fig. 4B). The colony formation assay results
demonstrated a significant reduction in the numbers and sizes of
colonies following knockdown of UCC in AGS cells (P=0.007; Fig. 4C) indicating that UCC promotes GC
cell proliferation in vitro.
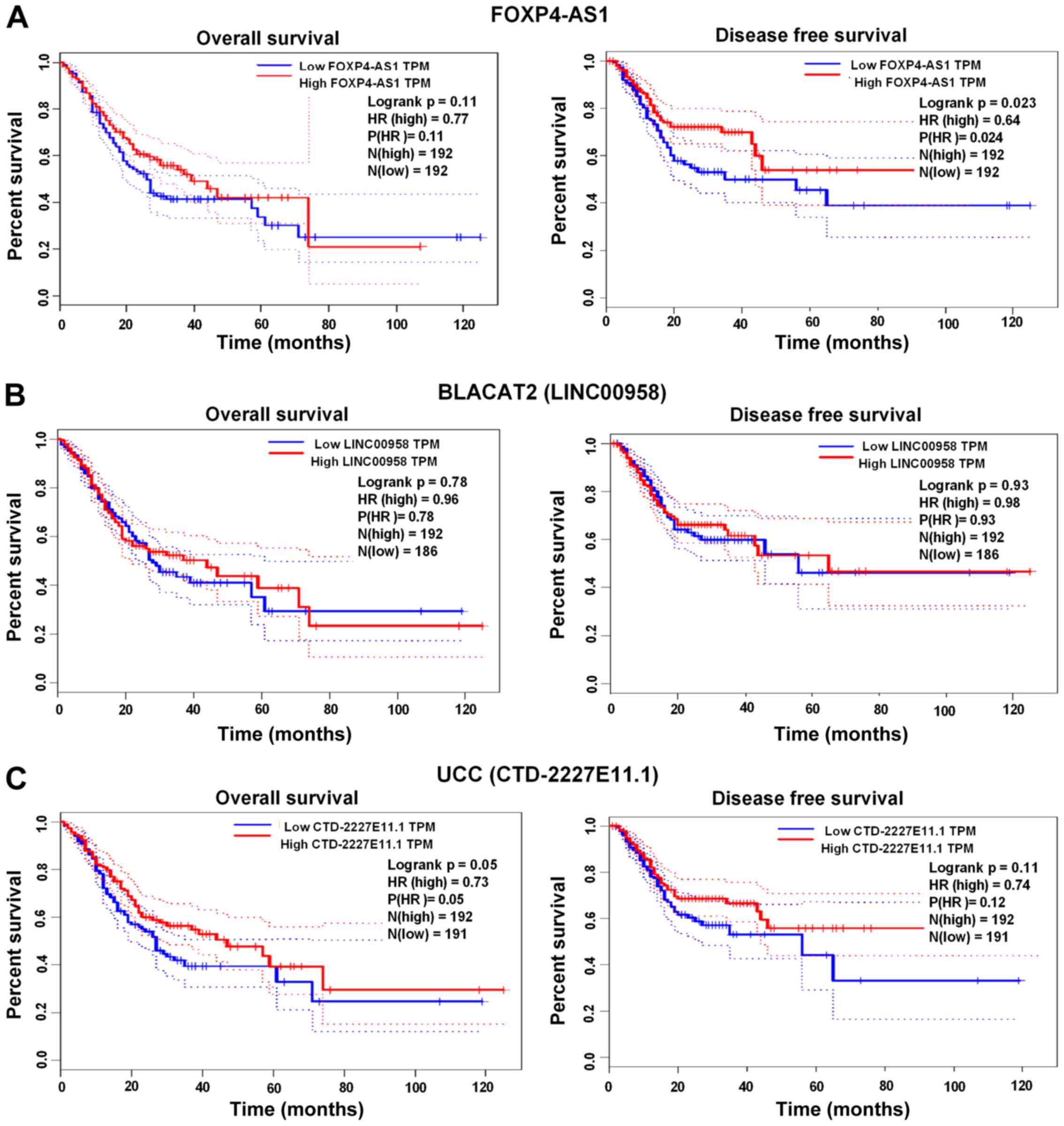
Prognostic efficacy of PTPRG-AS1,
FOXP4-AS1, BLACAT2, ZXF2 and UCC in GC
The prognostic efficacy of BLACAT2, FOXP4-AS1 and
UCC was assessed using the GEPIA web server (http://gepia.cancer-pku.cn), whereas the prognostic
efficacy of PTPRG-AS1 was examined using the Kaplan Meier Plot
database (www.kmplot.com) (Affymetrix ID:
232242_at). High FOXP4-AS1 expression was significantly associated
with shorter disease-free survival (DFS) rates compared with low
FOXP4-AS1 expression (P=0.023; Fig.
5A). No significant associations were found between increased
expression of BLACAT2 and UCC and the OS and DFS rates of patients
with GC (Fig. 5B and C). However,
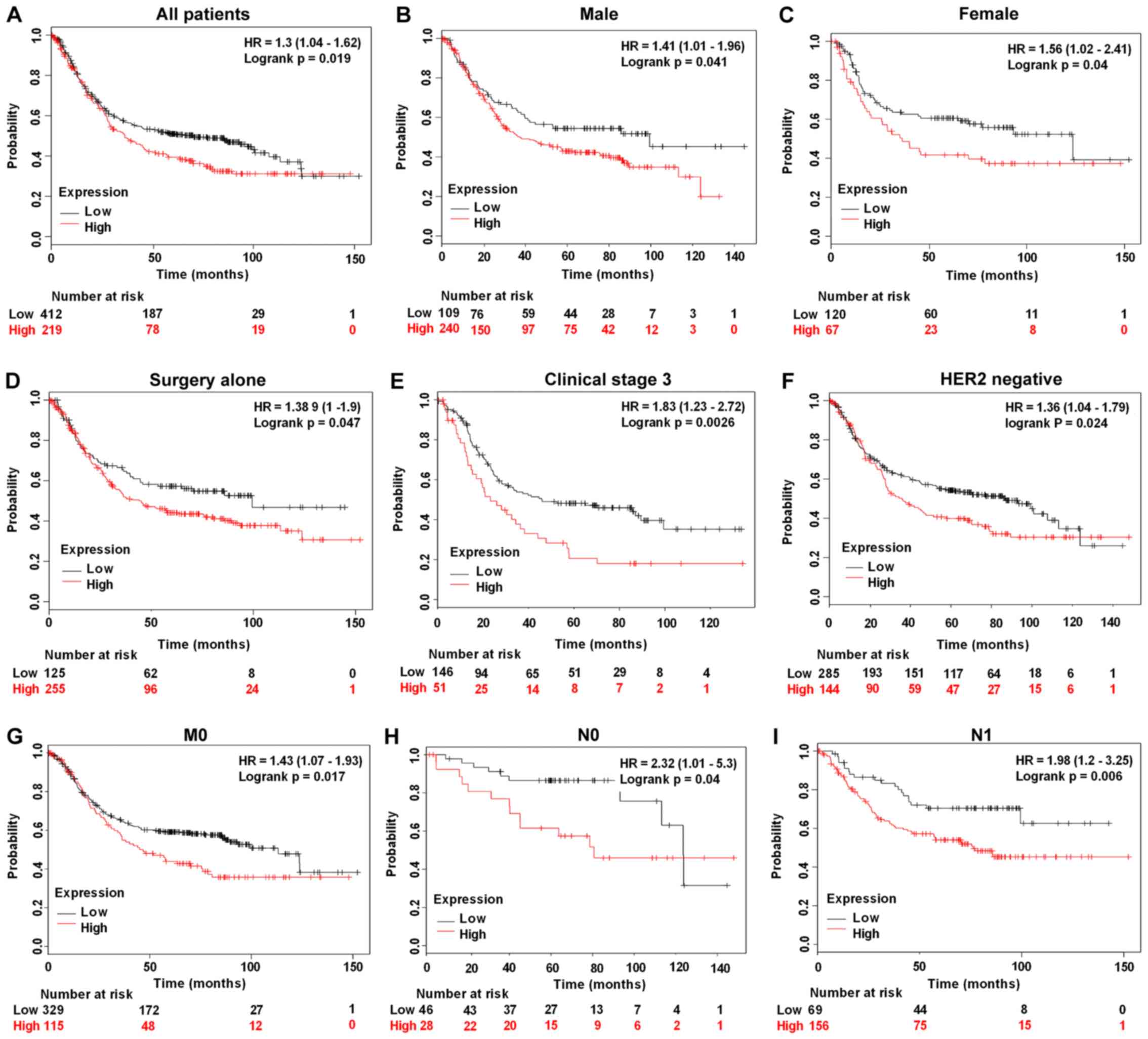
high PTPRG-AS1 was significantly associated with poor OS rates in
all patients (P=0.019; Fig. 6A),
male patients alone (P=0.041; Fig.
6B), female patients alone (P=0.040; Fig. 6C), patients treated with surgery
alone (P=0.047; Fig. 6D), patients
in clinical stage 3 (P=0.0026; Fig.
6E), HER2− patients (P=0.024; Fig. 6F), patients with stage M0 GC and no
metastasis (P=0.017; Fig. 6G),
patients with stage N0 GC and no lymph node metastasis (P=0.040;
Fig. 6H) and patients with stage N1
GC and few lymph node metastasis sites (P=0.006; Fig. 6I). Shorter survival rates were also
observed among patients treated with 5-fluorouracil adjuvant alone
who had high PTPRG-AS1 expression compared with patients with low
PTPRG-AS1 expression, although this was not statistically
significant (P=0.45; Table II).
Correlation analyses were also performed on other stages,
classifications and pathological grades. The results, which are
summarized in Table II, demonstrate
the prognostic efficacy of the lncRNAs in patients with GC that
have different clinicopathological characteristics.
 | Table II.Correlation of high PTPRG antisense
RNA 1 expression with other clinical characteristics of patients
with gastric cancer. |
Table II.
Correlation of high PTPRG antisense
RNA 1 expression with other clinical characteristics of patients
with gastric cancer.
|
Characteristics | n | HR 95% CI | P-value |
|---|
| Clinical
stages |
| 2 | 126 | 1.7 (0.89–3.26 | 0.11 |
| 4 | 140 | 0.72
(0.46–1.11) | 0.13 |
| Pathological
grades |
| Poorly
differentiated | 121 | 1.26
(0.78–2.06) | 0.34 |
|
Moderately differentiated | 67 | 0.47
(0.24–0.92) | 0.024 |
| Well
differentiated |
|
|
|
| HER2 status |
|
Positive | 202 | 1.43
(0.94–2.19) | 0.097 |
| Treatments |
| 5-FU
based adjuvant | 34 | 1.42
(0.57–3.54) | 0.45 |
| Other
adjuvants (e.g. irinotecan) | 76 | 0.52
(0.2–1.36) | 0.17 |
| T stage |
| 2 | 241 | 1.55
(0.99–2.44) | 0.054 |
| 4 | 38 | 0.61
(0.26–1.41) | 0.24 |
| N stage |
| 1 + 2 +
3 | 422 | 1.3
(0.97–1.73) | 0.074 |
| 2 | 121 | 0.71
(0.45–1.11) | 0.13 |
| 3 | 76 | 1.31
(0.76–2.26) | 0.33 |
| M stage |
| M1 | 56 | 0.63
(0.33–1.22) | 0.17 |
| Lauren
classification |
|
Intestinal | 269 | 1.47
(0.97–2.23) | 0.067 |
|
Mixed | 29 | 1.56 (0.5
−4.77) | 0.43 |
|
Diffuse | 240 | 1.35
(0.96–1.9) | 0.084 |
Genetic alteration of BLACAT2 is a
marker of poor outcome for patients with GC
Genetic alterations of the lncRNAs in GC were
analyzed using the cBioPortal online tool. A total of 1,365
patients from six datasets of patients with GC were evaluated. No
alterations were found in the FOXP4-AS1, ZXF2 and UCC genes in GC.
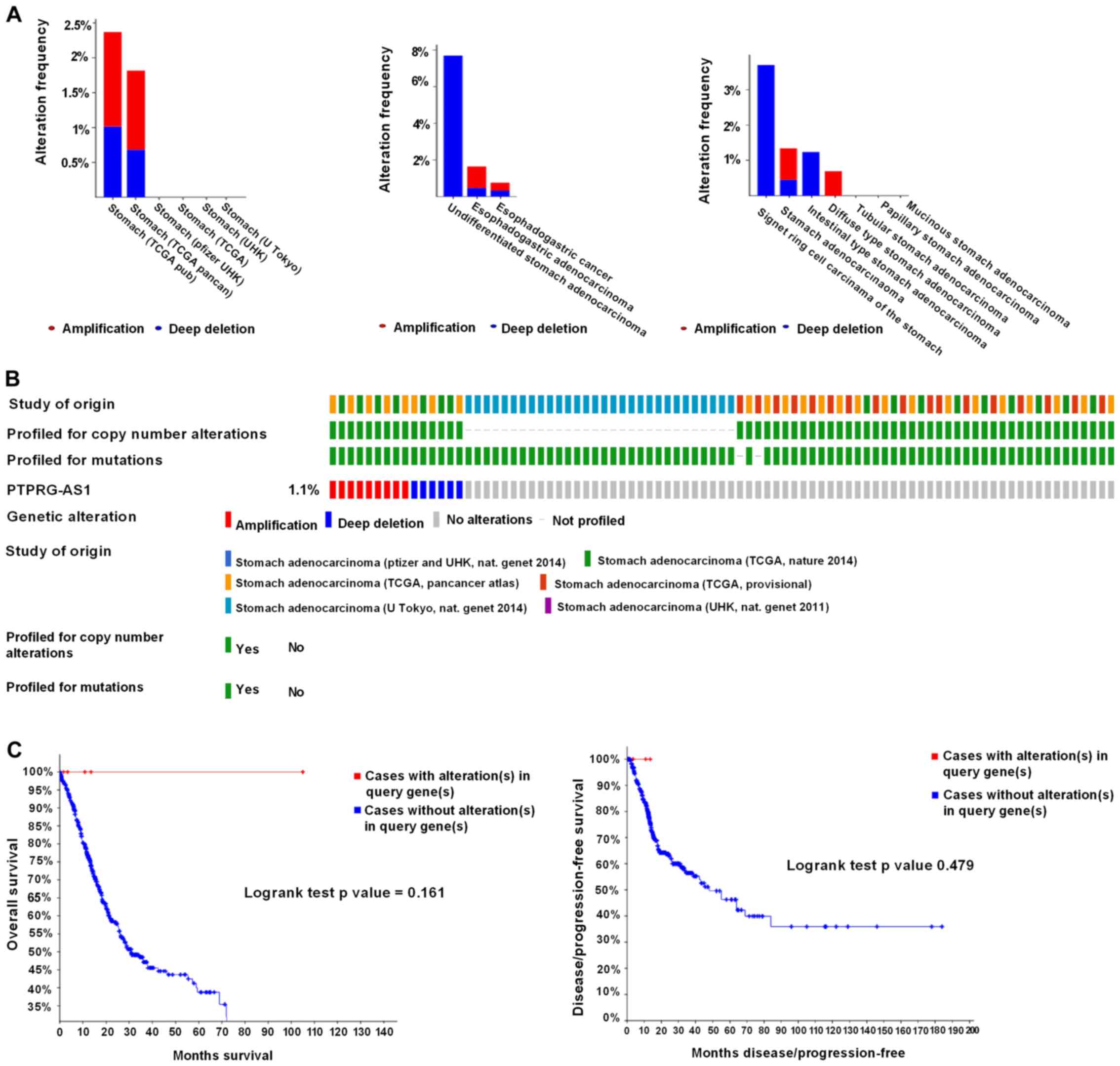
Regarding PTPRG-AS1, alterations ranging from 1.82 to 2.37% were
found for the gene sets submitted for analysis (Fig. 7A). The percentage of genetic
alterations in PTPRG-AS1 for GC was 1.1% (Fig. 7B). Following cBioPortal and
Kaplan-Meier plotter analyses and a log-rank test, the results
indicated that there was no significant difference in OS and DFS in
patients with or without alterations in PTPRG-AS1 (P=0.161 and
P=0.479 respectively; Fig. 7C).
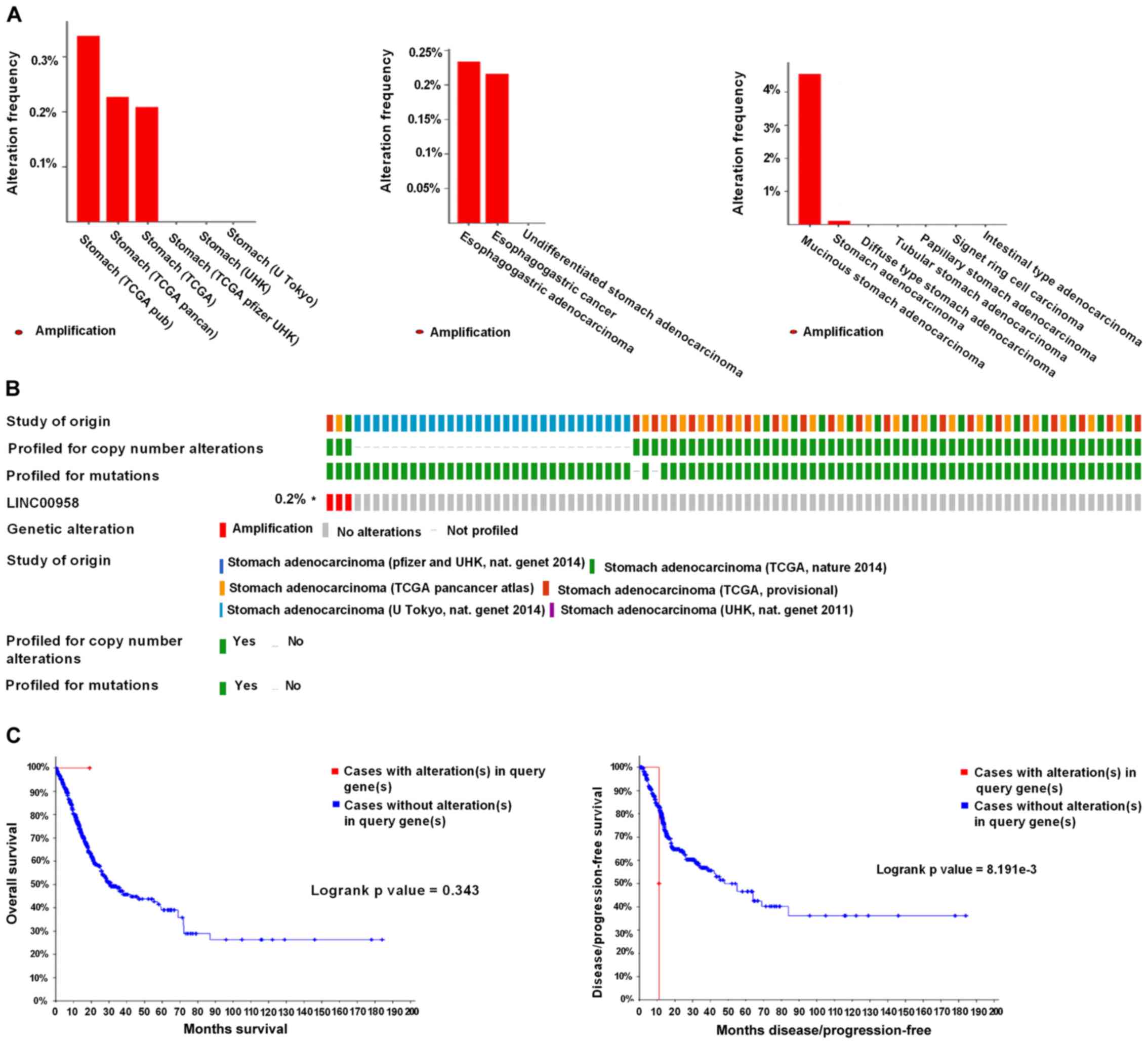
Regarding the BLACAT2 gene, alterations were found ranging from
0.21 to 0.34% (Fig. 8A) and the
percentage of genetic alterations was 0.2% (Fig. 8B). Following cBioPortal and
Kaplan-Meier plotter analyses and a log-rank test, the results
indicated that there was no significant difference in OS in cases
with or without alterations in BLACAT2 (P=0.239). By contrast,
cases with alterations in BLACAT2 had significantly poorer DFS
rates than those without alterations (P=0.0023; Fig. 8C).
Discussion
Therapeutic regimens available for GC, including
trastuzumab, a widely administered drug, have obvious limitations,
including their inability to be used for a wider range of patients
(22). Therefore, GC remains one of
the leading causes of cancer-related deaths (37). Studies have shown that upregulated
lncRNAs such as OCC-1, AK126698, SChLAP1, BORG and DANCR are
associated with poor prognosis in cancer patients (9,10,12–14).
Poor prognoses may also be attributed to late diagnosis, lack of
specific biomarkers and the metastatic and proliferative capacity
of cancer cells (38,39). Since specific biomarkers of GC are
currently limited, patients are more often diagnosed at a later,
usually metastatic stage. This increases the risk of disease
recurrence, GC-related deaths and poor prognoses. Consequently,
there is a necessity for further exploration of the genetic and
molecular modifications that characterize gastric carcinogenesis
and an urgent need for the identification of highly sensitive and
specific biomarkers.
In the present study, the expression profiles,
functions and prognostic efficacy of five lncRNAs in GC,
specifically PTPRG-AS1, FOXP4-AS1, BLACAT2, ZXF2 and UCC, were
investigated. Expression analyses revealed that all five lncRNAs
were upregulated in tumor tissues relative to ANTs, thus indicating
differential expression patterns of the lncRNAs between cancer and
healthy tissues. This was a significant finding since the aberrant
expression patterns of the lncRNAs suggest their application as
diagnostic markers for GC at the molecular level. The present study
results regarding the upregulation of PTPRG-AS1, FOXP4-AS1,
BLACAT2, ZXF2 and UCC are in accordance with the results of
Iranpour et al (23), Li
et al (25), He et al
(26), Yang et al (27) and Huang et al (15), which conducted studies on breast,
colorectal, bladder, lung and colorectal cancers, respectively. The
results of the current study were further verified in four GC cell
lines. As expected, all five lncRNAs were highly expressed in GC
cell lines relative to the healthy gastric epithelial cell line
GES-1. Significant associations were found between BLACAT2,
FOXP4-AS1, UCC and the sex of patients with GC. BLACAT2 and UCC
expression levels were higher in males than in females.
Nonetheless, higher FOXP4-AS1 expression was observed in females
than in males. Therefore, it was concluded that sex may be an
important predisposing factor for GC. Biological factors, including
age and sex, have been associated with the development of certain
cancer types and the sex differential in susceptibility may provide
insight into the etiology of cancer (40). The present study also reported new
findings on the relationship between BLACAT2, FOXP4-AS1 and UCC
expression and sex predisposition to GC.
PTPRG-AS1 expression was significantly associated
with the expression status of the cell proliferation marker Ki67.
In particular, the expression of ZXF2 was higher in Ki67-positive
patients than in Ki67-negative patients. Both PTPRG-AS1 and ZXF2
expression levels were found to increase in response to increased
Ki67 expression, hence indicating that the proliferative ability of
GC cells increases when the expression of PTPRG-AS1 and ZXF2 is
upregulated. These results suggest the involvement of PTPRG-AS1 and
ZXF2 in GC cell proliferation. As in other types of cancer, GC
develops due to genetic alterations that result in generalized loss
of growth control, which in turn leads to continual unregulated
proliferation of cancer cells (36).
Consequently, it was hypothesized that lncRNAs PTPRG-AS1 and ZXF2
are oncogenes that promote gastric tumorigenesis and/or progression
by promoting GC proliferation. This hypothesis was corroborated by
the findings of Iranpour et al (23) and Yang et al (27), which suggested the implication of
PTPRG-AS1 and ZXF2 in the oncogenicity of breast and lung cancer,
respectively. Patients with smaller tumors exhibited considerably
higher ZXF2 expression than patients with tumors of a larger size,
though the difference was not significant. Nevertheless, ZXF2 may
be a marker of the early stages of gastric carcinogenesis. This
possibility requires further investigation in large-scale
studies.
The human body comprises a network of lymphatic
vessels and lymph nodes mainly designed to serve protective roles
against infections by filtering harmful substances (41). LNM is defined as the spread of cancer
cells from a primary (original) tumor to the lymph nodes. It is a
well-known and clinically accepted strong prognostic factor for the
recurrence and survival of patients with most solid tumors
(41). Previous findings have
demonstrated the involvement of the unique lymph node
microenvironment in the formation of systemic metastasis suggesting
LNM as a source of further cancer spread and poor prognosis.
Adequate management of LNM could reduce further systemic metastasis
and improve patient outcomes (42).
In the current study, a higher UCC expression was observed in
patients with LNM than in those without LNM. This association
between UCC expression and LNM suggests that the overexpression of
UCC may be responsible for metastatic competence and disease
progression in patients with GC. This is an indication of the role
of UCC in GC metastasis and is in consonance with an aforementioned
study suggesting UCC upregulation in the metastasis of colorectal
cancer cells (15).
According to the CCK-8 assay results, UCC was
identified as a key driver of GC growth. Knockdown of UCC
significantly inhibited cell proliferation in AGS cells. This
finding was further confirmed by performing a colony formation
assay. In comparison with controls, the number and sizes of GC cell
colonies were found to be significantly reduced after UCC
knockdown, implicating UCC in GC cell proliferation. Similar
findings were reported in an aforementioned study on colorectal
cancer (15).
The prolonged survival times observed among patients
with high FOXP4-AS1 expression in the present study suggest that
FOXP4-AS1 is a marker of favorable outcome for patients with GC. No
associations were identified between high expression of BLACAT2 or
UCC with the OS or DFS of patients. This finding requires further
validation in studies with larger sample sizes. However, high
expression of PTPRG-AS1 was significantly associated with poor OS
probability in all patients; those with high expression of
PTPRG-AS1 had shorter survival rates than those with low PTPRG-AS1
expression. PTPRG-AS1 may be a novel predictor of prognosis for
patients with GC.
Surgical resection is usually the first line of
treatment for patients with GC. Thus far, it is regarded as the
only reliable possibility of a curative treatment and the only
effective therapy, though the locoregional recurrence and five-year
survival rates following resection remain poor (43). Shorter survival rates were observed
among patients with high PTPRG-AS1 expression treated with surgery
alone or 5-FU adjuvant alone than in those with low PTPRG-AS1
expression subjected to the same treatment. It is possible that
PTPRG-AS1 upregulation may be responsible for resistance to GC
therapy.
Over 80% of patients with GC are HER2−
(44). Although positive HER2 status
is generally associated with poor prognosis for patients with GC, a
recent study revealed that, when treated with trastuzumab,
HER2+ patients had better prognosis than
HER2− patients (44).
Therefore, the development of targeted therapies for
HER2− patients may prove to be beneficial. The results
from the survival analysis performed in the current study showed
that a high expression of PTPRG-AS1 was associated with poor
prognosis for HER2− patients, as OS significantly
decreased in patients with a high expression of this lncRNA
compared with those that had a low expression. PTPRG-AS1 may be a
novel therapeutic target for HER2− patients especially
since the trastuzumab regimen has been found to be ineffective in
these patients (22).
Of note, patients with advanced GC stage (stage
III), patients with earlier non-metastatic stages of GC (stage M0)
and no lymph node metastasis (stage N0) and those with metastasis
in few lymph nodes (stage N1) and high PTPRG-AS1 expression in the
current study, had poorer survival rates compared with patients
that had low expression. Poorer survival rates were also observed
among patients with other pathological stages of GC that had a high
PTPRG-AS1 expression compared with those that had downregulated
expression. Thus, it was concluded that PTPRG-AS1 may be a novel
prognostic predictor of both early and advanced GC and may serve as
a marker of patient outcomes and a therapeutic target for patients
with different pathological stages and subgroups of GC.
Mutations and gene amplifications are linked to
carcinogenesis; mutant genes cause a gain-of-function phenotype,
are involved in tumorigenesis and related to patient outcomes
(45). Herein, the percentages of
alterations in PTPRG-AS1 and BLACAT2 among patients were identified
as 1.1 and 0.2%, respectively. There were no significant
differences in OS and DFS in cases with or without alterations in
PTPRG-AS1 and no significant difference in OS rates in cases with
or without alterations in BLACAT2. However, cases with this type of
alteration had significantly poorer DFS rates than those without
alterations, indicating that BLACAT2 gene mutations are associated
with poor prognosis in GC. Upregulated lncRNAs may be more readily
applied as biomarkers than downregulated lncRNAs (15). A number of studies have reported an
association between lncRNA upregulation and patient outcomes and
have suggested their application as cancer biomarkers. For example,
Gao et al (46) found that
plasma orosomucoid 2 was significantly upregulated in patients with
stage II colorectal cancer and may be used as a biomarker in
diagnosis and prognostic evaluation. Li et al (47) revealed that high MALAT1 expression
was associated with decreased survival and poor response of
advanced colorectal cancer patients to oxaliplatin-based
chemotherapy. Du et al (48)
demonstrated that increased expression of urinary uc004cox.4 was
significantly associated with poorer prognosis in terms of
recurrence-free survival of non-muscle invasive bladder cancer and
may be combined with future non-invasive biomarkers for the
diagnosis and prognosis of bladder cancer, provided that it is
validated in a large-scale study. According to Li et al
(49), a high expression of
SPINT1-AS1 was associated with poor clinical outcome and SPINT1-AS1
may be a promising biomarker for the prognostic prediction and
treatment monitoring of patients with colorectal cancer. Moreover,
the present study reported new findings on the expression patterns,
functions and clinical significance of PTPRG-AS1, FOXP4-AS1,
BLACAT2, ZXF2 and UCC and their potential for application as
diagnostic, prognostic and therapeutic targets in GC.
Determination of target RNAs or proteins that
lncRNAs interact with is regarded as an important step in
characterizing the functions of lncRNAs (50). A recent study demonstrated that
lncRNAs regulate inflammation and reparative responses by
competitively sponging miRNAs and acting as competing endogenous
RNAs (51). Identification of
interactions and complex regulatory networks of lncRNAs, miRNAs and
other relevant genes may provide insight into the molecular
mechanisms involved in human cancer and the development of
diagnostic and therapeutic strategies (52). Certain molecular interactions of the
lncRNAs in our study have also been reported by a number of
studies. For instance, BLACAT2 was found to mediate the interaction
of WD repeat domain 5 (WDR5), a core component of the histone H3K4
methyltransferase complex that is required for vertebrate
development, with multiple promoter target sequences of vascular
endothelial growth factor C (26).
WDR5 is involved in the self-renewal of pluripotent stem cells and
cell differentiation, and interactions with WDR5 may induce
tumorigenesis by promoting target gene recognition (53). By contrast, UCC was identified as a
possible target of miR-143, which has been found to be
downregulated in a variety of cancer types (15). ZXF2 has been reported to interact
with c-myc and enhance lung cancer progression via the
c-myc/e-cadherin pathway (27) and
PTPRG-AS1 has been suggested to downregulate the expression of
PTPRG, a tumor suppressor gene (24), thereby enhancing disease progression
in breast cancer. The results from these studies confirm the
proto-oncogenic involvements of the studied lncRNAs in various
malignancies. However, their precise molecular interactions in GC
need to be investigated.
The present study had certain limitations. First,
the number of GC tissue samples used was relatively small. Although
significantly different expression levels of PTPRG-AS1, FOXP4-AS1,
BLACAT2 and UCC were observed, ZXF2 upregulation was not
statistically significant, probably due to this limitation.
Secondly, access to the follow-up data of patients was not
available, because the samples were collected from patients with
newly diagnosed GC at the time of analysis; consequently, the
prognostic efficacy of the lncRNAs could not be determined using
the results of RT-qPCR analysis. However, the effects of high
lncRNA expression on patient outcomes were examined using data from
publicly available online databases. Despite the above
shortcomings, the present study revealed that PTPRG-AS1, FOXP4-AS1,
BLACAT2, ZXF2 and UCC are upregulated in GC and have the potential
for application as biomarkers for GC detection at the molecular
level. The involvement of PTPRG-AS1 and ZXF2 in GC growth was also
revealed and ZXF2 was identified as a potential marker of the early
stages of gastric carcinogenesis. Furthermore, associations were
observed between BLACAT2, FOXP4-AS1, UCC and sex predisposition to
GC. An association was also identified between UCC overexpression
and lymph node metastasis and its role in promoting GC cell
proliferation in vitro. Finally, it was shown that BLACAT2
gene alterations were associated with worse DFS outcome and
PTPRG-AS1 overexpression was associated with worse OS outcome for
all patients with GC and the respective subgroups of GC patients.
Therefore, these lncRNAs are promising novel prognostic predictors
and therapeutic targets for different categories of patients with
GC.
In conclusion, PTPRG-AS1, FOXP4-AS1, BLACAT2, ZXF2
and UCC are potential biomarkers for GC detection at the molecular
level and they may be individually utilized as therapeutic targets
for GC and for prognostic predictions. Nonetheless, further in
vitro and in vivo research is required for each lncRNA
in order to ascertain their specific involvements in gastric
carcinogenesis.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to acknowledge The Second
Hospital of Shandong University for supporting the study and
providing data.
Funding
This work was supported by the Fundamental Research
Funds of Shandong University (grant nos. 2017BTS01, 2018JC002 and
2017JC031) and the Key Research and Development Program of Shandong
Province (grant no. 2018FYJH0505).
Availability of data and materials
The datasets used and analyzed in the present study
are available from the corresponding author upon reasonable
request.
Authors' contributions
HBB, YSW, LD and CW designed the study. HBB
performed the experiments and wrote the manuscript. HBB and MAT
analyzed and interpreted the data. HBB, MAT and AGAN revised the
paper. SS, NL and AGAN assisted with experimental protocols. YSW,
LD and CW supervised the study. All authors approved the final
version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The Second Hospital of Shandong University. All
experimental procedures were conducted in accordance with the
Declaration of Helsinki.
Patients consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ANTs
|
adjacent normal tissues
|
|
BLACAT2
|
bladder cancer-associated transcript
2
|
|
CCK-8
|
cell counting kit-8
|
|
DFS
|
disease free survival
|
|
GC
|
gastric cancer
|
|
HER2
|
human epidermal growth factor receptor
2
|
|
lncRNA
|
long non-coding RNA
|
|
LNM
|
lymph node metastasis
|
|
OS
|
overall survival
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
siRNA
|
small interfering RNA
|
|
UCC
|
upregulated in colorectal cancer
|
References
|
1
|
Xu MD, Wang Y, Weng W, Wei P, Qi P, Zhang
Q, Tan C, Ni SJ, Dong L, Yang Y, et al: A positive feedback loop of
lncRNA-PVT1 and FOXM1 facilitates gastric cancer growth and
invasion. Clin Cancer Res. 23:2071–2080. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhao Y, Liu Y, Lin L, Huang Q, He W, Zhang
S, Dong S, Wen Z, Rao J, Liao W and Shi M: The lncRNA MACC1-AS1
promotes gastric cancer cell metabolic plasticity via AMPK/Lin28
mediated mRNA stability of MACC1. Mol Cancer. 17:692018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang K, Shi H, Xi H, Wu X, Cui J, Gao Y,
Liang W, Hu C, Liu Y, Li J, et al: Genome-wide lncRNA microarray
profiling identifies novel circulating lncRNAs for detection of
gastric cancer. Theranostics. 7:213–227. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xia H, Chen Q, Chen Y, Ge X, Leng W, Tang
Q, Ren M, Chen L, Yuan D, Zhang Y, et al: The lncRNA MALAT1 is a
novel biomarker for gastric cancer metastasis. Oncotarget.
7:56209–56218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lv J, Qiu M, Xia W, Liu C, Xu Y, Wang J,
Leng X, Huang S, Zhu R, Zhao M, et al: High expression of long
non-coding RNA SBF2-AS1 promotes proliferation in non-small cell
lung cancer. J Exp Clin Cancer Res. 35:752016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Song X, Wang X, Hu J and Jiang L:
Silencing of LncRNA HULC enhances chemotherapy induced apoptosis in
human gastric cancer. J Med Biochem. 35:137–143. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gu Y, Chen T, Li G, Yu X, Lu Y, Wang H and
Teng L: LncRNAs: Emerging biomarkers in gastric cancer. Future
Oncol. 11:2427–2441. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu H, Hu Y, Liu X, Song W, Gong P, Zhang
K, Chen Z, Zhou M, Shen X, Qian Y and Fan H: LncRNA TRERNA1
function as an enhancer of SNAI1 promotes gastric cancer metastasis
by regulating epithelial-mesenchymal transition. Mol Ther Nucleic
Acids. 8:291–299. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lan Y, Xiao X, He Z, Luo Y, Wu C, Li L and
Song X: Long noncoding RNA OCC-1 suppresses cell growth through
destabilizing HuR protein in colorectal cancer. Nucleic Acids Res.
46:5809–5821. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fu X, Li H, Liu C, Hu B, Li T and Wang Y:
Long noncoding RNA AK126698 inhibits proliferation and migration of
non-small cell lung cancer cells by targeting Frizzled-8 and
suppressing Wnt/β-catenin signaling pathway. Onco Targets Ther.
9:3815–3827. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang S, Zhong G, He W, Yu H, Huang J and
Lin T: lncRNA Up-regulated in nonmuscle invasive bladder cancer
facilitates tumor growth and acts as a negative prognostic factor
of recurrence. J Urol. 196:1270–1278. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gooding AJ, Zhang B, Jahanbani FK, Gilmore
HL, Chang JC, Valadkhan S and Schiemann WP: The lncRNA BORG drives
breast cancer metastasis and disease recurrence. Sci Rep.
7:126982017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Prensner JR, Iyer MK, Sahu A, Asangani IA,
Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, et al:
The long noncoding RNA SChLAP1 promotes aggressive prostate cancer
and antagonizes the SWI/SNF complex. Nat Genet. 45:1392–1398. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mao Z, Li H, Du B, Cui K, Xing Y, Zhao X
and Zai S: LncRNA DANCR promotes migration and invasion through
suppression of lncRNA-LET in gastric cancer cells. Biosci Rep.
37:BSR201710702017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang FT, Chen WY, Gu ZQ, Zhuang YY, Li
CQ, Wang LY, Peng JF, Zhu Z, Luo X, Li YH, et al: The novel long
intergenic noncoding RNA UCC promotes colorectal cancer progression
by sponging miR-143. Cell Death Dis. 8:e27782017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao J, Du P, Cui P, Qin Y, Hu C, Wu J,
Zhou Z, Zhang W, Qin L and Huang G: LncRNA PVT1 promotes
angiogenesis via activating the STAT3/VEGFA axis in gastric cancer.
Oncogene. 37:4094–4109. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhan HX, Wang Y, Li C, Xu JW, Zhou B, Zhu
JK, Han HF, Wang L, Wang YS and Hu SY: LincRNA-ROR promotes
invasion, metastasis and tumor growth in pancreatic cancer through
activating ZEB1 pathway. Cancer Lett. 374:261–271. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi X, Wang X and Hua Y: LncRNA GACAT1
promotes gastric cancer cell growth, invasion and migration by
regulating MiR-149-mediated of ZBTB2 and SP1. J Cancer.
9:3715–3722. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang HM, Lu JH, Chen WY and Gu AQ:
Upregulated lncRNA-UCA1 contributes to progression of lung cancer
and is closely related to clinical diagnosis as a predictive
biomarker in plasma. Int J Clin Exp Med. 8:11824–11830.
2015.PubMed/NCBI
|
|
20
|
Wang YH, Ji J, Wang BC, Chen H, Yang ZH,
Wang K, Luo CL, Zhang WW, Wang FB and Zhang XL: Tumor-derived
exosomal long noncoding RNAs as promising diagnostic biomarkers for
prostate cancer. Cell Physiol Biochem. 46:532–545. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Elsayed ET, Salem PE, Darwish AM and Fayed
HM: Plasma long non-coding RNA HOTAIR as a potential biomarker for
gastric cancer. Int J Biol Markers. Apr 1–2018.(Epub ahead of
print). doi: 10.1177/1724600818760244. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu M, Wang Y, Liu X, Wen X, Liang C and
Tu J: LncRNAs act as prognostic biomarkers in gastric cancer: A
systematic review and meta-analysis. Front Lab Med. 1:59–68. 2017.
View Article : Google Scholar
|
|
23
|
Iranpour M, Soudyab M, Geranpayeh L,
Mirfakhraie R, Azargashb E, Movafagh A and Ghafouri-Fard S:
Expression analysis of four long noncoding RNAs in breast cancer.
Tumour Biol. 37:2933–2940. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao W, Luo J and Jiao S: Comprehensive
characterization of cancer subtype associated long non-coding RNAs
and their clinical implications. Sci Rep. 4:65912014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li J, Lian Y, Yan C, Cai Z, Ding J, Ma Z,
Peng P and Wang K: Long non-coding RNA FOXP4-AS1 is an unfavourable
prognostic factor and regulates proliferation and apoptosis in
colorectal cancer. Cell Prolif. 50:2017. View Article : Google Scholar
|
|
26
|
He W, Zhong G, Jiang N, Wang B, Fan X,
Chen C, Chen X, Huang J and Lin T: Long noncoding RNA BLACAT2
promotes bladder cancer-associated lymphangiogenesis and lymphatic
metastasis. J Clin Invest. 128:861–875. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang ZT, Li Z, Wang XG, Tan T, Yi F, Zhu
H, Zhao JP and Zhou XF: Overexpression of long non-coding RNA ZXF2
promotes lung adenocarcinoma progression through c-Myc pathway.
Cell Physiol Biochem. 35:2360–2370. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Thai P, Statt S, Chen CH, Liang E,
Campbell C and Wu R: Characterization of a novel long noncoding
RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer
cell lines. Am J Respir Cell Mol Biol. 49:204–211. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Beckedorff FC, Ayupe AC, Crocci-Souza R,
Amaral MS, Nakaya HI, Soltys DT, Menck CF, Reis EM and
Verjovski-Almeida S: The intronic long noncoding RNA ANRASSF1
recruits PRC2 to the RASSF1A promoter, reducing the expression of
RASSF1A and increasing cell proliferation. PLoS Genet.
9:e10037052013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Szász AM, Lánczky A, Nagy Á, Förster S,
Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A and Győrffy B:
Cross-validation of survival associated biomarkers in gastric
cancer using transcriptomic data of 1,065 patients. Oncotarget.
7:49322–49333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Győrffy B, Surowiak P, Budczies J and
Lánczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cooper MG: The development and causes of
cancer. The Cell: A Molecular Approach. 2nd. NCBI Bookshelf;
Sunderland, MA: 2000
|
|
37
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yao XM, Tang JH, Zhu H and Jing Y: High
expression of LncRNA CASC15 is a risk factor for gastric cancer
prognosis and promote the proliferation of gastric cancer. Eur Rev
Med Pharmacol Sci. 21:5661–5667. 2017.PubMed/NCBI
|
|
39
|
Uzan VR, Lengert Av, Boldrini É, Penna V,
Scapulatempo-Neto C, Scrideli CA, Filho AP, Cavalcante CE, de
Oliveira CZ, Lopes LF and Vidal DO: High expression of HULC is
associated with poor prognosis in osteosarcoma patients. PLoS One.
11:e01567742016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dorak MT and Karpuzoglu E: Gender
differences in cancer susceptibility: An inadequately addressed
issue. Front Genet. 3:2682012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Deng JY and Liang H: Clinical significance
of lymph node metastasis in gastric cancer. World J Gastroenterol.
20:3967–3975. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cho JK, Hyun SH, Choi N, Kim MJ, Padera
TP, Choi JY and Jeong HS: Significance of lymph node metastasis in
cancer dissemination of head and neck cancer. Transl Oncol.
8:119–125. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cuschieri A, Joypaul V, Fayers P, Cook P,
Fielding J and Craven J: Postoperative morbidity and mortality
after D1 and D2 resections for gastric cancer: Preliminary results
of the MRC randomised controlled surgical trial. Lancet.
347:995–999. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shitara K, Yatabe Y, Matsuo K, Sugano M,
Kondo C, Takahari D, Ura T, Tajika M, Ito S and Muro K: Prognosis
of patients with advanced gastric cancer by HER2 status and
trastuzumab treatment. Gastric Cancer. 16:261–267. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhou Q, Hou CN, Yang HJ, He Z and Zuo MZ:
Distinct expression and prognostic value of members of the
epidermal growth factor receptor family in ovarian cancer. Cancer
Manag Res. 10:6937–6948. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gao F, Zhang X, Whang S and Zheng C:
Prognostic impact of plasma ORM2 levels in patients with stage II
colorectal cancer. Ann Clin Lab Sci. 44:388–393. 2014.PubMed/NCBI
|
|
47
|
Li P, Zhang X, Wang H, Wang L, Liu T, Du
L, Yang Y and Wang C: MALAT1 is associated with poor response to
oxaliplatin-based chemotherapy in colorectal cancer patients and
promotes chemoresistance through EZH2. Mol Cancer Ther. 16:739–751.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Du L, Duan W, Jiang X, Zhao L, Li J, Wang
R, Yan S, Xie Y, Yan K, Wang Q, et al: Cell-free lncRNA expression
signatures in urine serve as novel non-invasive biomarkers for
diagnosis and recurrence prediction of bladder cancer. J Cell Mol
Med. 22:2838–2845. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li C, Li W, Zhang Y, Zhang X, Liu T, Zhang
Y, Yang Y, Wang L, Pan H, Ji J and Wang C: Increased expression of
antisense lncRNA SPINT1-AS1 predicts a poor prognosis in colorectal
cancer and is negatively correlated with its sense transcript. Onco
Targets Ther. 11:3969–3978. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Terai G, Iwakiri J, Kameda T, Hamada M and
Asai K: Comprehensive prediction of lncRNA-RNA interactions in
human transcriptome. BMC Genomics. 17 (Suppl 1):S122016. View Article : Google Scholar
|
|
51
|
Lei F, Zhang H and Xie X: Comprehensive
analysis of an lncRNA-miRNA-mRNA competing endogenous RNA network
in pulpitis. PeerJ. 7:e71352019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Tang XJ, Wang W and Hann SS: Interactions
among lncRNAs, miRNAs and mRNA in colorectal cancer. Biochimie.
163:58–72. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Thomas LR, Wang Q, Grieb BC, Phan J,
Foshage AM, Sun Q, Olejniczak ET, Clark T, Dey S, Lorey S, et al:
Interaction with WDR5 promotes target gene recognition and
tumorigenesis by MYC. Mol Cell. 440–452. 2015. View Article : Google Scholar : PubMed/NCBI
|