Introduction
Cervical cancer is responsible for 570,000 cases and
311,000 deaths in 2018 worldwide, ranking in the fourth most common
cancer affecting women (1). Although
early screening has aided in reducing the death rates, there is an
increased prevalence in patients aged between 20 and 40 years has
been observed (2). The age-adjusted
incidence rate in the cervical adenocarcinoma cases aged 39 or
younger has significantly increased from 1976 to 2012 (annual
percent change=5.0) in Japan (2). In
the United States, cervical cancer is currently most frequently
diagnosed among women aged 35 to 44 years compared with those aged
45 to 54 years in the 1990s (3).
Furthermore, the prognosis of advanced cervical cancer remains poor
(4). The 5-year survival rate in the
2018 FIGO staging system is 85.6% for stage I tumors. In contrast,
the 5-year survival rate for stage III and IV is 39.3 and 24.0%,
respectively (5). Therefore, novel
therapeutic interventions for advanced cervical cancer may confer
improved outcomes for patients.
Human telomere reverse transcriptase (hTERT) is a
catalytic subunit of telomerase that has been reported to regulate
telomerase activity and serve a critical role in the tumorigenesis
and the proliferation of cancer cells (6). Several recent studies demonstrated that
expression of hTERT is elevated in a variety of cancers such as
esophageal cancer (7) and thyroid
carcinoma (8). Notably,
downregulation of hTERT gene expression has been reported to
enhance radiosensitivity in cervical cancer cells (9). The c-Myc proto-oncogene is a key switch
for induced telomerase activity, including the upregulation of the
hTERT gene (10).
MicroRNAs (miRNAs/miRs) are single-stranded RNA
molecules of 21–25 base pairs. miRNA molecules bind to the
complementary 3′-untranslated regions (UTRs) of target mRNAs and
suppress gene expression via inhibition of the translation of its
target mRNA (11). Accumulating
evidence has indicated that miRNAs are implicated in a variety of
diseases, such as cancer (12),
cardiovascular disease (13) and
metabolic disorders (14). A recent
report revealed that decreased expression levels of miR-22 is
associated with a poor prognosis in patients with cervical cancer
(15). However, the role of miR-22
in the treatment of cervical cancer is poorly characterized. In a
previous study, miR-22 was identified as a tumor suppressor through
the direct repression of MYC-binding protein (MYCBP) and subsequent
reduction of downstream c-Myc-mediated molecules, which include
cyclin D2, cyclin-dependent kinase 4, ornithine decarboxylase,
lactate dehydrogenase-A, carbamoyl phosphate synthase-aspartate
transcarbamylase-dihydroorotase, nucleolin and eukaryotic
translation initiation factor 2A (16). However, the association between
miR-22 and hTERT expression is yet to be elucidated.
In the present study, the effect of miR-22
expression on its downstream target (MYCBP) was investigated in
cervical cancer cells. Moreover, the influence of miR-22 on the
subsequent hTERT repression was subsequently examined. In addition,
the biological role of miR-22 in radiosensitivity of cervical
cancer cells was also investigated.
Materials and methods
Cell line
The human cervical cancer cell lines C-4I, and SiHa
were purchased from the American Type Culture Collection. SKG-II
was provided by Keio University (Tokyo, Japan). Cells were cultured
in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% fetal bovine serum (FBS) (Equitech-Bio, Inc.) at 37°C in a
humidified incubator with 5% CO2.
Transfection of precursor miRNA
Pre-miR™ miRNA precursor molecules (pre-miR-22-3p;
cat. no. AM17101), negative (non-specific) control
(pre-miR-negative control #1; cat. no. AM17110) and inhibitor miRNA
(anti-miR-22-3p; cat. no. AM17001) were ordered from Ambion (Thermo
Fisher Scientific, Inc.). These were designed to mimic endogenous
mature miRNAs, but the sequences are not publicly available. The
mature miRNA sequence of miR-22-3p is 5′-AAGCUGCCAGUUGAAGAACUGU-3′.
Cervical cancer cells were transfected with pre-miR-22-3p,
anti-miR-22-3p or pre-miR-negative control (30 nM) for 24 h.
Oligonucleotide transfection was performed using siPORT NeoFX
Transfection Agent (Ambion; Thermo Fisher Scientific, Inc.).
3′UTR reporter assay
C-4I and SiHa cells were used for the 3′UTR reporter
assay. The full length MYCBP 3′UTR was inserted downstream of a
Gaussia luciferase (Gluc) reporter in the pEZX-MT05 vector
(GeneCopoeia, Inc.). The secreted alkaline phosphatase (seAP)
reporter gene was also present in the vector as an internal control
for transfection normalization. As a control (pEZX-MT05-CT), miRNA
target clone control vector (CmiT000001-MT05) was purchased from
GeneCopoeia, Inc. Cells (1×105/ml) seeded in 24-well
plates were co-transfected using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) complexed with the
pEZX-MT05 vector and pre-miR-22-3p, anti-miR-22-3p or
pre-miR-negative control (cont miR) according to the manufacturer's
protocol. Culture medium (DMEM, Gibco; Thermo Fisher Scientific,
Inc.) was collected after 48 h and the relative luciferase activity
(Gluc:seAP ratio) was analyzed using the Secrete-Pair Dual
Luminescence Assay kit (GeneCopoeia, Inc.) according to the
manufacturer's protocol.
Bioinformatic analysis
The putative human target genes of miR-22 were
analyzed using the TargetScan (version 6.0; targetscan.org/) and miRDB (version 5.0; mirdb.org/) web-based bioinformatics algorithms.
TargetScan predicts biological targets of miRNAs by searching for
the presence of conserved 8mer, 7mer and 6mer sites that match the
seed region of each miRNA (17). The
cumulative weighted context ++ score <-0.1 was applied as the
cut-off criteria (17). In miRDB,
the prediction of miRNA-mRNA pair is based on both the 3′UTR and
5′UTR regions of conserved and non-conserved genes, the base
composition in the regions flanking the seed pairing sites,
secondary structure, and the location of the site within the 3′UTR
(18).
RNA and miRNA extraction and reverse
transcription-quantitative (RT-q)PCR
Total RNA was extracted from C-4I, SKG-II and SiHa
cells using a RNeasy kit (Qiagen, Inc). A Super Script II Reverse
Transcriptase kit (Invitrogen; Thermo Fisher Scientific, Inc.) was
used to synthesize cDNA using random primers according to the
manufacturer's protocol. A total of 1 µg of RNA, 125 ng random
primers and 1 µl 10 mM dNTPs in 12 µl total volume were denatured
at 65°C for 5 min before 2 min on ice. Then, 4 µl 5× first strand
buffer and 2 µl of 0.1 M DTT were added followed by 1 µl
Superscript II. Reactions were incubated 10 min at 25°C, 42°C for
50 min and 70°C for 15 min. Subsequently, TaqMan qPCR was performed
in triplicate using the StepOne Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The mixed primers for
TaqMan qPCR used in the present study were purchased from Applied
Biosystems in the form of a probe mix (MYCBP: Hs 00429315_g1;
c-Myc: Hs00153408_m1; hTERT: Hs00972650_m1) and GAPDH
(Hs02786624_g1, Applied Biosystems) was used as a housekeeping
control gene. The sequences of these primers are not publicly
available. PCR conditions were 95°C for 10 min, followed by 60°C
for 1 min for 40 cycles following the manufacturer's protocol.
miRNA extraction was performed using a mirVana miRNA
isolation kit (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. miRNA was then
reverse transcribed using the microRNA reverse transcription kit
according to the manufacturer's protocol (Applied Biosystems;
Thermo Fisher Scientific, Inc.) in combination with the stem-loop
primer for miR-22-3p
(5′-GGCUGAGCCGCAGUAGUUCUUCAGUGGCAAGCUUUAUGUCCUGACCCAGCUAAAGCUGCCAGUUGAAGAACUGUUGCCCUCUGCC-3′)
and the endogenous control RNU48
(5′-GATGACCCCAGGTAACTCTGAGTGTGTCGCTGATGCCATCACCGCAGCGCTCTGACC-3′)
(Applied Biosystems; Thermo Fisher Scientific, Inc.) and served as
a template for the quantification of the expression of mature
miRNA. qPCR of miR-22 was performed according to the manufacturer's
instructions (Applied Biosystems; Thermo Fisher Scientific, Inc.).
PCR conditions were 95°C for 10 min, followed by 60°C for 1 min for
40 cycles. Data analyses were performed using the 2−ΔΔCq
method (19).
Western blot analysis
Western blot analysis was performed as described
previously (20). In brief, total
proteins from C-4I, SKG-II and SiHa were prepared using Pierce RIPA
Buffer (Thermo Fisher Scientific, Inc.). The protein concentration
was quantified by DC Protein Assay (Bio-Rad Laboratories, Inc.).
Protein samples (15 µg/lane) were separated using 4–15% gradient
gel electrophoresis (Bio-Rad Laboratories, Inc.) and transferred to
PVDF membranes. After being blocked with 10% bovine serum albumin
(New England BioLabs, Inc.) for 1 h at room temperature, the
membranes were incubated overnight at 4°C with primary antibodies
diluted at 1:200 (anti-MYCBP: Sigma-Aldrich, HPA041188) or 1:1,000
(anti-MYC: Cell Signaling, 13987 and anti-hTERT; LifeSpan
BioSciences, Inc., LS-B11086). After 1 h incubation with
horseradish peroxide-conjugated anti-rabbit secondary antibody
(1:4,000; goat anti-rabbit IgG; sc-2030; Santa Cruz Biotechnology,
Inc.) at room temperature, the blots were visualized using enhanced
chemiluminescence (ECL Plus; GE Healthcare Life Sciences).
Clonogenic assay
The clonogenic assay was performed using the
technique described previously by Franken et al (21). In brief, C-4I and SKG-II cells
(1.0×102/well for 2 Gy-2.4×103/well for 8 Gy)
transfected with miR-22, anti-miR-22 or cont miR were plated onto
6-well plates. Each group of cells was irradiated with various
doses of X-ray (0, 2, 4, 6 and 8 Gy) from an X-ray generator
(M-150WE; Softex Co., Ltd.) and incubated at 37°C in a humidified
incubator with 5% CO2 for 14 days. Fixation and staining
of colonies was performed using a mixture of 0.5% crystal violet in
methanol for 30 min at room temperature. Plates were rinsed with
water and left to dry at room temperature. Counting of colonies was
done on the following day. The cell survival was measured by
standard colony formation after radiation treatment. Colonies
containing >50 cells counted under a light microscope
(CK40-F100, Olympus) at ×40 magnification were defined as derived
from clonogenically viable cells. The survival fraction of the
cells was calculated by normalizing the plating efficiency of
treated cells by that of control cells as described previously
(21). Each experiment was performed
at least three times in triplicate wells.
Lentivirus infection
Lentivirus (1×107 plaque forming
units/ml) expressing LentimiRa-GFP-hsa-miR-22-3p (L-miR22-C-4I;
cat. no. mh15295) and Lnti-III-miR-GFP Control (L-cont-C-4I; cat.
no. m002) were purchased from Applied Biological Materials, Inc.
Lentiviral transduction was conducted at a multiplicity of
infection of 200 with a ViraDuctin Lentivirus Transduction kit
(Cell Biolabs, Inc.), according to the manufacturer's protocol. In
brief, 5.0×104 C-4I cells were seeded in 24-well plates
overnight at 37°C in a humidified incubator with 5% CO2.
LentimiRa-GFP-hsa-miR-22-3p or Lenti-III-miR-GFP Control was added
to the cells. After 48 h, purification was performed using
puromycin until antibiotic-resistant colonies were identified.
Post-transfection cells were further selected in DMEM (Gibco;
Thermo Fisher Scientific, Inc.) containing puromycin for 2 weeks to
establish stably transduced cells.
A tumor xenograft assay
Female 6-week-old athymic nude mice (BALB/c
nu/nu) (average body weight 16 g) were
purchased from Japan SLC, Int. A total of 10 animals were divided
into two groups, each consisting of 5 mice (n=5). Mice were housed
under standard environmental conditions at Osaka Medical College
Division of Research Animal Laboratory (temperature, 22°C;
humidity, 40–60%; light/dark cycle, 12 h light 12 h darkness) with
ad libitum access to food and water. All of the animal
studies were carried out in compliance with the guidelines of the
Osaka Medical College Animal Care and Use Committee, and followed
the institutional guidelines for animal welfare and experimental
conduct. Mice were monitored daily for signs of discomfort and pain
by laboratory personnel as well as by the staff at the Division of
Research Animal Laboratory. In addition to the pathological status,
the mice were monitored to ensure that a humane endpoint was
reached (defined as complete inability to ambulate). All mice
gained weight over the entire study period while appearing
generally healthy throughout the experiments. Under anesthesia with
2% isoflurane, C-4I cells infected with L-miR22-C-4I or L-cont-C-4I
were injected subcutaneously into the flanks of nude mice
(4×106 cells in 100 µl PBS per mouse).
The in vivo growth of C-4I xenografts was
monitored by measuring their volumes and calculated using the
modified ellipse formula (volume=length × (width)2/2).
When the xenograft volumes reached ~100 mm3, the tumor
was irradiated with X-rays (6 Gy) following the intraperitoneal
administration of a mixture of three anesthetic agents (0.3 mg/kg
medetomidine, 4 mg/kg midazolam and 5 mg/kg butorphanol). After
irradiation, the tumors were measured with calipers every 7 days. A
total of 35 days after irradiation, all mice were euthanized by
cervical dislocation under anesthesia with 5% isoflurane for sample
collection. Death was verified by the absence of a heart beat and
the onset of rigor mortis. The tumors were excised, weighed (the
maximum percentage of tumor weight of total body weight was
<0.15%) and fixed in 10% neutral buffered formalin for 24 h at
room temperature, and 4 µm-thick paraffin sections were prepared
for immunohistochemistry and the terminal deoxynucleotidyl
transferase dUTP nick-end labeling (TUNEL) assay.
Immunostaining
The aforementioned section from paraffin-embedded
xenograft tissues were subjected to immunostaining. Tissue samples
were formalin-fixed and embedded in paraffin. Deparaffinized and
rehydrated sections were autoclaved in 0.01 mol/l citrate buffer
(pH 6.0) for 15 min at 121°C for antigen retrieval. The endogenous
peroxidase activity was blocked with 0.3% hydrogen peroxide
solution in methanol for 30 min at room temperature, then sections
were incubated at room temperature for 30 min with rabbit anti-Ki67
antibody (1:300; AB9260; Merck KGaA). The sections were then washed
once with phosphate-buffered saline (PBS) and incubated with
secondary antibody Histofine Simple Stain MAX PO (MULTI) (ready to
use; cat. no. 414151F; Nichirei Corporation) for 30 min at room
temperature. Finally, the sections were washed once with PBS and
visualized by incubating with
H2O2/diaminobenzidine substrate solution for
5 min. The sections were counterstained with hematoxylin for 20 sec
at room temperature prior to dehydration and mounting. The Ki-67
index and percentage of apoptotic cells reflected the percentage of
the total number of tumor cells with nuclear staining in viable
regions per 5 high-power fields using a fluorescence microscope
(BZ-X700, KEYENCE) at ×400 magnification.
TUNEL assay
Apoptotic cell death was determined by TUNEL assay
using a in situ Apoptosis Detection kit (Wako Pure Chemical
Industries, Ltd.), according to the manufacturer's protocol. The
sections were deparaffinized for 10 min, dehydrated in 100% ethanol
for 10 min and proteins of the sections were digested using
pre-warmed protease solution for 5 min at 37°C. After washing, the
sections were incubated with 50 µl TdT reaction solution
(consisting of TdT Enzyme 1 µl + TdT Substrate Solution 49 µl) for
10 min at 37°C. After washing, the endogenous peroxidase activity
was blocked using 3% H2O2 for 5 min at room
temperature. After washing, the sections were reacted with 100 µl
of POD-conjugated antibody solution for 10 min at 37°C. After
removing the antibody solution and washing, immunoreactivity was
visualized using 3,3′-diaminobenzidine. The sections were
counterstained with 0.5% methyl green for 5 min at room
temperature. The TUNEL index was calculated as the percentage of
TUNEL-positive cells in 1,000 carcinoma cells in the areas of
highest nuclear labeling under a fluorescence microscope (BZ-X700,
KEYENCE) at ×400 magnification.
Statistical analysis
The statistical analyses were performed using the
StatView software program (version 5.0; SAS Institute, Inc.). The
data are presented as the mean ± standard deviation of three
independent experiments. The statistical analysis was performed
using the Student's paired t-test and P<0.05 was considered to
indicate a statistically significant difference.
Results
Overexpression of miR-22 suppresses
the expression of MYCBP in cervical cancer cell lines
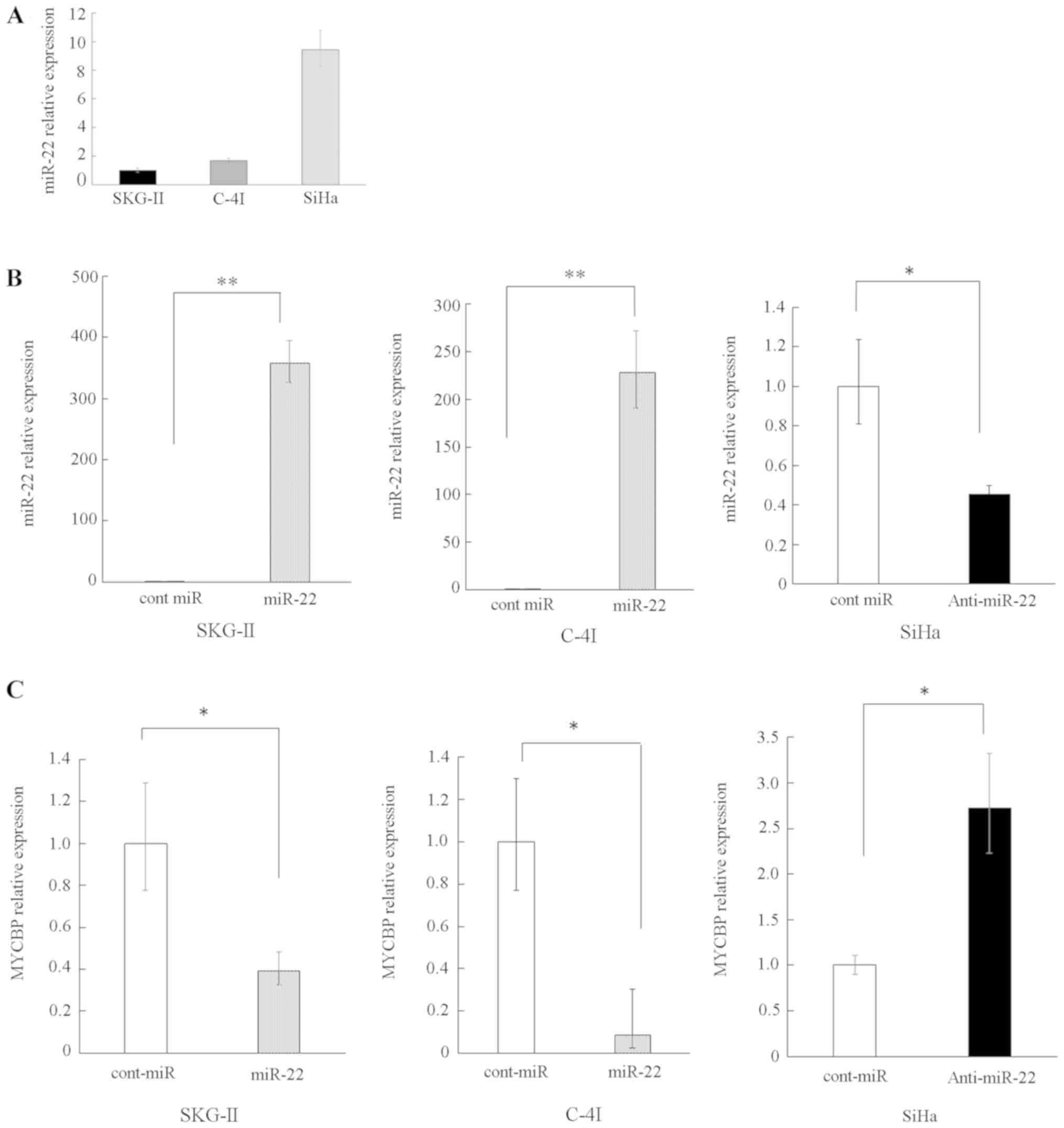
RT-qPCR was performed to evaluate the expression
profile of miR-22 in several cervical cancer cell lines. The
endogenous miR-22 expression was higher in the SiHa cell line
compared with the SKG-II and C-4I lines (Fig. 1A). Accordingly, SKG-II and C-4I were
used for overexpression experiments of miR-22, whereas SiHa was
used for reduced experiments of miR-22.
The overexpression of miR-22 was induced by the
transfection of pre-miR-22, and an increased level of miR-22 in
SKG-II and C-4I cells was confirmed (Fig. 1B). By contrast, the transfection of
anti-miR-22 reduced the miR-22 expression level in SiHa cells
(Fig. 1B).
Subsequently, it was determined whether or not
expression of MYCBP could be altered by the overexpression or
suppression of miR-22. RT-qPCR revealed that the level of MYCBP was
decreased under conditions of miR-22 overexpression, while MYCBP
mRNA was increased under conditions of miR-22 suppression (Fig. 1C).
miR-22 directly targets MYCBP
3′UTR
TargetScan and miRDB were utilized to predict that
the MYCBP gene has a putative miR-22 target site at its 3′UTR
region. To confirm that miR-22 directly targeted the MYCBP 3′UTR in
cervical cancer cells, a luciferase reporter assay was performed.
C-4I cells were co-transfected with either the precursor miRNA or
control and with either a plasmid containing a luciferase reporter
driven by the wild-type human MYCBP 3′UTR (pEZX-MT05-MYCBP;
Fig. 2A) or a control plasmid
(pEZX-MT05-CT). Treatment of C-4I with complexes of pre-miR-22 and
pEZX-MT05-MYCBP significantly reduced the luciferase activity
compared with that in the combination of cont miR and
pEZX-MT05-MYCBP (Fig. 2B). Knockdown
of miR-22 via the transfection of anti-miR22 significantly
increased the luciferase activity of MYCBP 3′UTR compared with
transfection of cont miR (Fig. 2C).
The current results indicate that miR-22 suppresses the expression
of MYCBP mRNA via direct targeting of the MYCBP 3′UTR in the
cervical cancer cell lines.
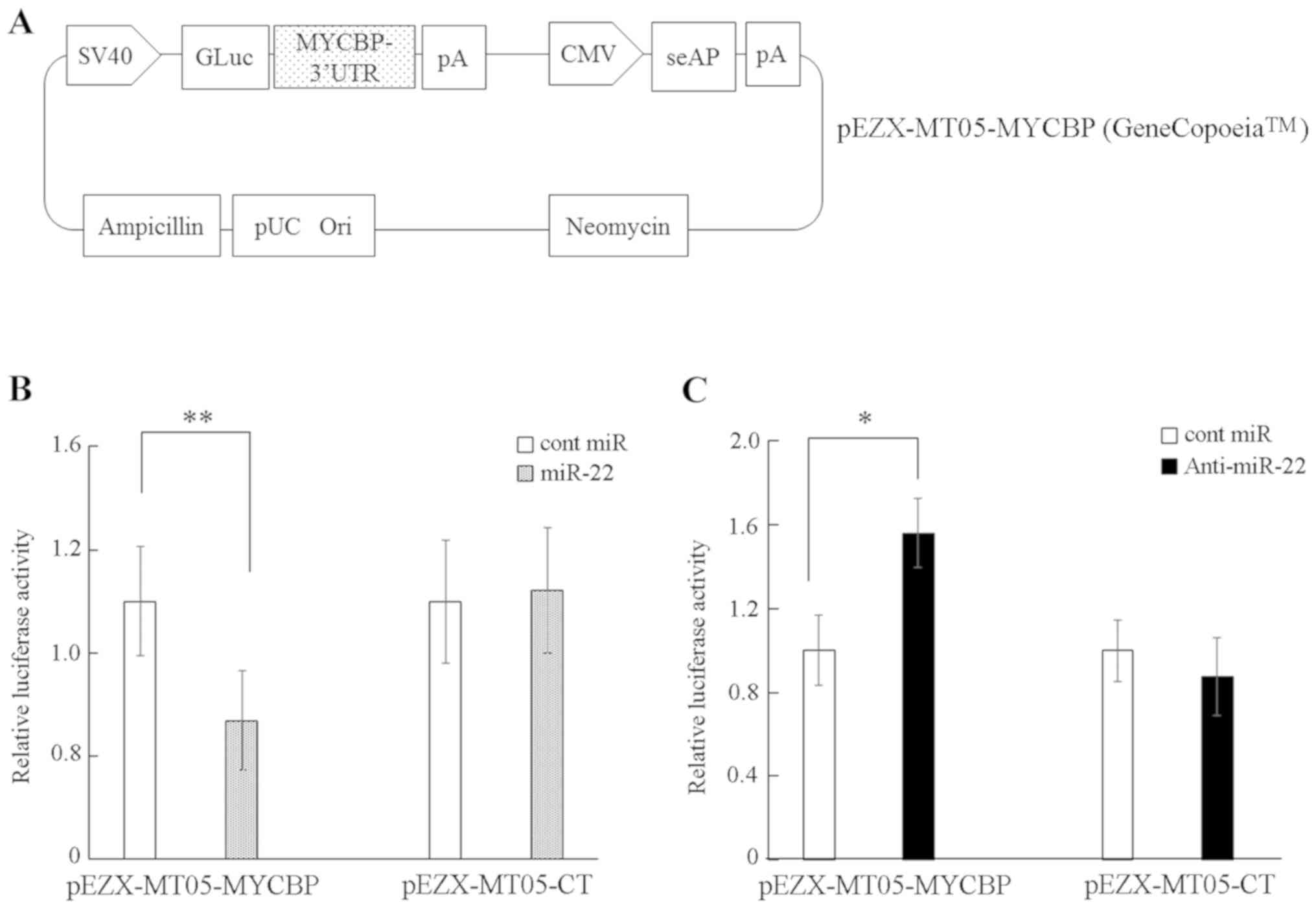 | Figure 2.MYCBP is a direct target of miR-22.
(A) A diagram of the MYCBP 3′UTR-containing reporter construct. (B)
C-4I cells were co-transfected with the MYCBP 3′UTR reporter
construct (pEZX-MT05-MYCBP) or control vector (pEZX-MT05-CT) and
with miR-22 or control miR. Relative gaussian luciferase-to-seAP
signal is shown. (C) SiHa cells were co-transfected with either
pEZX-MT05-MYCBP or pEZX-MT05-CT and with either anti-miR22 or
control. Data are exhibited as the mean ± SD of three independent
experiments. *P<0.05, **P<0.01 vs. control. Columns represent
the mean ± standard deviation. miR, microRNA; MYCBP, MYC binding
protein; 3′UTR, 3′untranslated region; cont/CT, control; seAP,
secreted alkaline phosphatase; CMV, cytomegalovirus promoter; GLuc,
Gaussia luciferase; pA, poly-A tail; pUC Ori, origin of
replication. |
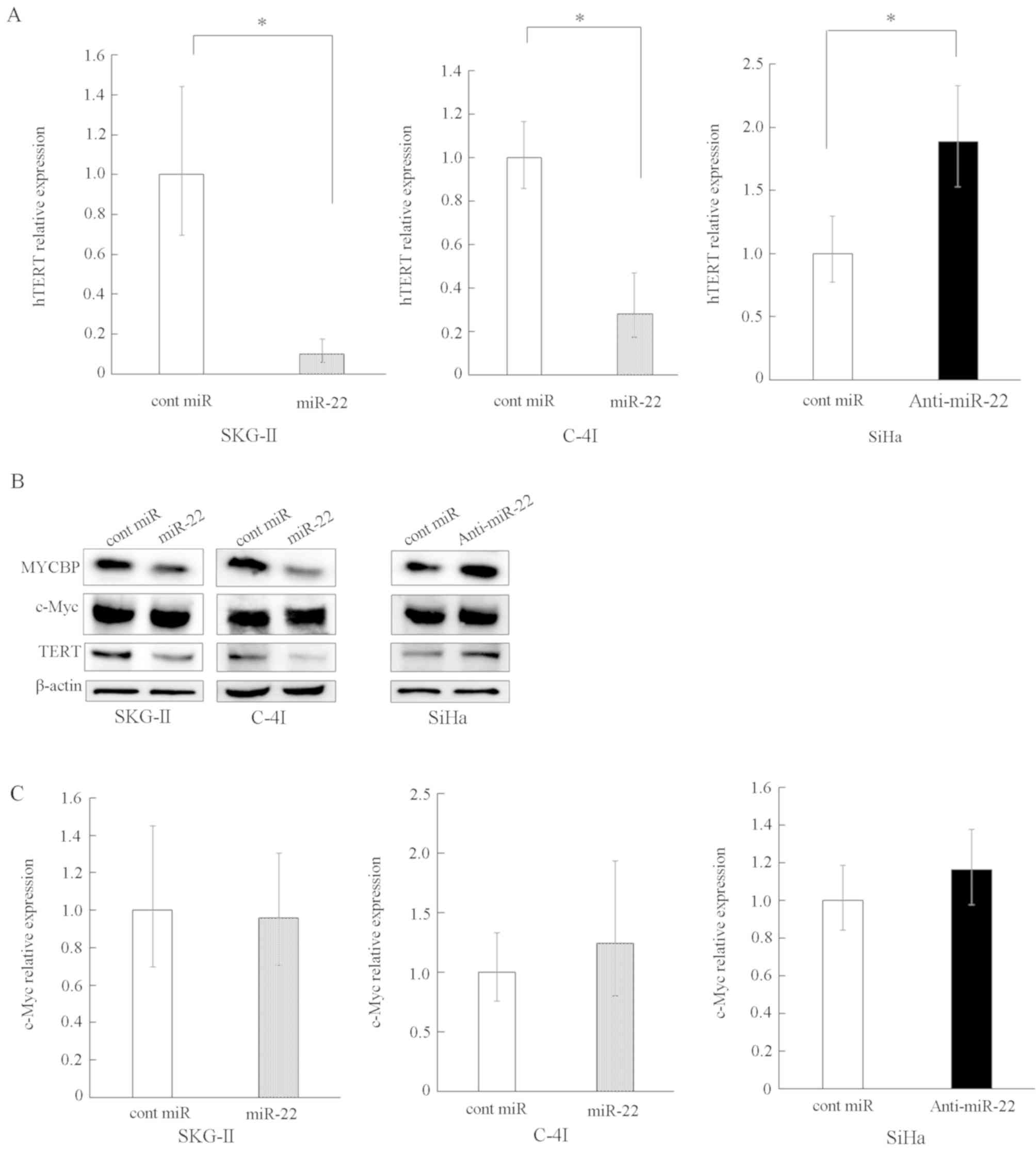
MYCBP regulates the expression of the
c-Myc target gene hTERT
MYCBP has been revealed to promote the activation of
the c-Myc target gene via E-box (22). Therefore, the current study
investigated whether or not the suppression of MYCBP by miR-22
results in subsequent suppression of the E-box-dependent c-Myc
target gene expression. The hTERT gene is an E-box-dependent target
gene (23) and does not contain a
predicted target site for miR-22 in its 3′UTR, according to
TargetScan and miRDB. Thus, the effect of the overexpression or
suppression of miR-22 on hTERT expression levels was examined. It
was revealed that the overexpression of miR-22 significantly
reduced the expression level of hTERT, whereas suppression of
miR-22 increased hTERT expression (Fig.
3A and B).
Subsequently, whether or not the miR-22-induced
hTERT suppression was dependent on the interaction of miR-22 with
c-Myc 3′UTR was investigated, and in silico analyses using
TargetScan and miRDB predicted that the c-Myc gene had no target
site for miR-22. Moreover, RT-qPCR and western blot analyses
revealed that neither the overexpression nor the suppression of
miR-22 affected c-Myc levels at the mRNA or protein level (Fig. 3B and C). The present results do not
support the possibility of a direct interaction between miR-22 and
c-Myc mRNA, thus indicating that the inhibition of MYCBP by miR-22
resulted in the subsequent reduction of the c-Myc target gene
hTERT.
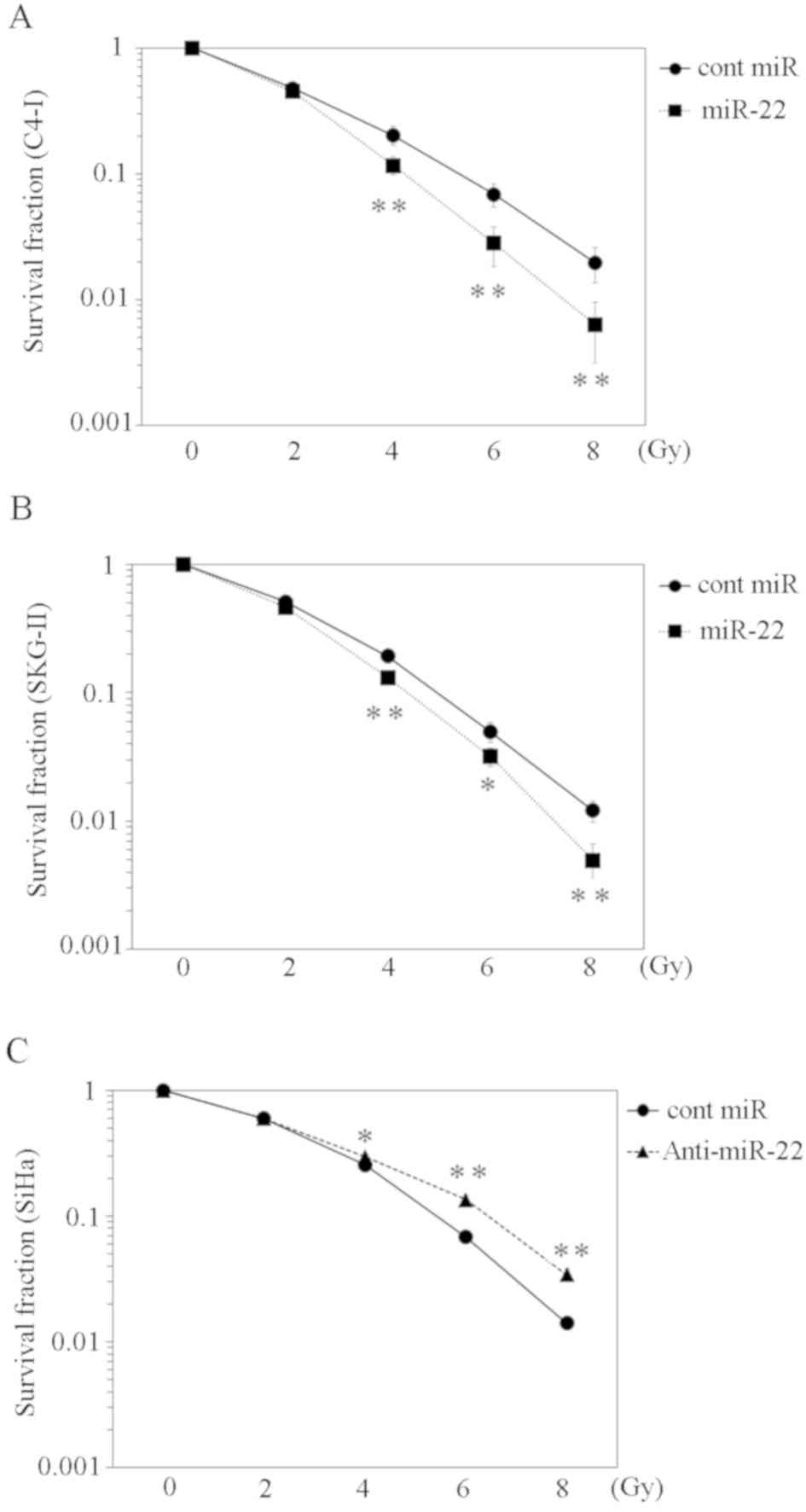
Increased miR-22 expression improves
the radiosensitivity of cervical cancer cell lines in vitro
Telomerase activity reportedly influences the
radiosensitivity of the cervical cancer cell line SiHa (24). To investigate the effect of miR-22 on
radiosensitivity, C-4I and SKG-II cells were transfected with
miR-22 or control miRNA and irradiated with various radiation doses
(2, 4, 6 and 8 Gy). Compared with the control miRNA, the survival
fraction of the miR-22-transfected group was significantly lower
(Fig. 4A and B). Conversely, SiHa
cells with a suppressed miR-22 expression exhibited a higher
survival fraction following irradiation compared with cells
transfected with the control miRNA (Fig.
4C).
A cervical cancer cell line transduced
with miR-22 exhibits an improved radiosensitivity in vivo
Given that the overexpression of miR-22 improved the
radiosensitivity of cervical cancer cells by the subsequent
reduction of hTERT, the therapeutic potential of miR-22 was
assessed in a cervical cancer xenograft model. C-4I cells were
stably transduced with lentiviruses containing precursor miR-22
(L-miR22) or control lentiviral vector (L-cont). Stable
transduction efficiency was confirmed by the expression of GFP
(Fig. 5A). Significant upregulation
of miR-22 in L-miR22-transduced C-4I cells (L-miR22-C-4I) was
confirmed using RT-qPCR (Fig. 5B).
Transduced C-4I cells (L-miR22-C-4I or L-cont-C-4I) were injected
subcutaneously, and then the tumor was irradiated with an X-ray
dose of 6 Gy once the xenograft volume reached 100 mm3.
The tumor growth was significantly inhibited in the L-miR22-C-4I
mice compared with that in the L-cont-C-4I mice (between 7 days and
21 days after irradiation; P<0.05, after 28 days or later;
P<0.01), indicating that miR-22 had a radiosensitizing effect
in vivo (Fig. 5C). A total of
35 days after irradiation, tumor nodules were excised and weighed
(Fig. 5D). It was revealed that the
L-miR22-C-4I tumors were significantly smaller compared with the
L-cont-C-4I tumors (P<0.05; Fig.
5D). In addition, paraffin sections were prepared from the
excised tumor and immunostaining of Ki-67 (Fig. 5E) was performed, in addition to a
TUNEL assay (Fig. 5F), to
investigate the effect on proliferation and apoptosis. The Ki-67
index was significantly lower in the L-miR22-C-4I tumor compared
with that in the L-cont-C-4I tumor group (P<0.05; Fig. 5E). Furthermore, the TUNEL index was
higher in the L-miR22-C-4I tumor group compared with that in the
L-cont-C-4I tumor group, suggesting that miR-22 may influence
apoptosis (P<0.05; Fig. 5F).
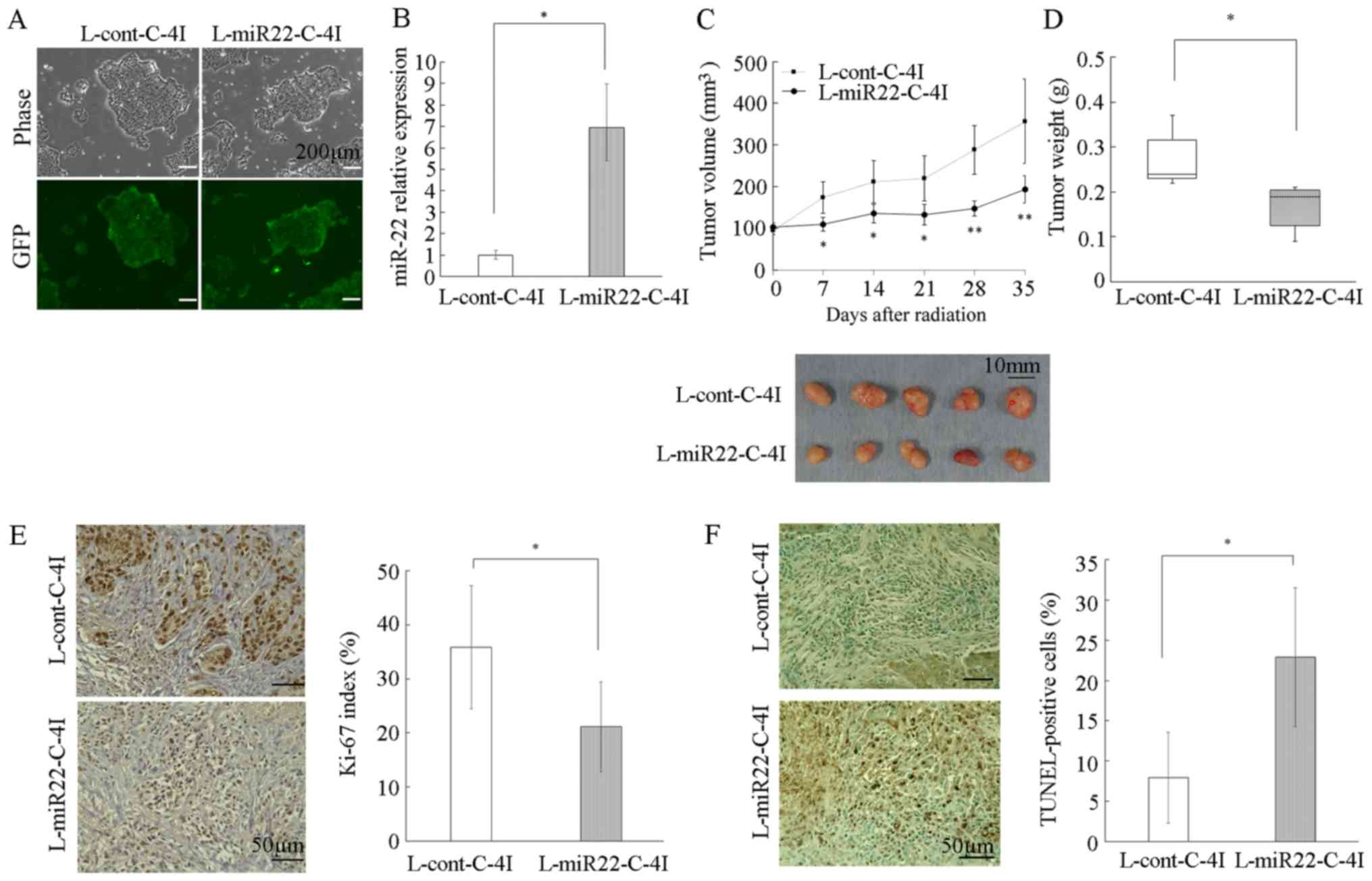 | Figure 5.Effect of miR-22 on the
radiosensitivity of cervical cancer cells in in vivo
xenografts. (A) C-4I cells were infected with miR-22 lentivirus or
control lentivirus, and selected using puromycin. Stably
transfected C-4I cells were examined by phase-contrast microscopy
(upper panel) and fluorescent microscopy (lower panel) (BZ-X700;
Keyence Corporation). Magnification, ×40. Scale bar=200 µm. (B)
miRNA was extracted from C-4I cells stably transfected with
L-miR22-C-4I or with L-cont L-cont-C-4I. The relative abundance of
miR-22 with respect to RNU48 was calculated using RT-qPCR, and
relative fold differences compared with L-cont-C-4I are presented.
(C) Transduced C-4I cells (L-miR22-C-4I or L-cont-C-4I) were
injected into the flanks of nude mice. When tumors reached ~100
mm3, the mice received irradiation treatment (6 Gy).
Tumor volume was measured once a week and each value represents the
mean volume ± SD. Tumor tissues removed from individual 5 mice in
each group are shown. (D) Tumors were excised and weighed five
weeks following irradiation treatment. Each column represent the
mean tumor weight of the five mice ± SD. (E) Immunostaining images
indicate the Ki67 expression of tumor tissue imaged on BZ-X700
microscope (Keyence Corporation). The percentage of Ki67-positive
nuclei in tumor cells was calculated from five fields arbitrarily
selected. Magnification, ×400. Bar=50 µm. (F) Apoptosis of tumors
was evaluated by TUNEL staining. Slides were imaged using a
fluorescence BZ-X700 microscope (Keyence Corporation). The
percentage of TUNEL-positive cells in tumor cells was calculated
from five fields arbitrarily selected. Magnification, ×400. Scale
bar=50 µm. *P<0.05, **P<0.01. miR, microRNA; GFP, green
fluorescent protein; cont, control. |
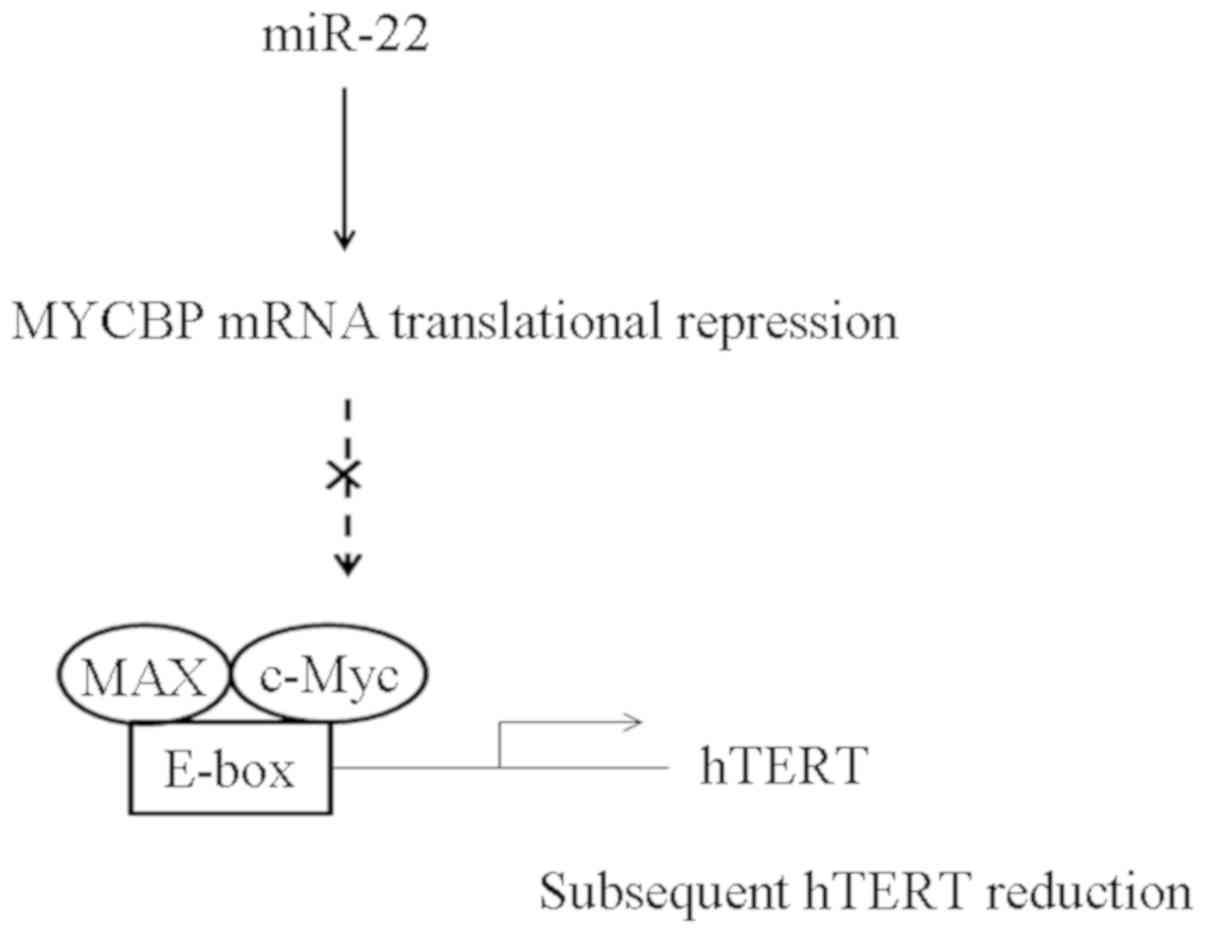
Direct inhibition of MYCBP by miR-22
subsequently reduced hTERT expression
The present study suggests a novel mechanism for
direct miR-22 mediated suppression of MYCBP expression resulting in
the subsequent reduction of c-Myc-mediated transactivation
(Fig. 6).
Discussion
In the present study, it was demonstrated that
miR-22 directly inhibited MYCBP mRNA expression by targeting the
3′UTR of MYCBP and subsequently reduced the hTERT expression level
in cervical cancer cells. Notably, the ectopic expression of miR-22
resulted in increased radiosensitivity both in vitro and
in vivo.
It has previously been indicated that certain miRNAs
serve as promoters of cancer progression such as miR-155 and
miR-221, while others serve as tumor suppressors, such as miR-34
and miR-200 (12,25). miR-22 was originally identified from
HeLa cells on chromosome 17p13 (26), and there is increasing evidence
indicating that miR-22 serves a tumor suppressive role in various
cancer types. For example, cervical cancer cell proliferation was
attenuated by miR-22 via the inhibition of ATP citrate lyase, which
is a key enzyme influencing metabolic activity (27). Furthermore, Li et al (28) reported a negative correlation between
miR-22 expression level and the metastatic potential of ovarian
cancer cells in vitro by analyzing the invasion of SKOV-3
cells. In gastric cancer, miR-22 suppressed invasion and metastasis
via the inhibiting of matrix metalloproteinase 14 and Snail
(29). Moreover, in colorectal
cancer cells, miR-22 promoted apoptosis in response to
5-fluorouracil treatment (30) and a
recent report indicated that a decrease in miR-22 expression in
cervical cancer cells was associated with a poorer prognosis
(15). These results highlight the
potential utility of miR-22 as a therapeutic target in cancers.
miRNA has also been revealed to serve a central role
in modulating the radiosensitivity of cervical cancer cells
(31,32). The overexpression of miR-29b in SiHa
and HeLa cells promoted radiosensitivity via the targeting of
phosphatase and tensin homolog deleted from chromosome 10 (31). Pedroza-Torres et al (32) reported that the overexpression of
miR-125 sensitized the SiHa, CaSki and HeLa cell lines to radiation
therapy, via the downregulation of cyclin-dependent kinase
inhibitor 1. Recently, Zhang et al (33) determined that miR-22 improved
radiosensitivity via targeting silent information regulator 1 in
breast cancer cells. In bone marrow mesenchymal stem cells, miR-22
expression level was increased following irradiation, and served an
important role in the generation of reactive oxygen species and
subsequent apoptosis (34). These
previous reports support the findings of the present study; miR-22
was revealed to enhance radiosensitivity and apoptosis following
irradiation in cervical cancer cells. Notably, the present results
indicated a novel mechanism by which miR-22 regulates the cellular
response to radiation via the modulation of MYCBP and hTERT
expression. It was observed that miR-22 suppressed MYCBP mRNA
expression levels without a change in c-Myc in the
MYCBP/c-Myc/hTERT axis. Bioinformatics analyses indicated a
potential binding site of miR-22 to MYCBP, which was validated by
luciferase reporter assays in the present study. On the other hand,
in silico prediction resources, such as TargetScan and
miRDB, indicated no potential binding site for miR-22 to c-Myc. In
addition, Xing et al revealed that the knockdown of MYCBP
using siRNAs had no significant impact on the c-Myc expression
(16), which supports the present
results.
The MYCBP gene encodes a protein of ~11 kDa, which
binds the N-terminal region of c-Myc via its C-terminal domain and
activates the E-box-dependent transcription activity of c-Myc
(22,35,36).
Previous studies have suggested that MYCBP is an important
regulator affecting the progression and development of tumors; for
example, in glioma cells, the MYCBP mRNA expression increased along
with the malignant grade (36).
Moreover, in gastric cancer, Gong et al (37) reported that MYCBP mRNA expression was
markedly increased compared with that in normal gastric tissues and
knockdown of MYCBP inhibited the metastatic capacity. However, the
influence of MYCBP on radiosensitivity is yet to be elucidated. A
limitation of the present study is that the association between
MYCBP and hTERT was not investigated to determine whether it was
direct or indirect.
In conclusion, the present findings not only
revealed the molecular mechanisms of miR-22 in cervical cancer
cells, but also highlighted a novel potential approach for
radiotherapy through miR-22 in cervical cancer cells. To elucidate
the mechanism underlying miR-22-mediated radio-sensitization in
greater detail, it would be necessary to determine whether the
association between MYCBP and hTERT is direct or indirect, and this
should be investigated in a future study.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
Japan Society for the Promotion of Science; (grant. nos. 25462619
and 17K11304).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
MNa and MH made substantial contributions to the
conception and design of the study, acquisition of the data and/or
statistical analyses and drafting of the manuscript. HK and KA
contributed to the xenograft model assay. MNu was involved in the
reporter assay experiments. YT, HS and MO preformed review and
editing of the manuscript and assisted with data analysis. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The animal welfare guidelines for the care and use
of laboratory animals were followed and experimental protocol was
approved by The Osaka Medical College Animal Care and Use
Committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 coutries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yagi A, Ueda Y, Kakuda M, Tanaka Y, Ikeda
S, Matsuzaki S, Kobayashi E, Morishima T, Miyashiro I, Fukui K, et
al: Epidemiologic and clinical analysis of cervical cancer using
data from the population-based Osaka cancer regstry. Cancer Res.
79:1252–1259. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fokom Domgue J and Schmeler KM:
Conservative management of cervical cancer: Current status and
obstetrical implications. Best Pract Res Clin Obstet Gynaecol.
55:79–92. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chohen PA, Jhingran A, Oaknin A and Denny
L: Cercvical cancer. Lancet. 393:169–182. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wright JD, Matsuo K, Huang Y, Tergas AI,
Hou JY, Khoury-Collado F, St Clair CM, Ananth CV, Neugut AI and
Hershman DL: Prognostic performance of the 2018 international
federation of gynecololgy and obstetrics cervical cancer staging
guidelines. Obstet Gynecol. 134:49–57. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pal J, Gold JS, Munshi NC and Shammas MA:
Biology of telomeres: Importance in etiology of esophageal cancer
and as therapeutic target. Transl Res. 162:364–370. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lord RV, Salonga D, Danenberg KD, Peters
JH, DeMeester TR, Park JM, Johansson J, Skinner KA, Chandrasoma P,
DeMeester SR, et al: Telomerase reverse transcriptase expression is
increased early in the Barrett's metaplasia, dysplasia,
adenocarcinoma sequence. J Gastrointest Surg. 4:135–142. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hoang-Vu C, Boltze C, Gimm O, Poremba C,
Dockhorn-Dworniczak B, Köhrle J, Rath FW and Dralle H: Expression
of telomerase genes in thyroid carcinoma. Int J Oncol. 21:265–272.
2002.PubMed/NCBI
|
|
9
|
Wang R, Lin F, Wang X, Gao P, Dong K, Wei
SH, Cheng SY and Zhang HZ: The therapeutic potential of survivin
promoter-driven siRNA on suppressing tumor growth and enhancing
radiosensitivity of human cervical carcinoma cells via
downregulating hTERT gene expression. Cancer Biol Ther.
6:1295–1301. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kyo S, Takakura M, Taira T, Kanaya T, Itoh
H, Yutsudo M, Ariga H and Inoue M: Sp1 cooperates with c-Myc to
activate transcription of the human telomerase reverse
transcriptase gene (hTERT). Nucleic Acids Res. 28:669–677. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ambros V: MicroRNAs: Tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kaur A, Mackin ST, Schlosser K, Wong FL,
Elharram M, Delles C, Stewart DJ, Dayan N, Landry T and Pilote L:
Systemativ review of microRNA biomarkers in acute coronary syndrome
and stable coronary artery disease. Cardiovasc Res. (pii):
cvz3022019.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ji C and Guo X: The clinical potential of
circulating microRNAs in obesity. Nat Rev Endocrinol. 15:731–743.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang L, Chen B and Ding D: Decreased
microRNA-22 is associated with poor prognosis in cervical cancer.
Int J Clin Exp Pathol. 10:9515–9520. 2017.PubMed/NCBI
|
|
16
|
Xiong J, Du Q and Liang Z:
Tumor-suppressive microRNA-22 inhibits the transcription of
E-box-containing c-Myc target genes by silencing c-Myc binding
protein. Oncogene. 29:4980–4988. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Grimson A, Farh KK, Johnston WK,
Garrett-Engele P, Lim LP and Bartel DP: MicroRNA targeting
specificity in mammals: Determinants beyond seed pairing. Mol Cell.
27:91–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wong N and Wang X: MiRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43((Database Issue)): D146–D152. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schimittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ono YJ, Hayashi M, Tanabe A, Hayashi A,
Kanemura M, Terai Y and Ohmichi M: Estradiol-mediated hepatocyte
growth factor is involved in the implantation of endometriotic
cells via the mesothelial-to-mesenchymal transition in the
peritoneum. Am J Physiol Endocrinol Metab. 308:E950–E959. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Franken NA, Rodermond HM, Stap J, Haveman
J and van Bree C: Clonogenic assay of cells in vitro. Nat Protoc.
1:2315–2319. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Taira T, Maeda J, Onishi T, Kitaura H,
Yoshida S, Kato H, Ikeda M, Tamai K, Iguchi-Ariga SM and Ariga H:
AMY-1, a novel C-MYC binding protein that stimulates transcription
activity of C-MYC. Genes Cells. 3:549–565. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu KJ, Grandori C, Amacker M, Simon-Vermot
N, Polack A, Lingner J and Dalla-Favera R: Direct activation of
TERT transcription by c-MYC. Nat Genet. 21:220–224. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang W and Xing L: RNAi gene therapy of
SiHa cells via targeting human TERT induces growth inhibition and
enhances radiosensitivity. Int J Oncol. 43:1228–1234. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nakatani F, Ferracin M, Manara MC, Ventura
S, Del Monaco V, Ferrari S, Alberghini M, Grilli A, Knuutila S,
Schaefer KL, et al: MiR-34a predicts survival of Ewing's sarcoma
patients and directly influences cell chemo-sensitivity and
malignancy. J Pathol. 226:796–805. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xin M, Qiao Z, Li J, Liu J, Song S, Zhao
X, Miao P, Tang T, Wang L, Liu W, et al: MiR-22 inhibits tumor
growth and metastasis by targeting ATP citrate lyase: Evidence in
osteosarcoma, prostate cancer, cervical cancer and lung cancer.
Oncotarget. 7:44252–44265. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li J, Liang S, Yu H, Zhang J, Ma D and Lu
X: An inhibitory effect of miR-22 on cell migration and invasion in
ovarian cancer. Gynecol Oncol. 119:543–538. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zuo QF, Cao LY, Yu T, Gong L, Wang LN,
Zhao YL, Xiao B and Zou QM: MicroRNA-22 inhibits tumor growth and
metastasis in gastric cancer by directly targeting MMP14 and Snail.
Cell Death Dis. 6:e20002015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang H, Tang J, Li C, Kong J, Wang J, Wu
Y, Xu E and Lai M: MiR-22 regulates 5-FU sensitivity by inhibiting
autophagy and promoting apoptosis in colorectal cancer cells.
Cancer Lett. 356:781–790. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang T, Xue X and Peng H: Therapeutic
delivery of miR-29b enhances radiosensitivity in cervical cancer.
Mol Ther. 27:1183–1194. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pedroza-Torres A, Campos-Parra AD,
Millan-Catalan O, Loissell-Baltazar YA, Zamudio-Meza H, Cantú de
León D, Montalvo-Esquivel G, Isla-Ortiz D, Herrera LA,
Ángeles-Zaragoza Ó, et al: MicroRNA-125 modulates radioresistance
through targeting p21 in cervical cancer. Oncol Rep. 39:1532–1540.
2018.PubMed/NCBI
|
|
33
|
Zhang X, Li Y, Wang D and Wei X: MiR-22
suppresses tumorigenesis and improves radiosensitivity of breast
cancer cells by targeting Sirt1. Biol Res. 50:272017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu Z, Li T, Deng S, Fu S, Zhou X and He
Y: Radiation induces apoptosis and osteogenic impairment through
miR-22-mediated intracellular oxidative stress in bone marrow
mesenchymal stem cells. Stem Cells Int. 2018:58454022018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sakamuro D and Prendergast GC: New
Myc-interacting proteins: A second Myc network emerges. Oncogene.
18:2942–2954. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang H, Yan X, Ji LY, Ji XT, Wang P, Guo
SW and Li SZ: MiR-139 functions as an antioncomir to repress glioma
progression through targeting IGF-1 R, AMY-1, and PGC-1β. Technol
Cancer Res Treat. 16:497–511. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gong L, Xia Y, Qian Z, Shi J, Luo J, Song
G, Xu J and Ye Z: Overexpression of MYC binding protein promotes
invasion and migration in gastric cancer. Oncol Lett. 15:5243–5249.
2018.PubMed/NCBI
|