Introduction
Colorectal cancer (CRC) is one of the most common
malignant tumors and is the fourth leading cause of cancer-related
deaths worldwide (1). While the
5-year survival rate of patients with localized CRC is 87–91%,
prognosis for patients in distant stages is markedly poor, with
5-year-survival rate of 10–14% (2).
Prognosis for patients with advanced stages of CRC has recently
been improved by newly-developed molecular-targeted drugs and
anticancer agents (3). However,
satisfactory outcomes have not yet been achieved in patients with
recurrent CRC. Novel therapeutic strategies targeting alternative
mechanisms of cancer proliferation and metastasis are urgently
needed to overcome advanced stages of CRC. For this, we need to
explore the molecular mechanisms driving the invasive front of CRC,
which likely plays a central role in the invasiveness of cancer
cells in advanced stages of cancer.
In our previous study, we have shown that expression
of C-X-C motif chemokine ligand (CXCL)9, 10 and 11, combined with
other factors related to epithelial mesenchymal transition (EMT),
is elevated in the invasive front of CRC tumor tissues (4). CXCL9-11 participate in numerous
biological processes including leukocyte trafficking, immune
response, and cellular proliferation. CXCL10 is particularly known
to promote the proliferation of several carcinoma cell lines
(5–7). Clinical studies have shown an
association between expression levels of CXCL10 and survival of
patients with colorectal cancer (8–11).
CXCL9-11 exert their biological effects via common receptor called
C-X-C motif chemokine receptor (CXCR)3. CXCR3 is a member of the G
protein-coupled receptor family, and is expressed in monocytes and
leukocytes (12). Previous studies
have reported that CXCR3 is expressed in CRC (9). CXCR3 is also involved in the
proliferation and metastasis of renal, melanoma, and breast cancer
cells. Three splice variants of CXCR3 (CXCR3A, CXCR3B, and
CXCR3-alt) have been identified in humans (13,14);
these splice variants play different roles in various cancer cells.
CXCR3A promotes the invasiveness and metastasis of gastric- and
renal-cancer cells (15,16), while CXCR3B inhibits the invasiveness
and migration of prostate-cancer cells (17). The function of CXCR3-alt remains
unknown, and functional diversity of CXCR3A and CXCR3B in CRC is
also undetermined.
In this study, we examined the diverse roles of
CXCR3A, CXCR3B, and the CXCL10-CXCR3 signaling pathway in the
proliferation and invasiveness of CRC cells in vitro.
Materials and methods
Cell culture
Human CRC cell line HCT116 was obtained from
American Tissue and Cell Culture. HCT116 cells were cultured in
Dulbecco's modified Eagles medium (DMEM; Gibco; Invitrogen; Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
(FBS) (Gibco; Thermo Fisher Scientific, Inc.) and
antibiotic-antimycotic solution (100X; Gibco) at 37°C and 5%
CO2.
Transfection of CXCR3
For transfection of CXCR3, HCT116 cells were seeded
into a 6-well plate at a density of 3×105 cells/well.
After 16 h, the cells were transfected with 500 ng CXCR3
(Myc-DDK-tagged) transcript variant A plasmid DNA or CXCR3
(Myc-DDK-tagged) transcript variant B plasmid DNA (both from
Origene Technologies) using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) per manufacturer's protocol. After 48 h of
transfection, cells were cultured in DME medium supplemented with
600 µg/ml G418 for 1 week. These cells were designated as
HCT116-CXCR3A and HCT116-CXCR3B, respectively. Similarly, HCT116
cells were transfected with pCMV6 mammalian vector, designated as
mock cells, and used as controls. Non-transfected cells, designated
as blank cells, were used as additional controls.
Sequence analysis of inserted
genome
After HCT116 cells were transfected with
CXCR3A or CXCR3B plasmid DNA, insertion was confirmed
by DNA sequencing. DNA was extracted from each cell line via QIAamp
DNA Mini Prep Kit (Qiagen) and labeled using Big Dye Terminator
v3.1 Cycle Sequencing kit (Thermo Fisher Scientific, Inc.); this
was followed by direct sequencing on an ABI Prism 3730×l Analyzer
(Thermo Fisher Scientific). All kits were used per instructions of
respective manufacturers.
Western blot analysis
To examine the expression of total CXCR3 in
transfected cells, we performed western blotting using antibodies
that recognize both CXCR3A and 3B. HCT116-CXCR3A, HCT116-CXCR3B,
mock, and blank cells were lysed in RIPA Lysis and Extraction
Buffer (Thermo Fisher Scientific, Inc.) and then centrifuged at
23,000 g for 15 min at 4°C. Supernatants were collected, and
protein concentration in the supernatants was assessed using Qubit
2.0 Fluorometer (Thermo Fisher Scientific, Inc.). Lysates (30 µg
total protein per lysate) were subjected to 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and proteins
were then transferred to 0.2 µm polyvinylidene difluoride (PVDF)
membranes (EMD Millipore,). The membranes were blocked in
Tris-buffered saline containing Tween-20 (TBST) and 5% skim milk at
25°C for 1 h. Subsequently, the membranes were incubated with
rabbit anti-human antibodies against CXCR3 (cat. ab154845, 1:1,500;
Abcam) overnight at 4°C, and then with horseradish
peroxidase-labeled goat anti-rabbit secondary antibody at 25°C for
1 h. The membranes were then washed thrice (15 min per wash) using
TBST. Signals were enhanced using the ECL Plus Western Blotting
System (Perkin-Elmer) and detected via LAS-4000 Luminescent Image
Analyzer. Anti-β-actin (cat. no. 8H10D10, 1:1,500; Cell Signaling
Technology) was used as loading control.
RNA preparation
Total RNA was extracted from each cell line using
RNA Mini Prep Kit (Qiagen) according to the manufacturer's
instructions. The concentration and purity of extracted RNA were
evaluated using NanoDrop (Thermo Fisher Scientific, Inc.). To
assess the quality of extracted RNA, RNA integrity number (RIN) was
calculated using Agilent 2100 Bioanalyzer (Agilent
Technologies).
Reverse transcription (RT)-PCR
RT-PCR was performed to examine the expression of
CXCR3A and CXCR3B mRNA using specific primers sets.
One microgram purified total RNA was used for cDNA synthesis via
Affinity Script QPCR cDNA Synthesis Kit (Agilent Technologies)
utilized per manufacturer's instructions. RT-PCR was performed
using Takara Ex Taq (Takara Bio, Inc.) using the following primers,
designed specifically for amplification of CXCR3A or
CXCR3B mRNA: CXCR3A forward, 5′-ccatggtccttgaggtgag-3′ and
reverse, 5′-tccatagtcataggaagagctgaa-3′; CXCR3B forward,
5′-ttgaggaagtacggccctg-3′ and reverse, 5′-tgagcagctcctcctataac-3′;
GAPDH forward, 5′-agccacatcgctcagacac-3′ and reverse,
5′-gcccaatacgaccaaatcc-3′.
Conditions for PCR were as follows: 95°C for 5 min,
and then 25 cycles at 95°C for 10 sec, 58°C for 30 sec, and 72°C
for 30 sec. Expression level of GAPDH mRNA was used as
internal control.
MTT assay
MTT
(3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide)
assay was used to examine the involvement of CXCL10/CXCR3 signaling
in the proliferation of CRC cells. While CXCR3 has three ligands
(CXCL9, 10 and 11), CXCL10 was most frequently examined in previous
studies. Furthermore, the level of CXCL10 expression is associated
with poor prognosis in patients with CRC. Therefore, CXCL10 was
used in our present study as a ligand for CXCR3.
In a preliminary study, we used qPCR to examine the
expression of CXCL10 mRNA in HCT116 cells; however, the
expression level of CXCL10 was below the detection
threshold. Therefore, we cultured the cell lines in the presence of
CXCL10 to enhance the activity of CXCR3A/3B. The four types of
cells were separately seeded into 24-well plate at a density of
5×104 cells/well. After 16 h, the cells were cultured
with or without CXCL10 (100 ng/ml) for 24 h, and MTT was added for
final 3 h before measuring. Prior to conducting measurements, the
media were removed and isopropanol was added to all the wells.
Absorbance was measured using a microplate reader (Hitachi
SH-9000Lab; Hitachi) at 570 nm. All experiments were repeated three
times.
Scratch wound healing assay
In order to examine CXCL10/CXCR3 signaling at the
invasive front of tumor tissue, we performed a scratch wound
healing assay as described previously with modifications (16). Each cell line was seeded into a 6-cm
dish at a density of 8×105 cells/well. After 36 h, the
cell sheets were scratched with a pipette tip (1,000-µl long filter
tip; Watson) and then treated with CXCL10 (100 ng/ml) for 24 h.
After 0 and 24 h, three different areas of the scratched cell
sheets were observed under microscopy. Wounded areas (the areas
with no attached cells) were measured using ImageJ software, and
rate of migration was calculated as a proportion of the areas at
24/0 h. All experiments were repeated three times.
Transwell assay
The role of CXCL10/CXCR3 signaling in cellular
invasiveness was examined using a Transwell assay performed via
CytoSelect 24-Well Cell Invasion Assay Kit (Cell Biolabs). A Boyden
chamber was used for the assay. Cells from each of the four cell
lines were added to the upper chamber and incubated overnight. The
medium in the upper chamber was then changed to Opti-minimum
essential media, and CXCL10 (100 ng/ml) was added to the medium
contained in the lower chamber. After 24 h, cells attached to the
lower surface of the membrane were stained with Cell Staining
Solution supplied with the CytoSelect 24-Well Cell Invasion Assay
Kit. Stained cells were lysed in elution buffer, and each lysate
was placed into 96-well plate at 200 µl/well. Absorbance of the
lysates was measured using a microplate reader (Hitachi SH-9000Lab)
at 560 nm. This assay was repeated six times.
Expression of MMP9 mRNA
CXCR3 potentiates the invasiveness and migration of
various cancer cells by upregulating the expression of MMP9, a
protein downstream of the ERK1/2 pathway via CXCL10-CXCR3A
signaling. In order to confirm the effect of CXCR3A on cancer cell
invasiveness and migration, we analyzed the expression of
MMP9 in HCT116-CXCR3A cells. Total RNA was extracted from
HCT116-CXCR3A cells and controls (blank and mock cells) and used to
synthesize cDNA via Affinity Script QPCR cDNA Synthesis Kit.
GAPDH was used as the housekeeping gene. Detection was
performed using LAS-4000 (GE Healthcare), and band intensity was
calculated using ImageQuant TL software (GE Healthcare). These
experiments were repeated three times.
Statistical analysis
All data were expressed as mean ± standard deviation
of the mean. Kruskal-Wallis test was used to evaluate differences
among the groups, and Bonferroni post hoc test was used for
multiple comparisons to evaluate the differences between two
groups. All statistical analyses were performed using SPSS version
2.0 (IBM, Corp.). P<0.05 was considered statistically
significant.
Results
Expression of CXCR3 in transfected
HCT116 cells
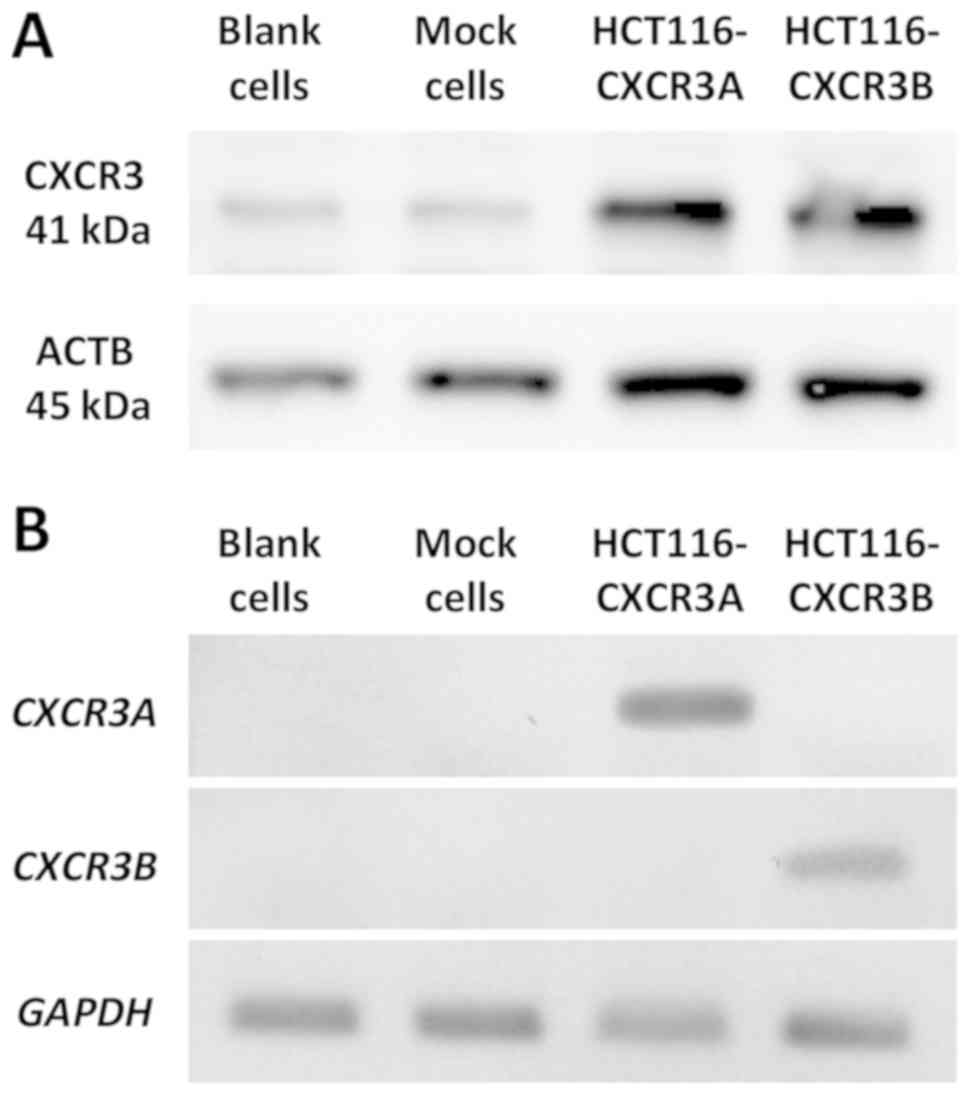
Using DNA sequencing, we confirmed that the plasmid
had been successfully inserted into each cell line (data not
shown). CXCR3B Expression of CXCR3A or CXCR3B in transfected cells
was then confirmed via western blotting and RT-PCR. Total CXCR3
expression at the protein level was markedly higher in
HCT116-CXCR3A and CXCR3B cells than in blank and mock cells
(Fig. 1A). RT-PCR was used to
examine gene expression of each isoform. Our results show that
CXCR3A mRNA was only expressed in HCT116-CXCR3A cells, but
not in blank, mock, or HCT116-CXCR3B cells (Fig. 1B). Similarly, CXCR3B mRNA was
only expressed in HCT116-CXCR3B cells. These four cell lines were
used in subsequent experiments.
Proliferation of CXCR3-transfected
cells
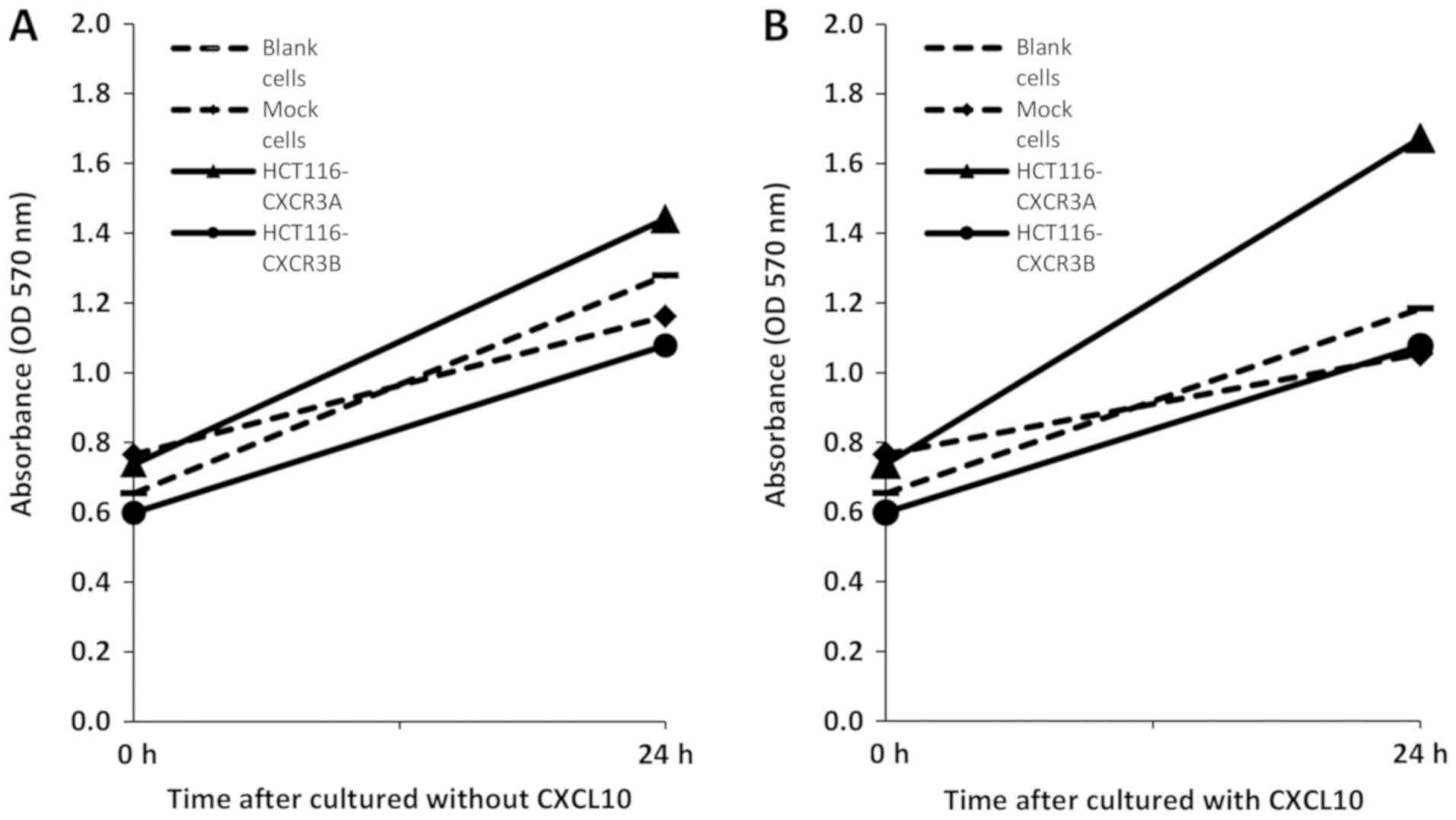
We first used an MTT assay to examine whether
overexpression of CXCR3A or CXCR3B can promote proliferation of
human colorectal cancer cells. After 24 h without CXCL10 treatment,
HCT116-CXCR3A cell proliferation was moderately enhanced by
approximately 1.1-fold compared to the proliferation of blank and
mock cells, whereas HCT116-CXCR3B cell proliferation was reduced by
approximately 20% compared to that in blank and mock control cells
(Fig. 2A). The proliferation of
HCT116-CXCR3A cells cultured in media containing CXCL10 for 24 h
was enhanced by approximately 1.5-fold compared with that of blank
cells and mock cells, and the difference from mock cells narrowly
missed the set threshold for statistical significance (P=0.076).
Conversely, the proliferation of HCT116-CXCR3B cells was similar to
that of blank and mock cells (Fig.
2B).
Scratch wound healing assay
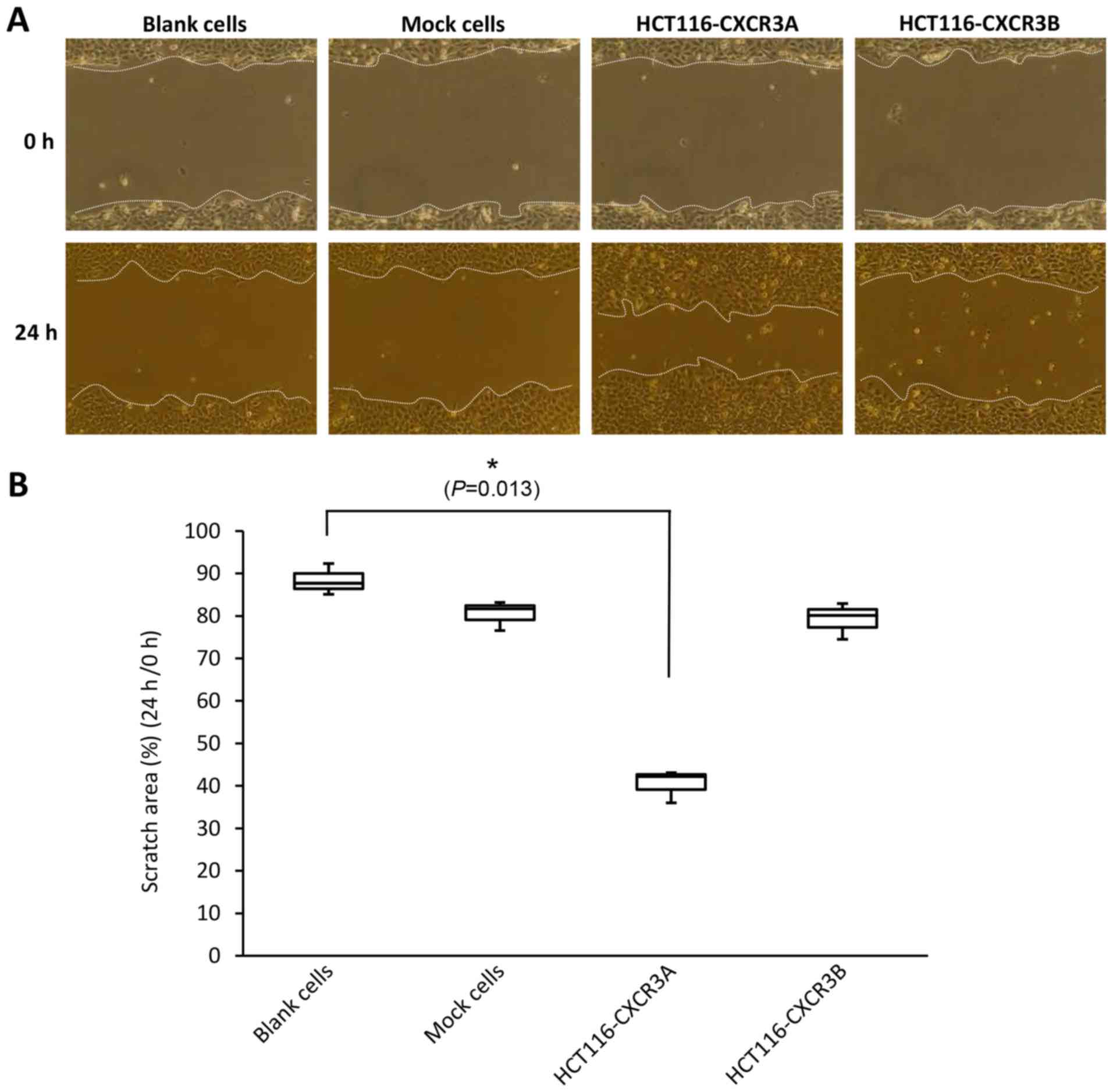
A scratch wound healing assay is used to assess
cellular invasiveness and proliferative ability by assessing the
state of the scratched areas 24 h after induction of the wound.
Scratched areas in the blank and mock cell sheets, assessed at 24 h
after scratching, were reduced to 88 and 80%, respectively,
compared with those assessed at 0 h. The scratched area of the
HCT116-CXCR3A cell sheet, assessed at 24 h, was reduced to 40% of
that assessed at 0 h; this reduction in the wounded area was
significantly greater than those shown by blank and mock cell
sheets, and the difference from blank cells was statistically
significant (P=0.013). The scratched area on the HCT116-CXCR3B cell
sheet was similar to that of blank and mock cells (Fig. 3).
Transwell assay
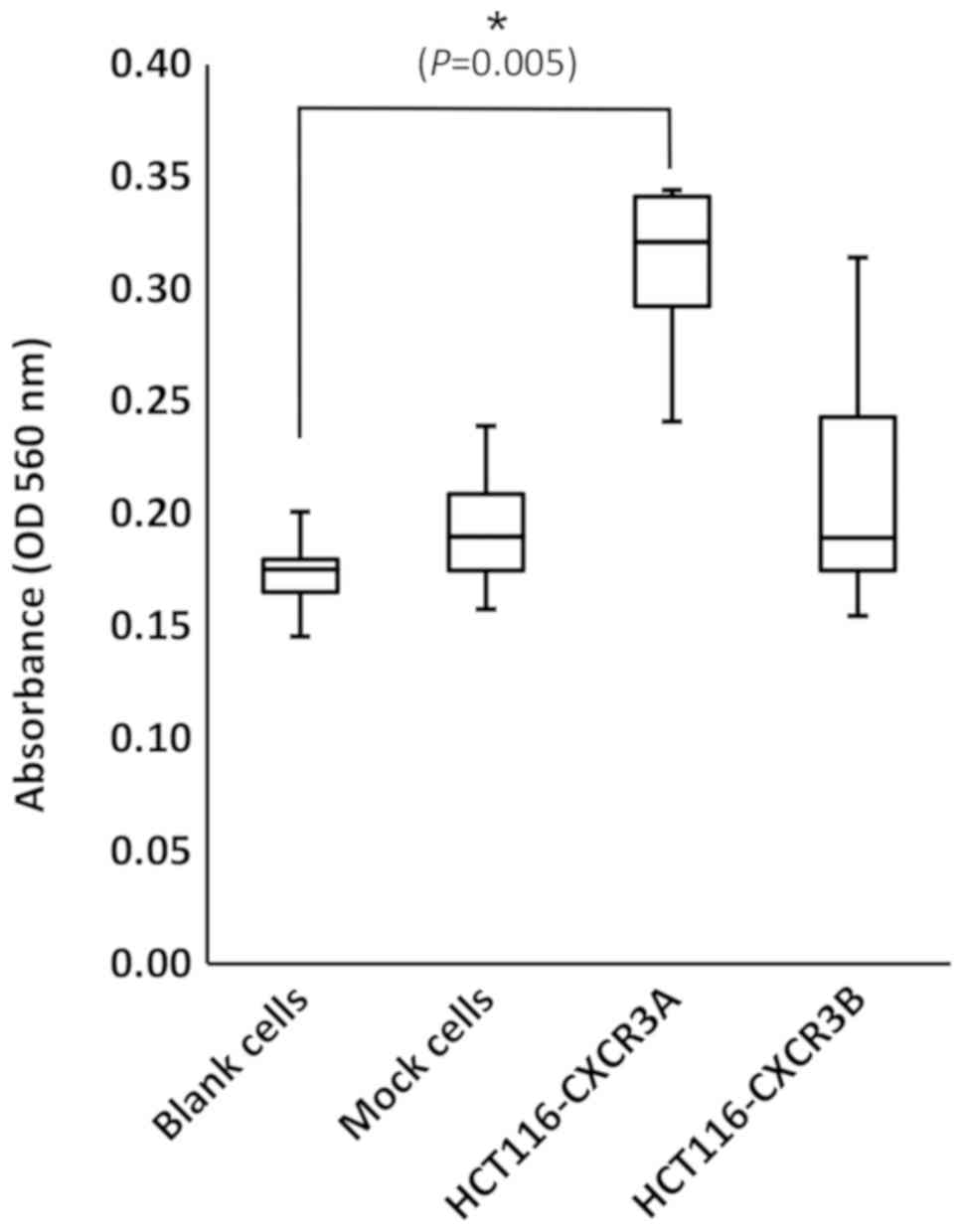
In the Transwell assay, the number of cells attached
to the lower surface of the membrane reflects the invasiveness of
the cells. The number of invasive HCT116-CXCR3A cells was increased
approximately 2-fold compared with those of blank and mock cells,
and the difference from blank cells was statistically significant
(P=0.005) (Fig. 4). In contrast, the
number of invasive HCT116-CXCR3B cells was similar to those of
blank and mock cells.
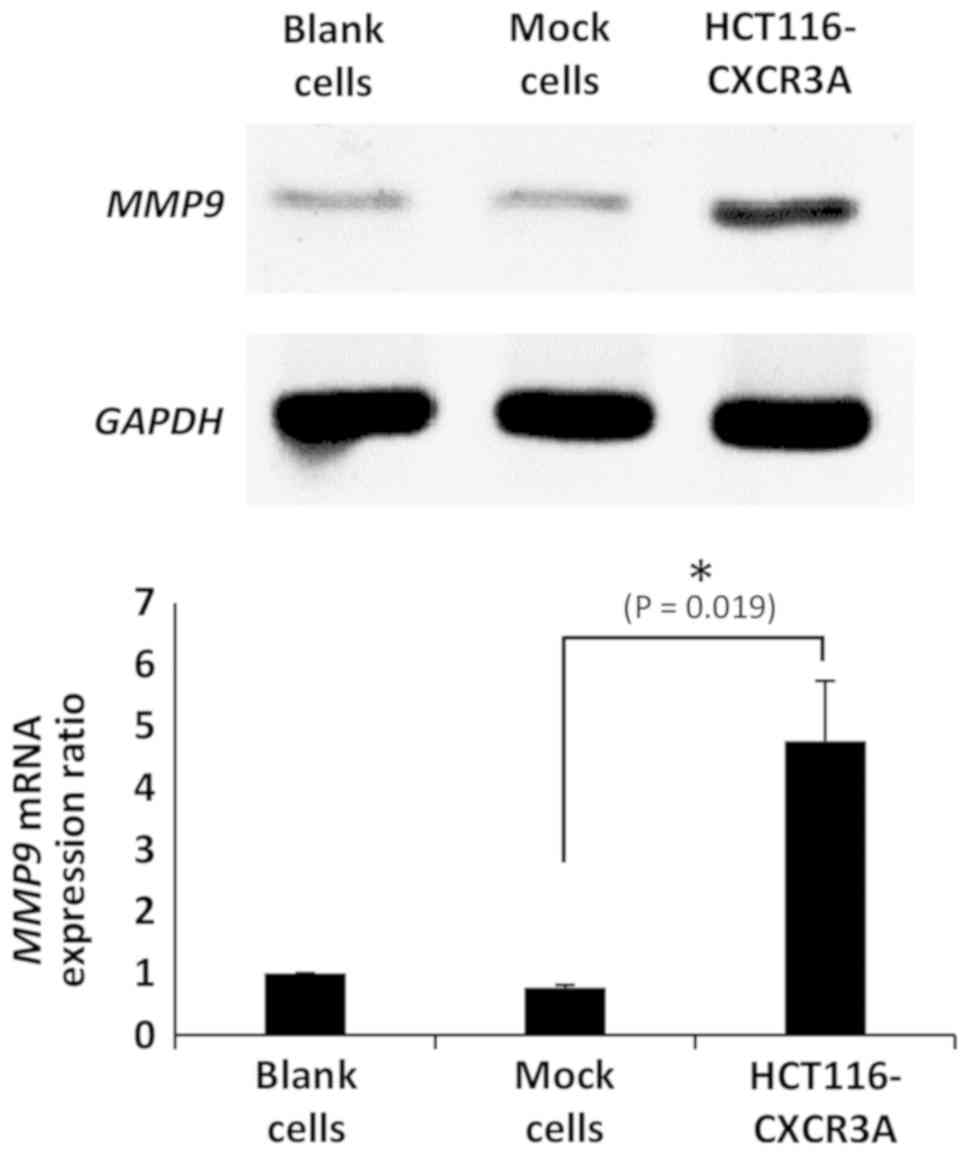
Expression of MMP9
To examine the status of CXCR3A downstream
molecules, we investigated mRNA expression of MMP9, which
was reported to be involved in ERK1/2 signaling in human CRC cell
lines. MMP9 mRNA expression in HCT116-CXCR3A cells was
4-fold higher than that in blank or mock cells, and the difference
from mock cells was statistically significant (P=0.019) (Fig. 5).
Discussion
In our previous study, we demonstrated that mRNA
expression of CXCL9, 10 and 11 was upregulated at the invasive
front of CRC tissue (4), suggesting
that these chemokines play crucial roles in tumor development
during CRC. Receptors, expressed by cancer cells, can determine how
these cells are affected by chemokines. The role of one such
receptor, CXCR3, has not been fully investigated in CRC
development, particularly with respect to differential properties
of CXCR3 subtypes. Therefore, in this study, we examined the
function of two CXCR3 variants (CXCR3A and CXCR3B) in CRC
cells.
CXCR3 is involved in the proliferation and
metastasis of renal, melanoma, and breast cancer cells. CXCR3
expression, which is upregulated in cancer tissues and metastatic
carcinoma compared with that in normal tissues, is correlated with
poor prognosis in patients with renal, melanoma, gastric, and
breast cancer (18–21). Molecular analysis has shown that
CXCR3 promotes tumor progression and metastasis by regulating
proliferative, migratory, and invasive abilities in these cancer
cells (6,16,20,22). Ma
et al has shown that in a murine model of metastatic breast
cancer, small-molecular-weight antagonists or siRNAs inhibit
metastatic spread into lungs; this anti-metastatic effect, however,
is compromised by depletion of natural killer cells in these mice
(21,23).
CXCR3A and CXCR3B play different roles in the growth
and invasiveness of several types of cancer cells. Wu et al
has shown that overexpression of CXCR3A promotes proliferation and
invasiveness, while overexpression of CXCR3B ameliorates
proliferation and invasiveness, of prostate cancer cells (17). Shen and Cao used siRNA interference
to show that proliferative and invasive abilities of prostate
cancer cells are reduced by suppressing the expression of CXCR3A
and enhanced by suppressing the expression of CXCR3B (24).
In our present study, we have shown that
overexpression of CXCR3A promoted the proliferation of HCT116
colorectal cancer cells. In particular, the proliferation of HCT116
cells exposed to CXCL10 was enhanced compared with that of
untreated control HCT116-CXCR3A cells. This indicates that
CXCL10-CXCR3A signaling plays an important role in the
proliferative ability of CRC cells. Conversely, overexpression of
CXCR3B had no effect on the proliferation of HCT116 cells. These
results suggest that CXCR3A, but not CXCR3B, was responsible for
CXCL10-mediated enhancement of proliferation in CRC cells. Scratch
wound healing assays showed that recovery of the wounded area was
promoted in HCT116-CXCR3A cells compared with that in
non-transfected cells. Transwell assays showed that the number of
invasive HCT116-CXCR3A cells was increased compared to those of
blank cells. These results indicate that overexpression of CXCR3A
contributed to the proliferation and invasiveness of HCT116 cells.
These results also agree with those obtained in previous studies
analyzing carcinomas, suggesting that CXCR3A, in conjunction with
CXCL10, plays crucial roles in the proliferation and invasiveness
of colorectal cancer cells. Conversely, previous studies indicate
that CXCR3B overexpression did not affect cellular invasiveness.
Whereas these previous studies indicate that CXCR3B inhibits the
migration of cancer cells, we did not observe this effect of CXCR3B
on CRC cells in our present study. This discrepancy may stem from
differences in the tumor microenvironment of different types of
cancer.
Previous studies have shown that CXCR3A induces
cancer cell proliferation and invasiveness by activating the ERK1/2
and PI3K pathway via Gαi or Gαq (15,25,26). In
addition, CXCR3A promotes cancer cell invasiveness and migration by
activating Gαq. CXCR3B activates Gαs by promoting enhanced cAMP
concentrations; this cascade inhibits the migratory ability of
cancer cells (17). Shen and Cao
reported that MMP activity is involved in CXCR3A signaling
downstream of PI3K in prostate cancer (24). Zipin-Roitman et al reported
that CXCL10 promotes the invasiveness of human colorectal cancer
cells by upregulating MMP9 expression (27). In our present study, we have shown
that CRC cells transfected with CXCR3A showed enhanced expression
of MMP9. This finding agrees with results obtained in
previous studies, showing that MMPs play crucial roles in promoting
the invasiveness and proliferation of cancer cells via CXCR3A.
Our present study has several limitations. In this
study, we used only CXCL10 as a ligand of CXCR3 for analysis, and
did not examine the functions of CXCR3 using CXCL9 and CXCL11.
These ligands may act differently on CXCR3 than did CXCL10.
Additionally, we used only the HCT116 line of CRC cells for
functional analysis. Therefore, our results may have been affected
by the individual characteristics of this cell line. Future studies
will need to determine the functions of CXCR3 using other ligands
and different CRC lines. Furthermore, among the molecules involved
in the CXCL10-CXCR signaling pathway in CXCR3A-transfected cells,
we only examined the expression of MMP9 mRNA. In our future
studies, we will examine the expression of other proteins, such as
Gαi and ERK1/2 involved in CXCR3-mediated proliferation and
invasiveness of CRC cells. Phosphorylation of ERK1/2 should be also
investigated to clarify the effect of ERK1/2-MMP9 signals. In
addition, it would be interesting to examine the roles of CXCR3
proteins by increasing or decreasing their expression. Actually, we
attempted to perform an siRNA-inhibition experiment with the
transfected cells; however, the results we obtained were highly
variable, probably due to insufficient effect of the synthesized
siRNAs. Future studies should include such experiments with a
modified experimental design.
In conclusion, our results indicate that
CXCL10-induced proliferation and invasiveness of colorectal cancer
cells was likely mediated by CXCR3A, but not by CXCR3B. Further
analysis of proteins will provide additional insights into the
molecular mechanisms involved in CRC development via CXCR3 and CXCL
pathways.
Acknowledgements
The authors would like to thank Dr Kanae Karita from
the Department of Public Health, Kyorin University School of
Medicine, for her assistance in statistical analysis.
Funding
The present work was supported by the Japan Society
for the Promotion of Science (grant no. 21791305).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
EN, TW and MS made substantial contributions to
conception and design. EN, TK and TM made substantial contributions
to acquisition of data. EN, TK, HO and KO made substantial
contributions to analysis and interpretation of data. In addition,
all authors have been involved in drafting the manuscript or
revising it critically for intellectual content, given final
approval of version to be published, and agreed to be accountable
for all aspects of the work in ensuring that questions related to
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer CXC
|
|
C-X-C motif chemokine ligand CXCR
|
C-X-C motif chemokine receptor
|
|
siRNA
|
small interfering RNA
|
|
MMP
|
matrix metalloproteinase
|
References
|
1
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kopetz S, Chang GJ, Overman MJ, Eng C,
Sargent DJ, Larson DW, Grothey A, Vauthey JN, Nagorney DM and
McWilliams RR: Improved survival in metastatic colorectal cancer is
associated with adoption of hepatic resection and improved
chemotherapy. J Clin Oncol. 27:3677–3683. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kobayashi T, Masaki T, Nozaki E, Sugiyama
M, Nagashima F, Furuse J, Onishi H, Watanabe T and Ohkura Y:
Microarray analysis of gene expression at the tumor front of colon
cancer. Anticancer Res. 35:6577–6581. 2015.PubMed/NCBI
|
|
5
|
Datta D, Flaxenburg JA, Laxmanan S, Geehan
C, Grimm M, Waaga-Gasser AM, Briscoe DM and Pal S: Ras-induced
modulation of CXCL10 and its receptor splice variant CXCR3-B in
MDA-MB-435 and MCF-7 cells: Relevance for the development of human
breast cancer. Cancer Res. 66:9509–9518. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lasagni L, Francalanci M, Annunziato F,
Lazzeri E, Giannini S, Cosmi L, Sagrinati C, Mazzinghi B, Orlando
C, Maggi E, et al: An alternatively spliced variant of CXCR3
mediates the inhibition of endothelial cell growth induced by
IP-10, Mig, and I-TAC, and acts as functional receptor for platelet
factor 4. J Exp Med. 197:1537–1549. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bodnar RJ, Yates CC and Wells A: IP-10
blocks vascular endothelial growth factor-induced endothelial cell
motility and tube formation via inhibition of calpain. Circ Res.
98:617–625. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wightman SC, Uppal A, Pitroda SP, Ganai S,
Burnette B, Stack M, Oshima G, Khan S, Huang X, Posner MC, et al:
Oncogenic CXCL10 signalling drives metastasis development and poor
clinical outcome. Br J Cancer. 113:327–335. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bai M, Chen X and Ba YI: CXCL10/CXCR3
overexpression as a biomarker of poor prognosis in patients with
stage II colorectal cancer. Mol Clin Oncol. 4:23–30. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Toiyama Y, Fujikawa H, Kawamura M,
Matsushita K, Saigusa S, Tanaka K, Inoue Y, Uchida K, Mohri Y and
Kusunoki M: Evaluation of CXCL10 as a novel serum marker for
predicting liver metastasis and prognosis in colorectal cancer. Int
J Oncol. 40:560–566. 2012.PubMed/NCBI
|
|
11
|
Jiang Z, Xu Y and Cai S: CXCL10 expression
and prognostic significance in stage II and III colorectal cancer.
Mol Biol Rep. 37:3029–3036. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Groom JR and Luster AD: CXCR3 ligands:
Redundant, collaborative and antagonistic functions. Immunol Cell
Biol. 89:207–215. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Billottet C, Quemener C and Bikfalvi A:
CXCR3, a double-edged sword in tumor progression and angiogenesis.
Biochim Biophys Acta. 1836:287–295. 2013.PubMed/NCBI
|
|
14
|
Ehlert JE, Addison CA, Burdick MD, Kunkel
SL and Strieter RM: Identification and partial characterization of
a variant of human CXCR3 generated by posttranscriptional exon
skipping. J Immunol. 173:6234–6240. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang C, Zheng W and Du W: CXCR3A
contributes to the invasion and metastasis of gastric cancer cells.
Oncol Rep. 36:1686–1692. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Utsumi T, Suyama T, Imamura Y, Fuse M,
Sakamoto S, Nihei N, Ueda T, Suzuki H, Seki N and Ichikawa T: The
association of CXCR3 and renal cell carcinoma metastasis. J Urol.
192:567–574. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu Q, Dhir R and Wells A: Altered CXCR3
isoform expression regulates prostate cancer cell migration and
invasion. Mol Cancer. 11:32012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jöhrer K, Zelle-Rieser C, Perathoner A,
Moser P, Hager M, Ramoner R, Gander H, Höltl L, Bartsch G, Greil R
and Thurnher M: Up-regulation of functional chemokine receptor CCR3
in human renal cell carcinoma. Clin Cancer Res. 11:2459–2465. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jacquelot N, Enot DP, Flament C, Vimond V,
Blattner C, Pitt JM, Yamazaki T, Roberti MP, Daillère R, Vétizou M,
et al: Chemokine receptor patterns in lymphocytes mirror metastatic
spreading in melanoma. J Clin Invest. 126:921–97. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou H, Wu J, Wang T, Zhang X and Liu D:
CXCL10/CXCR3 axis promotes the invasion of gastric cancer via
PI3K/AKT pathway-dependent MMPs production. Biomed Pharmacother.
82:479–488. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ma X, Norsworthy K, Kundu N, Rodgers WH,
Gimotty PA, Goloubeva O, Lipsky M, Li Y, Holt D and Fulton A: CXCR3
expression is associated with poor survival in breast cancer and
promotes metastasis in a murine model. Mol Cancer Ther. 8:490–498.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kawada K, Sonoshita M, Sakashita H,
Takabayashi A, Yamaoka Y, Manabe T, Inaba K, Minato N, Oshima M and
Taketo MM: Pivotal role of CXCR3 in melanoma cell metastasis to
lymph nodes. Cancer Res. 64:4010–4017. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Walser TC, Rifat S, Ma X, Kundu N, Ward C,
Goloubeva O, Johnson MG, Medina JC, Collins TL and Fulton AM:
Antagonism of CXCR3 inhibits lung metastasis in a murine model of
metastatic breast cancer. Cancer Res. 66:7701–7707. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shen D and Cao X: Potential role of CXCR3
in proliferation and invasion of prostate cancer cells. Int J Clin
Exp Pathol. 8:8091–8098. 2015.PubMed/NCBI
|
|
25
|
Aksoy MO, Yang Y, Ji R, Reddy PJ,
Shahabuddin S, Litvin J, Rogers TJ and Kelsen SG: CXCR3 surface
expression in human airway epithelial cells: Cell cycle dependence
and effect on cell proliferation. Am J Physiol Lung Cell Mol
Physiol. 290:L909–L918. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ji R, Lee CM, Gonzales LW, Yang Y, Aksoy
MO, Wang P, Brailoiu E, Dun N, Hurford MT and Kelsen SG: Human type
II pneumocyte chemotactic responses to CXCR3 activation are
mediated by splice variant A. Am J Physiol Lung Cell Mol Physiol.
294:L1187–L1196. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zipin-Roitman A, Meshel T, Sagi-Assif O,
Shalmon B, Avivi C, Pfeffer RM, Witz IP and Ben-Baruch A: CXCL10
promotes invasion-related properties in human colorectal carcinoma
cells. Cancer Res. 67:3396–3405. 2007. View Article : Google Scholar : PubMed/NCBI
|