Introduction
Hepatocellular carcinoma (HCC) was the fifth most
common malignancy and was the second-most frequent cause of
cancer-associated mortality worldwide in 2012 (1). Every year, ~780,000 new cases of HCC
are diagnosed and 745,000 mortalities are due to HCC tumor
progression (2). Despite the
advancements in therapeutic strategies, including improvements in
surgery and radio-chemotherapy regimens for patients with HCC, the
5-year survival rate remains low due to the high recurrence and
metastasis of the disease (3).
Therefore, identification of novel and effective therapies is of
prime importance.
Cancer cells exhibit reprogrammed metabolism, and
present aerobic glycolysis, which is a phenomenon known as the
Warburg effect (4,5). The Warburg effect is one of the major
metabolic changes in tumor cells, which is characterized by an
increased glucose uptake and lactate production in the presence of
oxygen (4,5). MicroRNAs (miRNAs) have previously been
demonstrated to be involved in energy metabolism (6), for example, the mineralocorticoid
receptor suppresses the progression of cancer and the Warburg
effect via modulation of the miRNA (miR)-338-3p/pyruvate kinase
axis in HCC (7). miR-199a maturation
is suppressed by RNA-binding protein human antigen R, which is
crucial for the hypoxia-induced glycolytic switch in HCC (8). STAT3-mediated activation of miR-23a
suppresses gluconeogenesis in HCC by downregulating
glucose-6-phosphatase and peroxisome proliferator-activated
receptor γ, coactivator 1 α (9).
However, the roles of miR-125 in HCC progression and glycolysis
remain unknown.
The present study suggested that miR-202 expression
was lower in HCC tissues, and lower miR-202 expression was
associated with poor prognosis in patients with HCC. In
vitro, miR-202 was found to suppress cell glucose uptake,
lactate production and cell proliferation. Furthermore, it was
identified that hexokinase 2 (HK2) was a target of miR-202. miR-202
inhibited cell proliferation and cells glycolysis by targeting HK2
in HCC. Therefore, these results suggested that miR-202 may serve
as a potential therapeutic target for treating HCC.
Materials and methods
Clinical tissue samples
A total of 56 patients with HCC including 43 males
and 13 females with a mean age of 55±8.17 years (range, 32–78
years) were recruited in to the present study. Paired fresh HCC
tissues and corresponding adjacent (2 cm) non-cancerous tissue
samples (the distance between the samples of the HCC tissues and
the adjacent non-cancerous tissues is 2 cm) were obtained from
patients with HCC undergoing hepatectomy at the Department of Liver
Disease, Affiliated Hospital of Shaoxing University (Shaoxing,
China) from March 2008 to February 2015. The tissue samples were
confirmed as HCC by histopathological examination. None of the
patients in the present study received chemotherapy or radiation
therapy prior to surgery. Following surgery, the sample was stored
at −80°C until further use. The present study was approved by the
Institutional Ethics Committee of Affiliated Hospital of Shaoxing
University. Written informed consent was obtained from all patients
that participated in the present study.
Cell line culture
Human liver cancer cells, including HepG2, 97-L,
Hep3B, Huh-7 and a hepatocellular cell line (THLE-3) were purchased
from the cell bank of type culture collection at the Chinese
Academy of Sciences (Shanghai, China). All cells were cultured in
DMEM (HyClone; GE Healthcare Life Sciences) and supplemented with
10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml
penicillin and 100 mg/ml streptomycin (Gibco; Thermo Fisher
Scientific, Inc.) at 37°C in a humidified incubator with 5%
CO2.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNA isolation from HCC tissues and liver
cancer cell lines was performed using TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.). The cDNA was reversed
transcribed using RNA with a reverse transcription kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) using the following
cnditions: 18°C for 30 min, 42°C for 30 min and 90°C for 5 min.
RT-qPCR was performed using an ABI 7500 real-time system with SYBR
Premix Ex Taq kit (Takara Biotechnology Co., Ltd.) according to the
manufacturer's protocol. The following conditions were used:
Initial denaturation at 95°C for 5 min, 30 cycles of denaturation
at 94°C for 30 sec, annealing at 56°C (58°C was used for GAPDH) for
30 sec, and extension at 72°C for 30 sec. Data were analyzed using
the 2−ΔΔCq method (10).
GAPDH was used as an internal control. The forward and reverse
sequences were as follows: GAPDH: forward,
5′-GACTCATGACCACAGTCCATGC-3′; reverse, 5′-AGAGGCAGGGATGATGTTCTG-3′;
HK2: forward: 5′-CCACAGGTCATCATAGTTCC-3′; reverse,
5′-GGCACCCAGCACAATGAAG-3′; miR-202: forward,
5′-CTCCAGAGAGAUAGUAGAGCCT-3′; reverse,
5′-CTCAACCAATCACCTGGCACAGA-3′; U6: forward,
5′-CTCGCTTCGGCAGCACA-3′; reverse, 5′-AACGCTTCACGAATTTGCGT-3′. GAPDH
was used as the endogenous control for normalization of the mRNA
quantification and U6 was used as the endogenous control for
normalization of the miRNA quantification.
Cell transfection
The miR-202 mimic (50 nM), miR-202 inhibitor (50 nM)
and miRNA negative control (NC; 50 nM) and small interfering
(si)-negative control (50 nM) or si-HK2 (50 nM) were designed by
and purchased from Shanghai GenePharma Co., Ltd. Cells were seeded
in 6-well plates until they reached 80–90% confluence and were then
transfected using Lipofectamine® 3,000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Cells were harvested 48 h after cell
transfection.
Cell proliferation assay
Cells were seeded into 96-well plates (3,000
cells/well) and then transfected with miR-202 mimic, miR-202
inhibitor or miR-NC. Following cell transfection at 0, 24, 48, 72
and 96 h, cell proliferation was detected using a Cell Counting Kit
8 (CCK8; Dojindo Molecular Technologies, Inc.) according to the
manufacturer's protocol. Briefly, cells were incubated with 10 µl
CCK8 reagents for 2 h at 37°C. The optical density value was
detected at 450 nm using an ELISA reader (Bio-Rad Laboratories,
Inc.).
Target prediction
The predicted binding sites between miR-202 and HK2
were obtained using Miranda online software 2010 (http://www.microrna.org/microrna/home.do; August 2010
release).
Glucose uptake assay
Cells were seeded in 6-well plates and transfected
with miR-202 mimic (50 nM), miR-202 inhibitor (50 nM) or miR-NC (50
nM). At 48 h, 97-L and Huh7 cells were cultured with serum-starved
and glucose-free medium (DMEM; HyClone; GE Healthcare Life
Sciences). Cells were then treated with 50 mM
2-[N-(7-nitrobenz-2-oxa-1,b 3-diazol-4-yl) amino]-2-deoxy-D-glucose
(Invitrogen; Thermo Fisher Scientific, Inc.) for 1 h at 37°C.
Glucose uptake was quantified using a flow cytometer (Pathway 855
Bioimaging system; BD Pathway™ 800 Series Software; BD
Bioscience).
Lactate production assay
Cells were seeded in 6-well plates and transfected
with miR-202 mimic, miR-202 inhibitor or miR-NC, as aforementioned.
At 48 h, culture medium was removed from cells and lactate
concentration was determined using lactate test strips and an
Accutrend Lactate analyzer (Accutrend Lactate; Roche Diagnostics).
Results were normalized to the cell number in the negative control
group.
Western blot analysis
Proteins were extracted using a RIPA lysis buffer
(Beyotime Institute of Biotechnology). The protein concentration
was determined using a bicinchoninic acid assay protein assay kit
(Pierce; Thermo Fisher Scientific, Inc.). A total of 50 µg
extracted protein samples were separated via SDS-PAGE on 10% gels
and transferred to a PVDF membrane (EMD Millipore). The membranes
were blocked in 5% skimmed milk at room temperature for 1 h and
incubated with primary antibodies anti-HK2 (1:1,000; cat. no.
sc-6521; Santa Cruz Biotechnology, Inc.) at 4°C overnight. The
membranes were then incubated with the corresponding horseradish
peroxidase-conjugated goat anti-rabbit IgG secondary antibodies
(1:1,000; cat. no. A0208; Beyotime Institute of Biotechnology) for
1 h at room temperature. The protein bands were detected using an
ECL kit (Beyotime Institute of Biotechnology).
Dual luciferase reporter assay
The 97L and Huh7 (10,000/well) cells were incubated
in 24-well plates for the dual-luciferase reporter assay (Promega
Corporation) at 37°C. The wild- or mutant-type of 3′-untranslated
region (3′-UTR) of HK2 was inserted into the psiCHECK2 vector
(Invitrogen; Thermo Fisher Scientific, Inc.). Then, wild- or
mutant-type 3-UTR HK2 and miR-202 mimic were transfected with
Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol into 97L
and Huh7 cells. After 48 h, the dual luciferase reporter assay
system (Promega Corporation) was applied to measure luciferase
activities and the results were normalized to Renilla
luciferase activity.
Statistical analysis
Data are presented as the mean ± SD. Differences
were compared using a paired Student's t-test or one-way ANOVA and
Student-Newman-Keuls was used as a post hoc test following the
ANOVA. The patients were divided into two groups: i) High miR-202
expression; and ii) low miR-202 expression groups, according to the
median expression of miR-202 in HCC tissue samples. The association
between the clinicopathological features of HCC and miR-202 was
calculated using the χ2 test. The overall survival rates
and survival differences were detected using univariate analysis
and Kaplan-Meier method with the log-rank test. P<0.05 was
considered to indicate a statistically significant result.
Results
miR-202 expression is downregulated in
HCC tissue samples and liver cancer cells
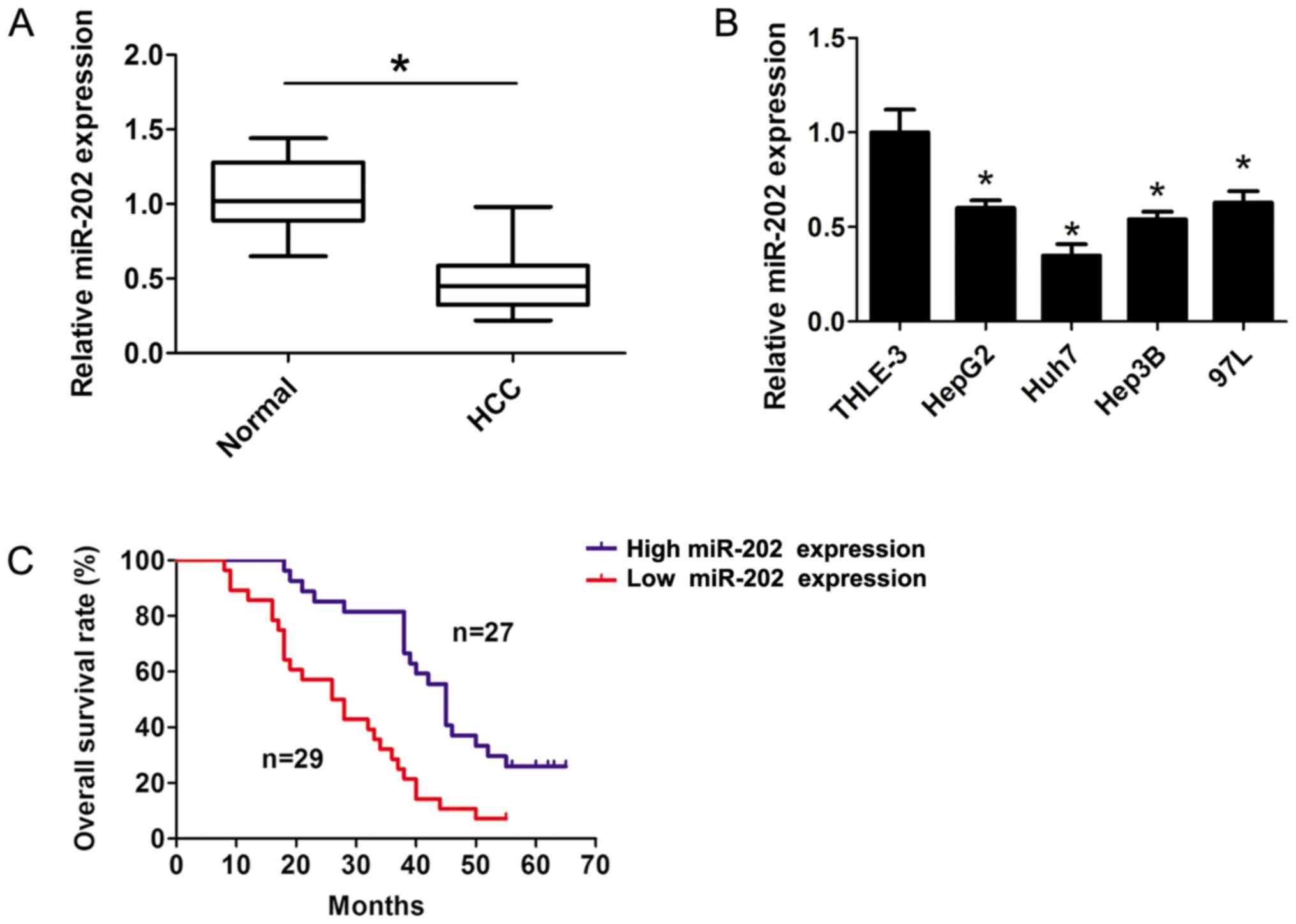
In the present study, the expression levels of
miR-202 were detected in HCC tissue samples and the corresponding
adjacent non-cancerous tissue samples using RT-qPCR analyses. The
results indicated that miR-202 expression was significantly
downregulated in HCC tissue samples compared with the corresponding
adjacent non-cancerous tissue samples (P<0.05; Fig. 1A). Furthermore, the RT-qPCR results
revealed that miR-202 expression was dramatically downregulated in
several liver cancer cells compared with THLE-3 cells (P<0.05;
Fig. 1B). The patients were divided
into two groups: i) High miR-202 expression; and ii) low miR-202
expression groups, according to the median expression (0.45-fold)
of miR-202 in HCC tissue samples. The χ2 analysis was
applied to detect the potential associations between miR-202
expression and the clinical characteristics. The results suggested
that miR-202 expression was significantly associated with tumor
size, vascular invasion and Tumor, Node and Metastasis (TNM) stage
(11) of the patients with HCC (all
P<0.05; Table I). However, there
was no association with age, sex, differentiation and AFP level
(all P>0.05; Table I).
Furthermore, a survival plot was calculated using the Kaplan-Meier
method and Log-rank test between the high (n=27) and low (n=29)
median miR-202 expression groups. The results indicated that
patients with higher miR-202 expression levels exhibited longer
survival rates compared with patients with lower miR-202 expression
levels (P<0.05; Fig. 1C),
suggesting that lower expression levels of miR-202 contributed to
the development of HCC, and the expression level of miR-202 may
serve as a predictor of HCC.
 | Table I.Association between miR-202 expression
and clinicopathological parameters in 56 patients with
hepatocellular carcinoma. |
Table I.
Association between miR-202 expression
and clinicopathological parameters in 56 patients with
hepatocellular carcinoma.
|
|
| miR-202
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
parameters | Total | High (n=27) | Low (n=29) | P-value |
|---|
| Age, years |
|
|
| 0.643 |
| ≤55 | 41 | 19 | 22 |
|
|
>55 | 15 | 8 | 7 |
|
| Sex |
|
|
| 0.422 |
| Male | 43 | 22 | 21 |
|
|
Female | 13 | 5 | 8 |
|
| Tumor size, cm |
|
|
| 0.015 |
|
<5 | 30 | 19 | 11 |
|
| ≥5 | 26 | 8 | 18 |
|
| Differentiation |
|
|
| 0.672 |
| Well and
moderately | 40 | 20 | 20 |
|
| Poor | 16 | 7 | 9 |
|
| AFP (ng/ml) |
|
|
| 0.336 |
|
<400 | 18 | 7 | 11 |
|
| ≥400 | 38 | 20 | 18 |
|
| Vascular
invasion |
|
|
| 0.012 |
|
Negative | 34 | 21 | 13 |
|
|
Positive | 22 | 6 | 16 |
|
| TNM stage |
|
|
| 0.035 |
| I–II | 38 | 22 | 16 |
|
|
III–IV | 18 | 5 | 13 |
|
miR-202 inhibits cell proliferation
and cell glycolysis in HCC
The Warburg effect is characterized by an increase
in glucose uptake and lactate production in the presence of oxygen
(6). Whether miR-202 expression
affected cell proliferation and glycolysis in HCC cells was further
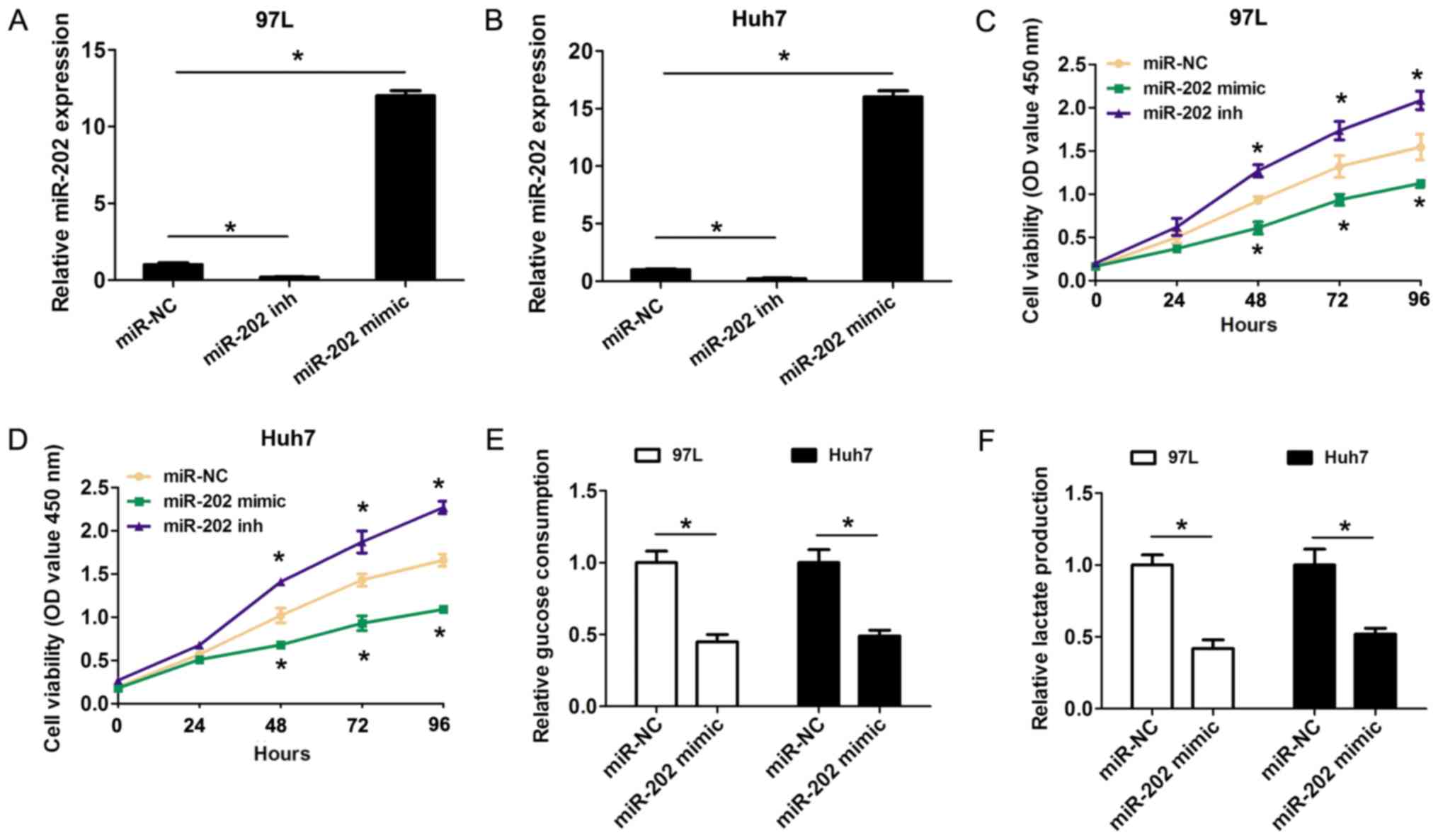
investigated in the present study. The gain-of-function and
loss-of-function assays were performed by transfecting miR-202
mimic or miR-202 inhibitor into 97-L and Huh7 cells due their
higher and lower miR-202 expression levels in numerous liver cancer
cell lines. Importantly, both miR-202 mimic and inhibitor
significantly affected the expression level of miR-202 (Fig. 2A and B). Compared with the negative
controls, the CCK8 results revealed that transfection of miR-202
mimic in 97-L and Huh7 cells significantly inhibited cell
proliferation, while transfection of miR-202 inhibitor promoted
cell proliferation (Fig. 2C and D).
Furthermore, the effects of increased miR-202 expression on cell
glycolysis were detected in HCC cells. The present results
suggested that glucose consumption was significantly decreased
after 97-L and Huh7 cells were transfected with miR-202 mimic,
compared with the control groups (Fig.
2E). Furthermore, lactate production was also decreased
transfection with miR-202 mimic, compared with the control groups
(Fig. 2F). The present results
indicated that increased expression levels of miR-202 inhibited
cell proliferation and cell glycolysis in HCC cells.
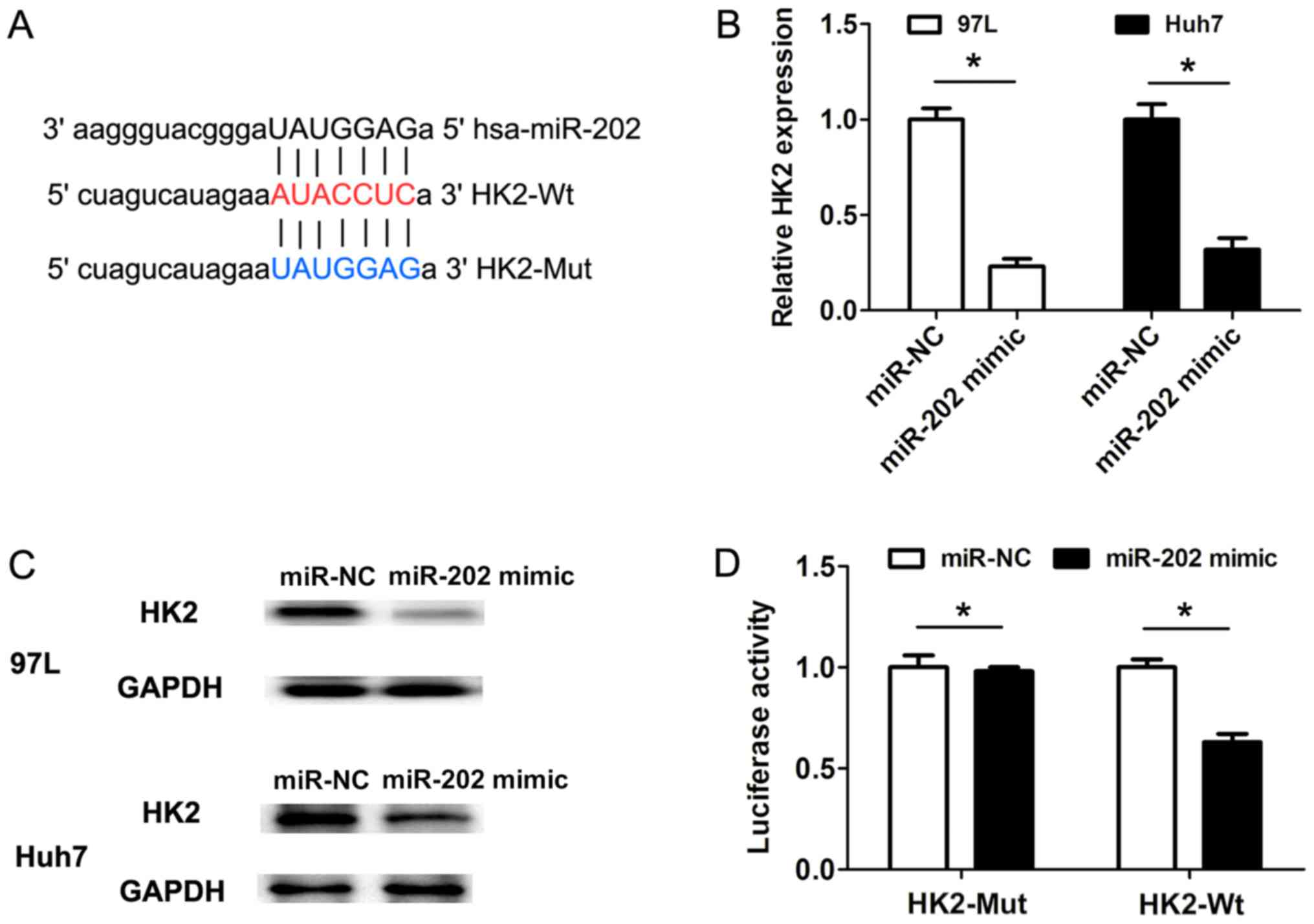
HK2 is a direct target of miR-202
To uncover the molecular mechanisms underlying
miR-202 expression affecting cell proliferation and cell
glycolysis, the software miRanda was used to predict the potential
targets of miR-202. Bioinformatics analysis revealed that HK2 was a
potential target of miR-202 (Fig.
3A). Furthermore, the RT-qPCR results suggested that
transfection of miR-202 mimic in 97 L and Huh7 cells could decrease
the HK2 mRNA expression levels when compared with miR-NC groups
(Fig. 3B). Consistent with the
changes in HK2 mRNA expression levels, the protein expression
levels of HK2 were also downregulated following miR-202 mimic
transfection in 97 L and Huh7 cells, compared with the
corresponding control groups (Fig.
3C). Furthermore, HK2 3′-untranslated region (3′UTR) sequences
containing the wild-type or mutated binding site of miR-202 were
cloned into the psiCHECK2 vectors, respectively (Fig. 3A). The dual-luciferase reporter gene
assays revealed that transfection of miR-202 mimic in 97 L cells
markedly suppressed the luciferase activity of the HK2 3′-UTR
wild-type reporter construct, but not the mutant HK2 3′-UTR
construct (Fig. 3D). Therefore, the
present results suggested that HK2 was a direct target of
miR-202.
miR-202 inhibits cell proliferation
and cell glycolysis by targeting HK2 in HCC cells
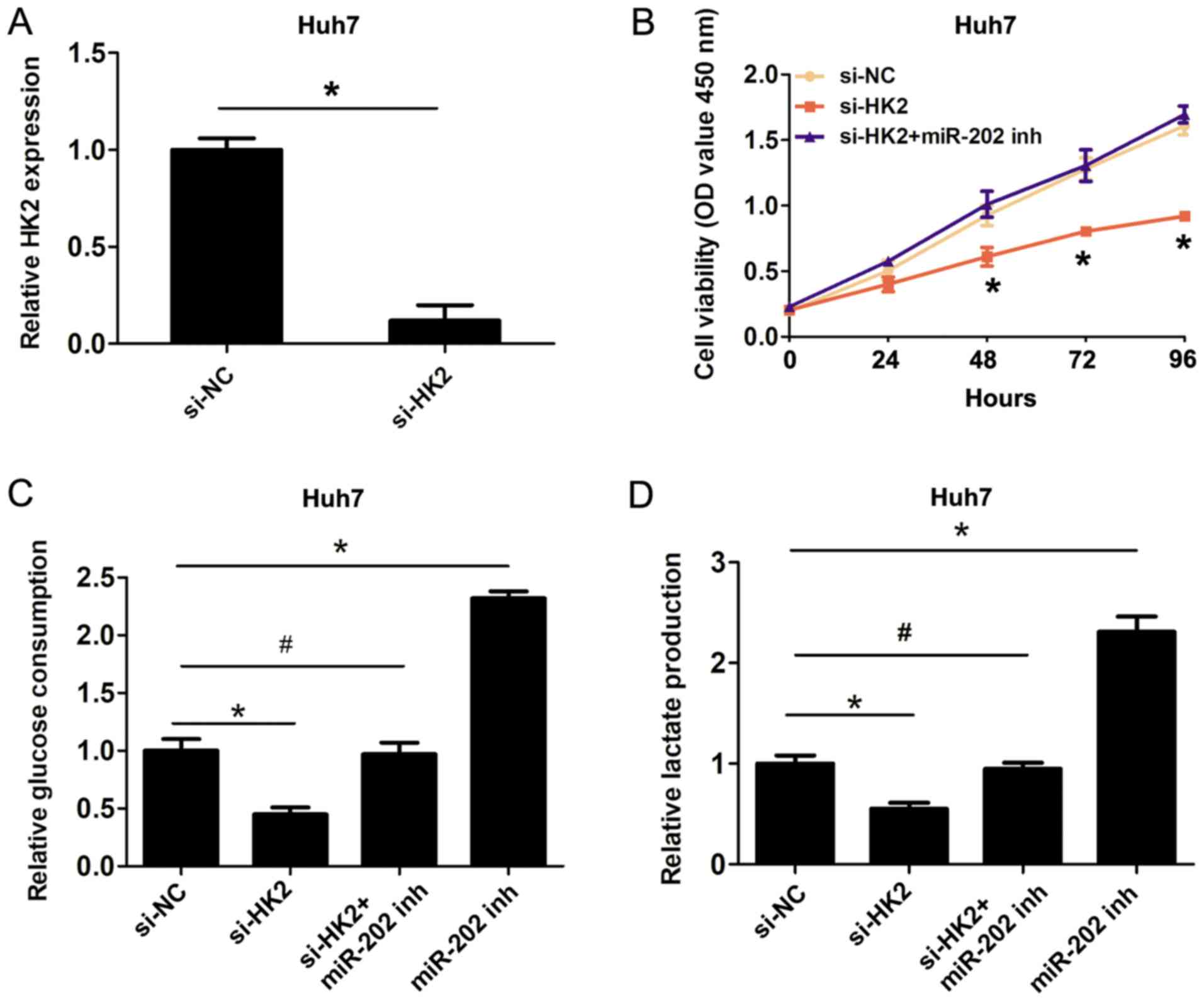
To confirm the involvement of the miR-202/HK2 axis
in the regulation of cell proliferation and cell glycolysis in HCC
cells, HK2 was silenced using si-HK2 (Fig. 4A). The CCK8 cell proliferation assays
revealed that knockdown of HK2 inhibited cell proliferation
compared with the si-NC group, whereas co-transfection of miR-202
inhibitor plus si-HK2 reversed the inhibitory effects of HK2
silencing on cell proliferation (Fig.
4B). Furthermore, knockdown of HK2 inhibited cell glucose
consumption and lactate production, while co-transfection of
miR-202 inhibitor and si-HK2 reversed the inhibitory effects of HK2
knockdown on cell glucose consumption and lactate production
(Fig. 4C and D). Therefore, the
present results indicated that miR-202 inhibited cell proliferation
and cell glycolysis by targeting HK2 in HCC cells.
Discussion
miR-202 has been described as a tumor-suppressing
miRNA that is dysregulated in several different types of tumor. In
a previous study, miR-202 was demonstrated to inhibit tumor
progression by targeting laminin subunit α 1 in esophageal squamous
cell carcinoma (12). miR-202
inhibits the progression of human cervical cancer through
inhibition of cyclin D1 (13).
miR-202 functions as a tumor suppressor in non-small cell lung
cancer by targeting STAT3 (14).
miR-202 inhibits the cell proliferation, migration and invasion of
glioma by directly targeting metadherin (15). In HCC, miR-202 was demonstrated to
suppress cell proliferation by downregulating low-density
lipoprotein receptor-related protein 6 post-transcriptionally
(16). Therefore, the underlying
molecular mechanism of miR-202 in the Warburg effect was
investigated in the present study. The present results indicated
that miR-202 expression was markedly downregulated in HCC tissue
samples compared with the corresponding adjacent non-cancerous
tissue samples. miR-202 expression levels were significantly
associated with tumor size, vascular invasion and TNM stage.
Furthermore, higher expression levels of miR-202 revealed a longer
survival rate compared with lower levels of miR-202 expression.
Therefore, the present results suggested that lower miR-202 may
contribute to the development of HCC. However, the number of
patients used in the present study was small, and so this sample
size should be expanded in future studies.
Furthermore, the functional effects of miR-202
expression on HCC cells were detected in the present study. The
results indicated that miR-202 could inhibit cell proliferation and
cell glycolysis in HCC cells. The glycolytic enzyme HK2 is crucial
for the Warburg effect, and elevated levels of HK2 had been
revealed in a number of different types of human tumor, as well as
being associated with cell glycolysis in tumor cells. As such,
STAT3 regulates cell glycolysis via targeting HK2 in HCC (17). It has previously been demonstrated
that miR-143 suppresses oral squamous cell carcinoma cell growth,
invasion and glucose metabolism through targeting HK2 (18). forkhead box M1 promotes reprogramming
of glucose metabolism in epithelial ovarian cancer cells via
activation of GLUT1 and HK2 transcription (19). The miR-125a/HK2 axis regulates the
reprogramming of cancer cell energy metabolism in HCC (20). In the present study, it was
identified that HK2 was a direct target of miR-202 in HCC cells.
miR-202 overexpression could decrease the HK2 mRNA and protein
expression levels in HCC cells. Knockdown of HK2 inhibited cell
proliferation, cell glucose consumption and lactate production,
while co-transfection of miR-202 inhibitor and si-HK2 reversed the
inhibitory effects of HK2 knockdown on cell glucose consumption and
lactate production. Thus, these results indicated that miR-202
could inhibit cell proliferation and cell glycolysis by targeting
HK2 in HCC cells.
Collectively, in the present study, the miR-202
expression levels were identified to be lower in HCC tissues and
liver cancer cells. In addition, lower miR-202 expression levels
were found to be associated with a poor prognosis in patients with
HCC. Furthermore, miR-202 was found to function as a tumor
suppressor in liver cancer cells by targeting HK2 to inhibit cell
proliferation and cell glycolysis. Therefore, the present results
suggested that miR-202 may represent a novel target for treating
HCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
JW, JC and FS conceived and designed the study, and
drafted the manuscript. ZW, WX, YY, FD and HS collected, analyzed
and interpreted the data, and critically revised the manuscript for
intellectual content. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Affiliated Hospital of Shaoxing University. Written
informed consent was obtained from all patients in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Aravalli RN, Steer CJ and Cressman EN:
Molecular mechanisms of hepatocellular carcinoma. Hepatology.
48:2047–2063. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mathupala SP, Ko YH and Pedersen PL:
Hexokinase-2 bound to mitochondria: Cancer's stygian link to the
‘Warburg Effect’ and a pivotal target for effective therapy. Semin
Cancer Biol. 19:17–24. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Singh PK, Mehla K, Hollingsworth MA and
Johnson KR: Regulation of aerobic glycolysis by microRNAs in
cancer. Mol Cell Pharmacol. 3:125–134. 2011.PubMed/NCBI
|
|
7
|
Nie H, Li J, Yang XM, Cao QZ, Feng MX, Xue
F, Wei L, Qin W, Gu J, Xia Q and Zhang ZG: Mineralocorticoid
receptor suppresses cancer progression and the Warburg effect by
modulating the miR-338-3p-PKLR axis in hepatocellular carcinoma.
Hepatology. 62:1145–1159. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang LF, Lou JT, Lu MH, Gao C, Zhao S, Li
B, Liang S, Li Y, Li D and Liu MF: Suppression of miR-199a
maturation by HuR is crucial for hypoxia-induced glycolytic switch
in hepatocellular carcinoma. EMBO J. 34:2671–2685. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang B, Hsu SH, Frankel W, Ghoshal K and
Jacob ST: Stat3-mediated activation of microRNA-23a suppresses
gluconeogenesis in hepatocellular carcinoma by down-regulating
glucose-6-phosphatase and peroxisome proliferator-activated
receptor gamma, coactivator 1 alpha. Hepatology. 56:186–197. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)). Method. 25:402–408. 2001. View Article : Google Scholar
|
|
11
|
Zhong JH, Xiang BD, Gong WF, Ke Y, Mo QG,
Ma L, Liu X and Li LQ: Comparison of long-term survival of patients
with BCLC stage B hepatocellular carcinoma after liver resection or
transarterial chemoembolization. PLoS One. 8:e681932013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Meng X, Chen X, Lu P, Ma W, Yue D, Song L
and Fan Q: MicroRNA-202 inhibits tumor progression by targeting
LAMA1 in esophageal squamous cell carcinoma. Biochem Biophys Res
Commun. 473:821–827. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yi Y, Li H, Lv Q, Wu K and Zhang W, Zhang
J, Zhu D, Liu Q and Zhang W: miR-202 inhibits the progression of
human cervical cancer through inhibition of cyclin D1. Oncotarget.
7:72067–72075. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao Z, Lv B, Zhang L, Zhao N and Lv Y:
miR-202 functions as a tumor suppressor in non-small cell lung
cancer by targeting STAT3. Mol Med Rep. 16:2281–2289. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang J, Fan B, Zhao Y and Fang J:
MicroRNA-202 inhibits cell proliferation, migration and invasion of
glioma by directly targeting metadherin. Oncol Rep. 38:1670–1678.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Y, Zheng D, Xiong Y, Xue C, Chen G,
Yan B and Ye Q: miR-202 suppresses cell proliferation in human
hepatocellular carcinoma by downregulating LRP6
post-transcriptionally. FEBS Lett. 588:1913–1920. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li M, Jin R, Wang W, Zhang T, Sang J, Li
N, Han Q, Zhao W, Li C and Liu Z: STAT3 regulates glycolysis via
targeting hexokinase 2 in hepatocellular carcinoma cells.
Oncotarget. 8:24777–24784. 2017.PubMed/NCBI
|
|
18
|
Sun X and Zhang L: MicroRNA-143 suppresses
oral squamous cell carcinoma cell growth, invasion and glucose
metabolism through targeting hexokinase 2. Biosci Rep. 37(pii):
BSR201604042017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Y, Yun Y, Wu B, Wen L, Wen M, Yang H,
Zhao L, Liu W, Huang S, Wen N and Li Y: FOXM1 promotes
reprogramming of glucose metabolism in epithelial ovarian cancer
cells via activation of GLUT1 and HK2 transcription. Oncotarget.
7:47985–47997. 2016.PubMed/NCBI
|
|
20
|
Jin F, Wang Y, Zhu Y, Li S, Liu Y, Chen C,
Wang X, Zen K and Li L: The miR-125a/HK2 axis regulates cancer cell
energy metabolism reprogramming in hepatocellular carcinoma. Sci
Rep. 7:30892017. View Article : Google Scholar : PubMed/NCBI
|