Introduction
Gastric cancer (GC) is the fourth most commonly
occurring cancer in the world, and the second cause of
cancer-associated deaths worldwide. Approximately 990,000 new cases
of GC occur every year, with ~738,000 associated deaths worldwide
(1). The high mortality rate of GC
is mainly due to limited diagnosis at the earlier phases of the
disease, which consequently means that gastric carcinoma is
detected when is established at a more advanced stage (2). Hence, the identification of GC-specific
tumor biomarkers is warranted for the diagnosis and therapy of
patients with GC.
A non-coding RNA (ncRNA) is a type of RNA that is
not translated into a protein (3).
Due to its structural characteristics, an ncRNA can regulate the
expression of an mRNA to promote or inhibit the production of its
encoded protein and indirectly exert its biological effects. The
ncRNA family includes long ncRNAs (lncRNAs), microRNAs
(miR/miRNAs), small interfering RNAs, small nucleolar RNAs and
ribosomal RNAs (4). lncRNAs
correspond to a class of ncRNAs, which lack open reading frames and
are typically longer than 200 base pairs (5). A number of studies have demonstrated
that lncRNAs are involved in the pathogenesis of many malignancies
and, therefore, could be used as putative biomarkers for the
prognosis of these various conditions. The function of lncRNAs can
be either cancer-promoting or tumor-suppressing, depending on the
level of regulation towards protein-coding genes and/or the
chromosome structure (6,7). lncRNAs play roles in various cellular
processes, mainly regulating cell growth, differentiation and
apoptosis (8,9). miRNA is another type of RNA, ~22
nucleotides in length, that can also control the expression of
protein-coding genes (10). This
type of RNA can modulate gene expression at the
post-transcriptional level, by attaching to miRNA response elements
along the target transcript (11,12). In
2011, the competing endogenous RNA (ceRNA) theory emerged,
introducing novel regulatory mechanisms driven by distinct
non-coding RNAs (12). The main idea
behind this theory involves the competitive cross-regulation of
lncRNAs, mRNAs and miRNAs (13).
Recent studies have revealed that several lncRNAs
are associated with the occurrence, metastasis, prognosis and drug
resistance of GC (14–16). Notwithstanding this, the roles of
lncRNA and miRNA in early and advanced GC are not yet fully
understood. Therefore, further studies on the roles of lncRNA in GC
could be conducive to the diagnosis and therapy of patients with
GC.
In conclusion, the purpose of the present study was
to identify novel and specific lncRNAs associated with GC, and to
provide insight into the features of lncRNAs as biomarkers for the
diagnosis and therapy of patients with GC, thus further improving
the prognosis.
Materials and methods
Data acquisition from The Cancer
Genome Atlas (TCGA)
The expression data of mRNA, lncRNA and miRNA in
patients with GC were downloaded from TCGA database (https://cancergenome.nih.gov). Specifically, the
expression profiles of 393 patients with GC were retrieved from the
TCGA-Stomach Adenocarcinoma (STAD) dataset (17) via the University of California Santa
Cruz Xena database (https://xenabrowser.net/datapages/). The inclusion
criteria were as follows: i) Tissues samples with completed data
for analysis and ii) histologic diagnosis is STAD without other
malignancies. The exclusion criteria were set as follows: i)
Tissues samples without completed data for analysis; ii) histologic
diagnosis is not STAD; iii) patients with additional malignancies
other than GC and iv) patients who received preoperative
chemoradiation. For GC samples containing detailed RNA expression
data, the corresponding clinical details were also accessed,
including survival time, survival status, sex, staging and other
clinical features. A total of 361 cases of GC (233 males and 128
females; mean age, 65.66 years; age range, 30–90 years) and 32
cases of normal (22 males and 10 females; mean age, 68.78 years;
age range, 50–88 years) gastric tissue were included in the present
study. The detailed clinical features of all patients with GC are
summarized in Table I.
 | Table I.Clinicopathological characteristics
of patients with gastric cancer. |
Table I.
Clinicopathological characteristics
of patients with gastric cancer.
|
| Patients
(n=361) |
|---|
|
|
|
|---|
| Characteristic | n | % |
|---|
| Age category,
years |
|
|
|
≤65 | 161 | 44.6 |
|
>65 | 200 | 55.4 |
| Sex |
|
|
|
Male | 233 | 64.5 |
|
Female | 128 | 35.5 |
| Pathological
stage |
|
|
| I | 63 | 17.5 |
| II | 112 | 31.0 |
|
III | 152 | 42.1 |
| IV | 34 | 9.4 |
| Pathological T
stage |
|
|
|
Tis | 8 | 2.2 |
| T1 | 18 | 5.0 |
| T2 | 75 | 20.8 |
| T3 | 164 | 45.4 |
| T4 | 96 | 26.6 |
| Pathological N
stage |
|
|
| N0 | 123 | 34.1 |
| N1 | 96 | 26.6 |
| N2 | 66 | 18.3 |
| N3 | 76 | 21.1 |
| Pathological M
stage |
|
|
| M0 | 322 | 89.2 |
| M1 | 23 | 6.4 |
| Mx | 16 | 4.4 |
| Survival
status |
|
|
|
Alive | 219 | 60.7 |
|
Dead | 142 | 39.3 |
In silico RNA screening
The differentially expressed mRNAs, lncRNAs and
miRNAs (DEmRNAs, DElncRNAs and DEmiRNAs, respectively), in both GC
and non-tumor tissues, were independently selected using the edgeR
package (R language; version 3.28.0; http://bioconductor.org/packages/2.4/bioc/html/edgeR.html)
with a threshold of |log2fold-change
(log2FC)|>2 and false discovery rate (FDR)<0.01
(18). Hierarchical clustering
analysis was completed using the pheatmap package in R (version
1.0.8), in order to estimate the divergence among the
differentially expressed RNAs (19).
Differential mRNA enrichment
analysis
DEmRNAs were added into the Database for Annotation,
Visualization and Integrated Discovery (https://david.ncifcrf.gov/), which takes advantage of
GO (http://geneontology.org/) to determine
the gene function described in the respective gene profiles. The
Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.kegg.jp/) was also utilized as a platform
to investigate the likely functions of the aforementioned genes and
their pathways.
Construction of lncRNAs and mRNA
co-expression networks
lncRNA-mRNA networks were designed as the basis for
lncRNA-mediated control of mRNA abundance. For this, DElncRNAs and
DEmRNAs were selected by comparing GC with normal control tissues.
To determine the respective RNA contents, a standard selection
criteria was set at P<0.05 and fold change >2. The lncRNA and
mRNA co-expression networks were constructed by correlating
DElncRNAs and their respective target mRNAs.
Construction of ceRNA network
TargetScan (http://www.targetscan.org/vert_72/), miRTarBase
(http://mirtarbase.mbc.nctu.edu.tw/php/index.php) and
miRDB (http://mirdb.org/) databases were used to predict
target mRNAs (combined with DEmiRNAs). The prediction analysis of
lncRNA binding to DEmiRNAs was performed using miRcode software
(version 11; http://www.mircode.org). Datasets
were intersected in order to strengthen the correlations between
mRNAs and DEmRNAs. The lncRNA-miRNA-mRNA ceRNA network was formed
on the basis of the DEmiRNA-DElncRNA and DEmiRNA-DEmRNA
interactions, and further visualized using Cytoscape software
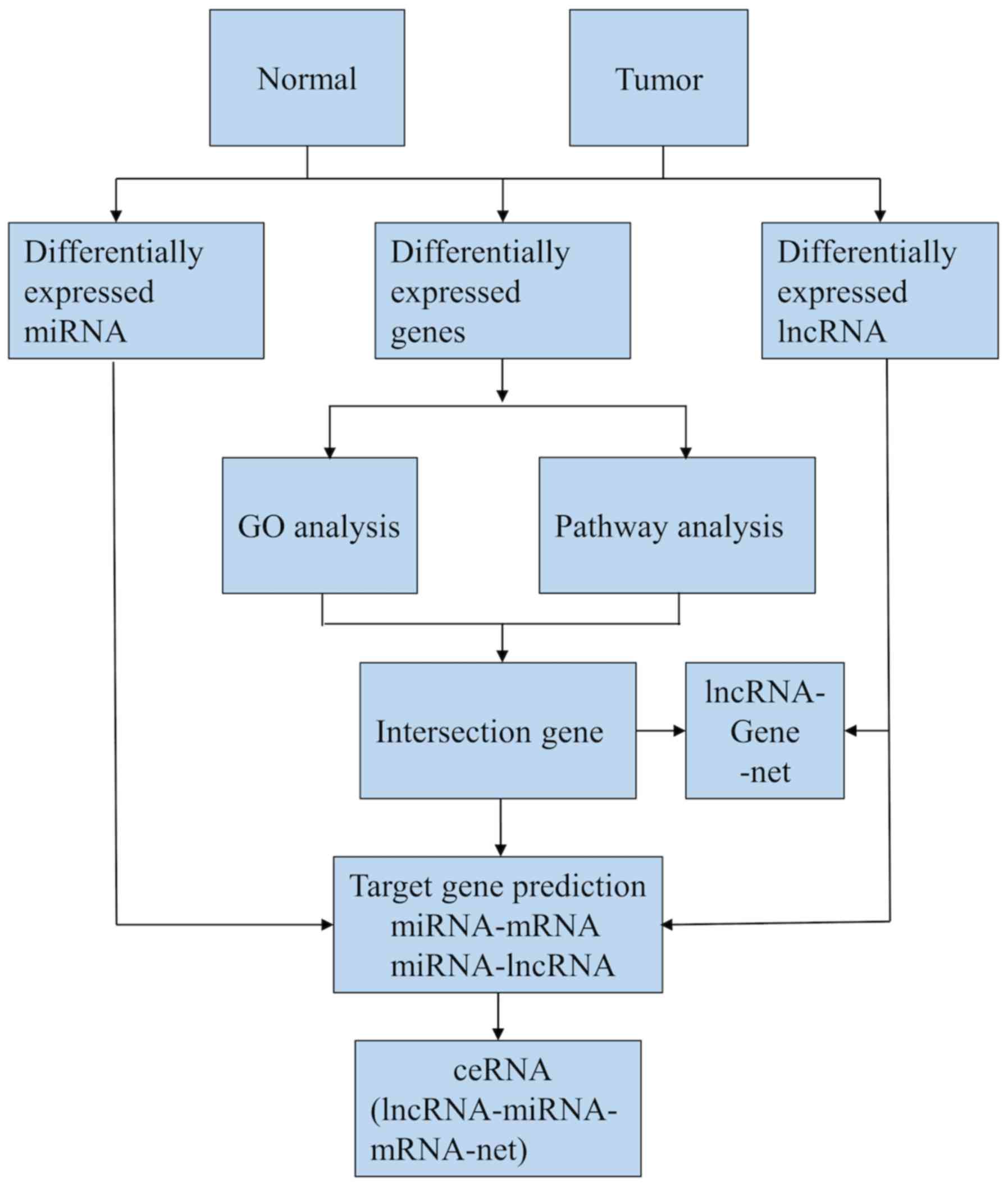
(version 3.7.2; http://cytoscape.org). A flow diagram
of the lncRNA-miRNA-mRNA ceRNA network analysis is shown in
Fig. 1.
Survival analysis
Using the median expression of the differentially
expressed RNAs as the cutoff value, patients with GC were divided
into high and low expression groups. The difference in survival
time between the two groups was evaluated by Kaplan-Meier survival
curve and log-rank test analysis via the SPSS Statistics software,
version 21 (IBM Corp.). As a result, lncRNAs were found
accordingly. P<0.05 was used as a threshold for statistical
significance.
Results
Differential expression of mRNAs,
lncRNAs and miRNAs in GC
Based on the cutoff threshold, a total of 1,370
DEmRNAs (627 upregulated and 743 downregulated RNAs) was selected
from both GC and normal tissues. A total of 393 DElncRNAs (197
upregulated and 196 downregulated RNAs) and 46 DEmiRNAs (24
upregulated and 22 downregulated RNAs) were differentially
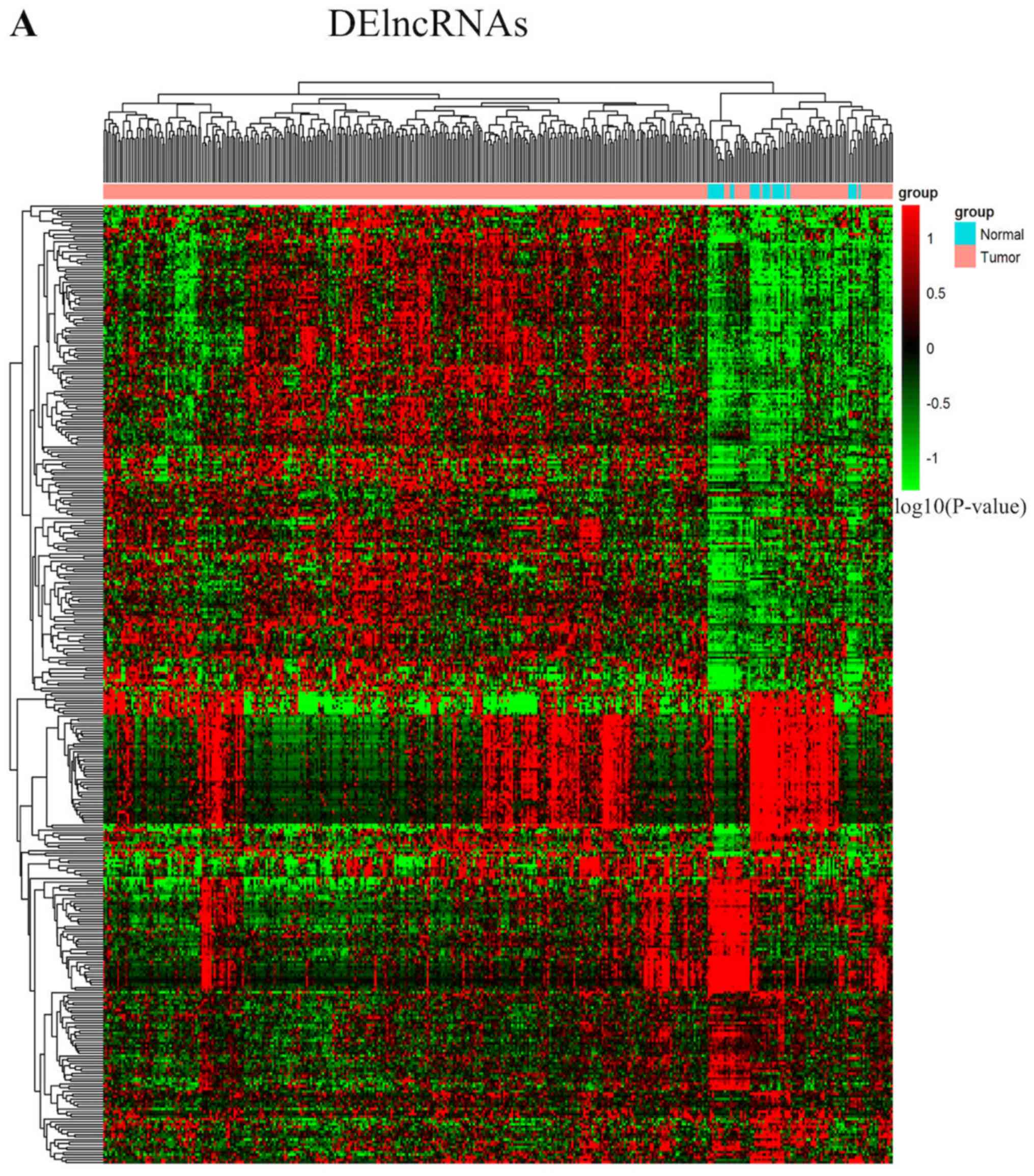
expressed in GC compared with normal tissues. Unsupervised
hierarchical clustering analysis was achieved for all selected
DEmRNAs, DElncRNAs and DEmiRNAs. A heatmap was constructed to
display the distinctive pattern of RNAs expressed in GC compared
with non-tumor samples (Fig. 2A-C).
Thereafter, all differentially expressed RNAs were used for further
analyses.
GO and KEGG pathway analysis
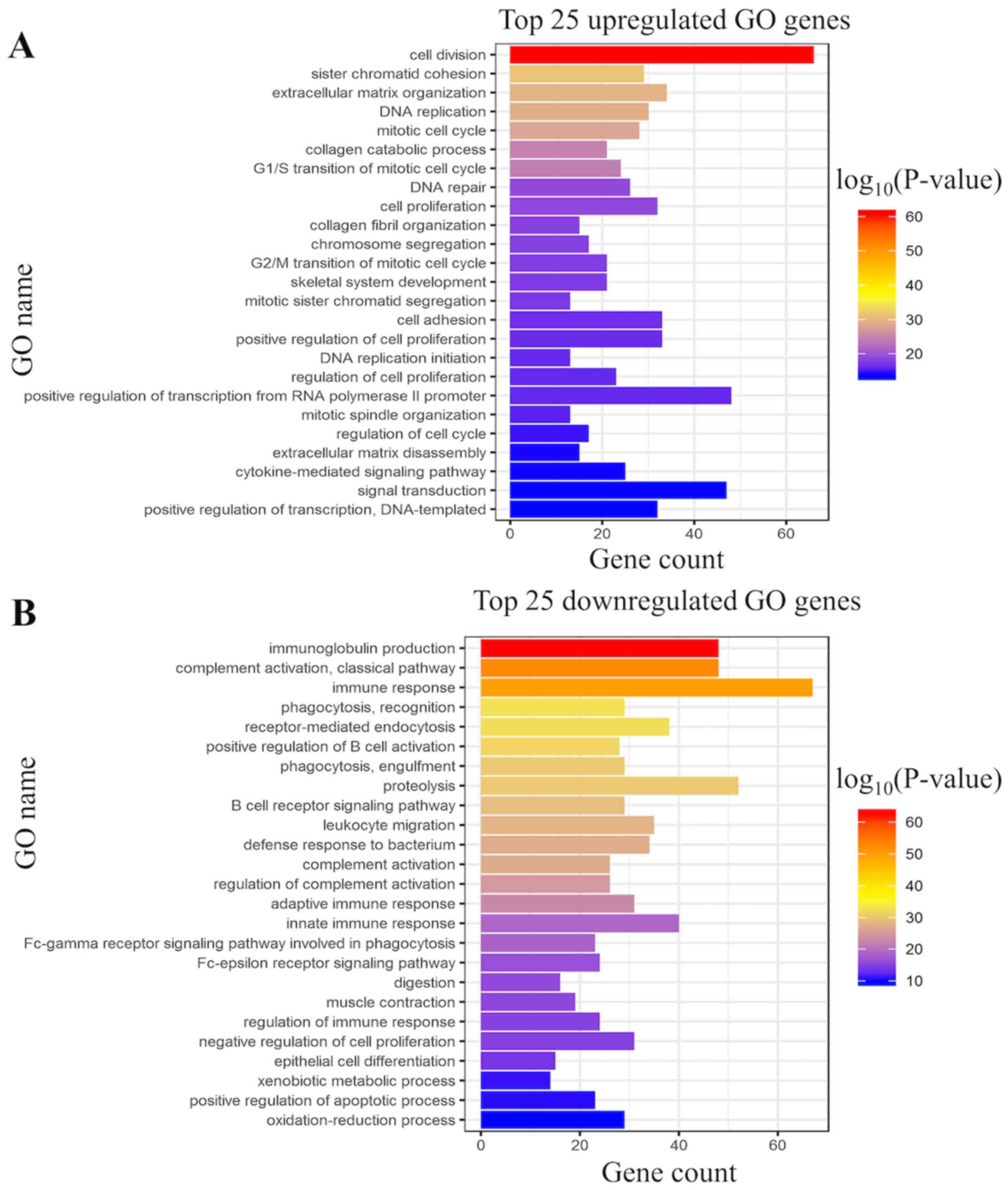
GO and KEGG pathway analysis uncovered 25
upregulated and downregulated GO and KEGG terms. The most
significant GO terms were associated with ‘cell division’,
‘proliferation’ and ‘mitotic cell cycle’ (Fig. 3A and B). KEGG pathway enrichment
analysis was also performed using the selected DEmRNAs. The leading
25 upregulated and downregulated KEGG pathways were found to be
permanently enriched by DEmRNAs in the ceRNA network, among which
the ‘PI3K-Akt signaling pathway’, ‘cell cycle’, ‘pathways in
cancer’ and the ‘p53 signaling pathway’ were most closely
associated with the pathogenesis of cancer (Fig. 3C and D).
Construction of lncRNA-mRNA
co-expression network
Subsequently, an lncRNA-mRNA co-expression network
was established using the GO terms and pathways compiled by
respective target genes and DElncRNAs. The statistical basis of the
co-expression network was Pearson correlation index |r|>0.9 in a
network where both lncRNA-mRNA and mRNA-mRNA connections were
mutually regulated. The co-expression network in GC and normal
gastric tissues was constructed according to the Pearson's
correlation coefficient, which measured values of lncRNA and mRNA.
Based on preceding conclusions (Figs.
2A and 3), the lncRNA-mRNA
co-expression network was created among differentially expressed
lncRNAs and mRNAs in both GC and normal gastric tissues. The
results revealed network associations between 12 lncRNAs and 57
mRNAs (Fig. S1).
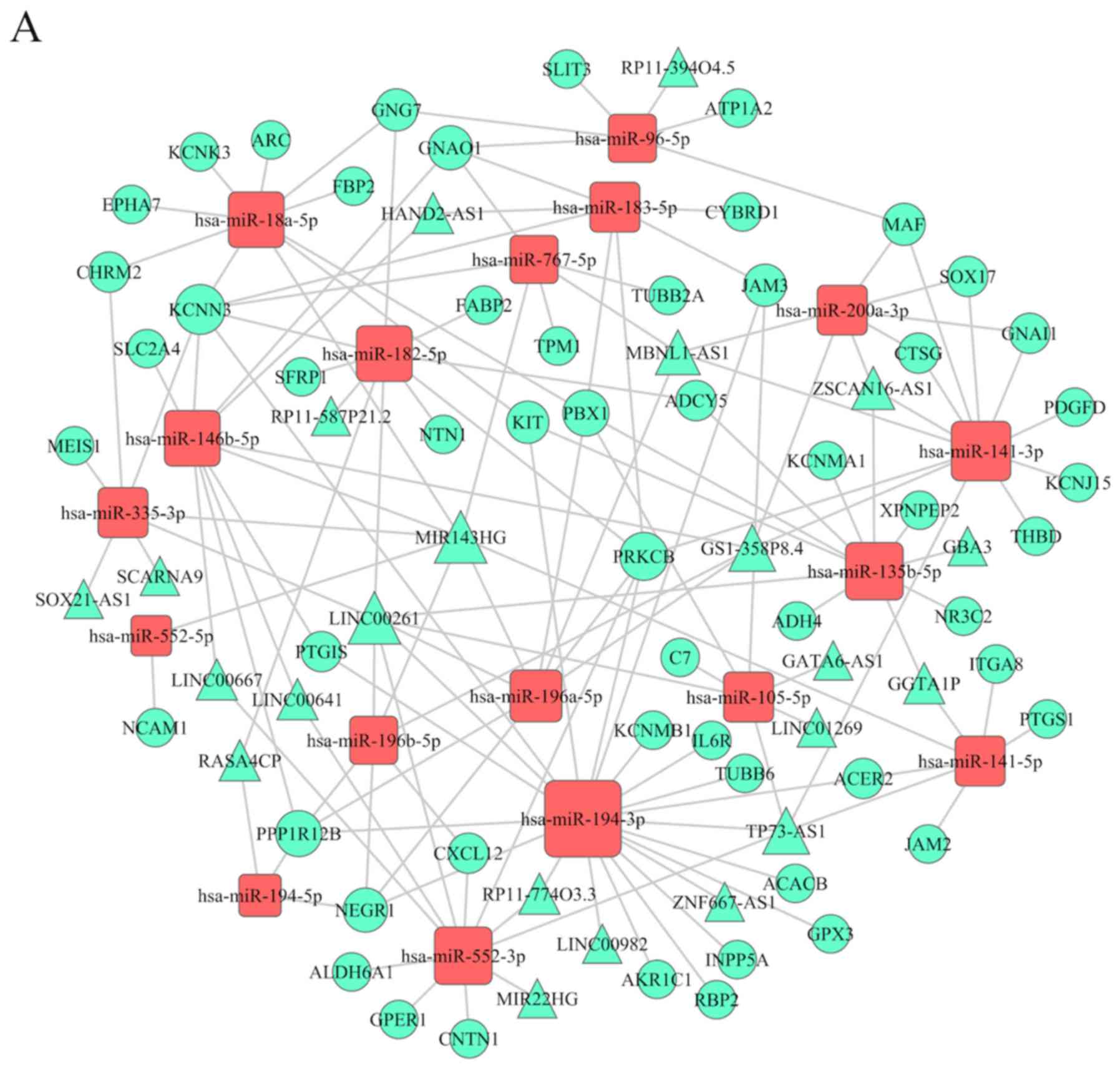
DElncRNA-DEmiRNA interactions
predicted by miRcode
The prediction analysis of lncRNA binding to
DEmiRNAs was performed using miRcode. As such, predicted lncRNAs
and DElncRNAs were intersected, and the intersecting lncRNAs were
included into the ceRNA network. Correspondingly, 342 interactions
between 64 DElncRNAs (including PVT1, RP11-284F21.7,
RP11-284F21.10, HAND2-AS1 and ZNF667-AS1) and 35 DEmiRNAs
(including hsa-miR-194-3p, hsa-miR-196b-5p, hsa-miR-218-5p and
hsa-miR-145-3p) were annotated and further analyzed. The top 10
miRNA-mediated lncRNAs are shown in Table II.
 | Table II.Top 10 DElncRNAs putatively targeted
by most DEmiRNAs in the competing endogenous RNA network. |
Table II.
Top 10 DElncRNAs putatively targeted
by most DEmiRNAs in the competing endogenous RNA network.
| DElncRNAs | DEmiRNAs |
|---|
| MIR143HG | hsa-miR-141-5p,
hsa-miR-146b-5p, hsa-miR-18a-5p, hsa-miR-196a-5p, hsa-miR-196b-5p,
hsa-miR-335-3p, hsa-miR-552-5p, hsa-miR-767-5p |
| LINC00261 | hsa-miR-105-5p,
hsa-miR-135b-5p, hsa-miR-182-5p, hsa-miR-194-3p, hsa-miR-196a-5p,
hsa-miR-335-3p, hsa-miR-552-3p |
| TP73-AS1 | hsa-miR-105-5p,
hsa-miR-141-3p, hsa-miR-141-5p, hsa-miR-194-3p, hsa-miR-552-3p |
| GS1-358P8.4 | hsa-miR-141-3p,
hsa-miR-146b-5p, hsa-miR-196a-5p, hsa-miR-196b-5p,
hsa-miR-200a-3p |
| MBNL1-AS1 | hsa-miR-141-3p,
hsa-miR-196a-5p, hsa-miR-200a-3p, hsa-miR-767-5p |
| AFAP1-AS1 | hsa-miR-129-5p,
hsa-miR-145-3p, hsa-miR-149-5p, hsa-miR-30c-2-3p |
| NSUN5P1 | hsa-miR-129-5p,
hsa-miR-139-3p, hsa-miR-30c-2-3p, hsa-miR-5683 |
| RP11-242D8.1 | hsa-miR-145-3p,
hsa-miR-149-5p, hsa-miR-30c-2-3p |
| MIR4435-2HG | hsa-miR-1-3p,
hsa-miR-145-5p, hsa-miR-149-5p |
| TUG1 | hsa-miR-144-3p,
hsa-miR-29c-3p, hsa-miR-5683 |
Prediction of DEmiRNA-interacting
DEmRNAs
Previously annotated DEmiRNAs were mapped into
TargetScan, miRTarBase and miRDB databases to search for their
target mRNAs. A total of 357 mRNAs were converged by these DEmiRNAs
in all three databases. As a result, 118 mRNAs were determined as
the DEmiRNA-targeted DEmRNAs. To underline the ceRNA
characteristics of the network, DEmiRNAs that were capable of
combining with both DEmRNAs and DElncRNAs were introduced into the
ceRNA network. Ultimately, 37 DEmiRNAs, 118 DEmRNAs and 65
DElncRNAs were selected for ceRNA network construction. The top 27
miRNAs, which were predicted to modulate most of the mRNAs, are
shown in Table III.
 | Table III.DEmiRNAs (n=27) with corresponding
target DEmRNAs in the competing endogenous RNA network. |
Table III.
DEmiRNAs (n=27) with corresponding
target DEmRNAs in the competing endogenous RNA network.
| DEmiRNAs | DEmRNAs |
|---|
| hsa-miR-194-3p | ACACB, ACER2,
AKR1C1, GPX3, IL6R, INPP5A, JAM3, KCNMB1, KCNN3, KIT, NEGR1,
PPP1R12B, PRKCB, PTGIS, RBP2, TUBB6 |
|
hsa-miR-30c-2-3p | LYN, APLN, CDC25A,
CDH1, CTSB, CXCL10, CXCL9, E2F3, HEYL, ITGA11, LIF, MYBL2, OAS3,
PAICS, SERPINB5 |
| hsa-miR-195-5p | APLN, BIRC5, CDK6,
CHEK1, HOXA10, MYB, RUNX1, THBS2 |
| hsa-miR-141-3p | CTSG, GNAI1,
KCNJ15, MAF, PDGFD, PRKCB, SOX17, THBD |
| hsa-miR-145-5p | ACTG1, CDK6, CXCL3,
F11R, OAS3, SERPINE1, TNFRSF10B |
|
hsa-miR-135b-5p | ADCY5, ADH4,
KCNMA1, KIT, NR3C2, PBX1, XPNPEP2 |
| hsa-miR-18a-5p | ARC, CHRM2, EPHA7,
FBP2, GNG7, KCNK3, KIT, PBX1 |
| hsa-miR-182-5p | ADCY5, FABP2, GNG7,
KCNN3, NTN1, PRKCB, SFRP1 |
| hsa-miR-183-5p | CYBRD1, GNAO1,
JAM3, KCNN3, PBX1, PRKCB |
| hsa-miR-5683 | BID, CCNB1, CDK6,
HNF4A, IDO1, ITGA2, OAS3 |
| hsa-miR-552-3p | ALDH6A1, CNTN1,
CXCL12, GPER1, PRKCB |
|
hsa-miR-146b-5p | GNAO1, KCNN3,
PPP1R12B, PTGIS, SLC2A4 |
| hsa-miR-204-5p | CDC7, CDK6, INHBA,
RAD51, TNFRSF12A |
| hsa-miR-149-5p | CXCL5, F2RL2,
NOTCH3, OAS2, PLAU |
| hsa-miR-96-5p | ATP1A2, GNAO1,
GNG7, MAF, SLIT3 |
|
hsa-miR-196a-5p | NEGR1, PBX1,
PPP1R12B, PRKCB |
| hsa-miR-29c-3p | CCNA2, CLDN1,
COL6A3, HNF4G |
| hsa-miR-767-5p | GNAO1, KCNN3, TPM1,
TUBB2A |
| hsa-miR-133b | CHDH, CXCL11,
EFNA3, MMP14 |
| hsa-miR-490-3p | CDC6, MTHFD1L,
SKP2, TFRC |
| hsa-miR-141-5p | ACER2, ITGA8, JAM2,
PTGS1 |
|
hsa-miR-196b-5p | CXCL12, NEGR1,
PPP1R12B |
|
hsa-miR-200a-3p | CTSG, GNAI1, MAF,
SOX17 |
| hsa-miR-335-3p | CHRM2, KCNN3,
MEIS1 |
|
hsa-miR-133a-3p | CDH3, CXCL11,
EFNA3 |
| hsa-miR-139-5p | EFNA3, HNF4G,
RFC3 |
| hsa-miR-144-3p | COL11A1, NDC1 |
Profile of the ceRNA network in
GC
Based on data from Tables II and III, and Fig.
S1, an miRNA-lncRNA-mRNA ceRNA network was constructed. The
miRNA-lncRNA-mRNA associations were integrated into the ceRNA
network, according to their mutual regulation roles. The ceRNA
network was constructed with 220 modules (including 37 DEmiRNAs,
118 DEmRNAs and 65 DElncRNAs) and 717 interactions. The network was
profiled using Cytoscape (Fig. 4A and
B). This network demonstrated the possibility of indirect
associations between DElncRNAs and DEmRNAs.
lncRNAs associated with the prognosis
of patients with GC
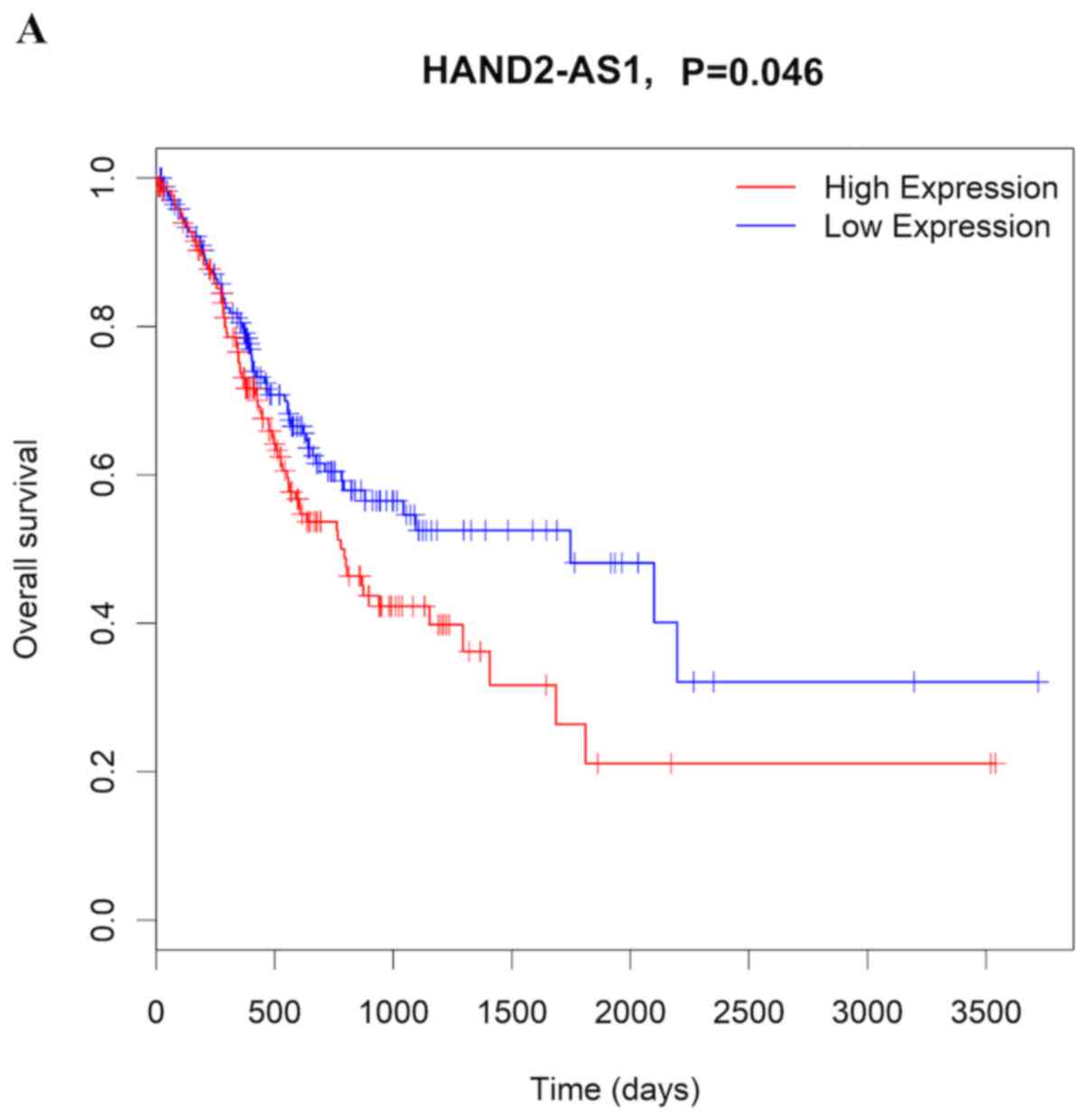
In order to discover lncRNAs that are significantly
correlated with the prognosis of patients with GC, Kaplan-Meier
survival curves and log-rank test analyses were conducted for each
of the DElncRNAs comprising the ceRNA network. The results
demonstrated that three distinct DElncRNAs (PVT1, ZNF667-AS1 and
HAND2-AS1) were associated with survival (P<0.05; Fig. 5A-C). ZNF667-AS1 and HAND2-AS1
appeared to be pro-cancerous, since patients with higher levels of
these RNAs presented with shorter overall survival time (Fig. 5A and C). By contrast, PVT1 appeared
to have a protective role, since its high expression was associated
with longer overall survival time in patients with GC (Fig. 5C).
Discussion
GC is one of the most common types of cancer
worldwide, and it largely affects the quality of life and the
health of Asian populations. In China, the incidence and mortality
rates of GC have escalated to be the second highest among all
cancer types (20). Although the
diagnosis and treatment of GC have significantly improved, the
5-year overall survival rate of patients with advanced GC is still
~20%, mostly due to late diagnosis, rapid progression and
metastatic characteristics (21,22).
Therefore, the discovery and characterization of early GC
biomarkers is imperative for a more efficient diagnosis and
treatment of the disease.
The role of lncRNAs in cancer has become
increasingly important. Moreover, lncRNAs appear to play a vital
role in cancer proliferation, development and prognosis. For
example, MEG3 inhibits GC metastasis by activating the p53
signaling pathway (23). The lncRNA
AWPPH enhances the growth and proliferation of triple-negative
breast cancer by upregulating FZD7 (24), whereas cancer progression and
shortened survival in patients with colon cancer are associated
with the overexpression of the lncRNA DANCR (25).
A number of studies have shown that lncRNAs can
regulate diverse biological activities in tumor cells, including
transcriptional, post-transcriptional and phenotype regulation
(26–28). lncRNAs influence the stability of
target-encoding genes by modulating the activity of miRNAs
(29). In the context of GC, a
complicated regulatory network involving lncRNAs and miRNAs or
mRNAs plays a vital role in the occurrence and development of the
disease (30,31). Therefore, the concept of ceRNAs was
proposed in order to explain the mutual regulatory mechanism
between non-coding and coding RNAs during cancer progression, as
well as its influence on the survival of patients with cancer
(11). Compared with the coding
RNAs, lncRNAs have a greater advantage as an effective biomarker
for the diagnosis and prognosis of early GC (32).
A number of lncRNAs have been found to be associated
with GC. It is also well known that Helicobacter pylori
infection plays a role in the pathogenesis of GC. In vitro
and in vivo studies have revealed that infection by H.
pylori can, for example, drive the downregulation of lncRNA
AF147447 in GC. lncRNA AF147447 can suppress the proliferation and
invasion of GC by repressing MU2 and upregulating the expression of
miR-34c (33). The lncRNA NORAD is
highly expressed in GC tissues and cells, and is strongly
correlated with poor clinical outcomes in GC. Furthermore, NORAD
may act as a ceRNA to promote the outgrowth of GC cells, by
regulating the miR-608/FOXO6 signaling cascade (34). The lncRNA LINC00460 has also been
suggested as an independent risk factor for the prognosis of GC,
since it can promote the proliferation and migration of cancer
cells by stimulating the Wnt/β-catenin pathway (35). In addition, the lncRNA FLVCR1-AS1
sponges miR-155 to further promote the development of GC (36).
Following TCGA analysis of the DElncRNA content in
GC and paracancerous tissue samples, MIR100HG was found to be
highly expressed in GC and, thus, could be considered as a
potential prognostic marker of GC (37). AKT protein kinase has been widely
studied as a cancer-promoting factor. A recent study reported that
lncRNA STXBP5-AS1 can inhibit the activation of AKT in GC, which
leads to the inhibition of proliferation and migration of GC cells
(38).
The present study attempted to identify a highly
specific lncRNA in GC, and to further understand its impact during
the pathogenesis and progression of the disease. In order to
achieve this aim, the expression profiles of lncRNAs, miRNAs and
mRNAs were first assessed in 361 GC and 32 adjacent and normal
tissues from TCGA database. A detailed bioinformatics analysis of
GC compared with normal tissues uncovered some DElncRNAs (PVT1,
RP11-284F21.7 and RP11-284F21.10) that were significantly
upregulated, and at least two DElncRNAs (HAND2-AS1 and ZNF667-AS1)
that were significantly downregulated. Moreover, a number of
DEmiRNAs and DEmRNAs were upregulated and downregulated. In
particular, enriched DEmRNAs were closely associated to processes
that impact carcinogenesis, including ‘cell division’,
‘proliferation’ and the ‘mitotic cell cycle’. Moreover, a number of
DEmRNAs were associated with the ‘PI3K-Akt signaling pathway’
(39), ‘pathways in cancer’ and the
‘p53 signaling pathway’ (40) in GC,
and abnormalities in these pathways are also associated with
cancer. Subsequently, the association between the three RNAs was
analyzed. Furthermore, a ceRNA network map was constructed, and
Kaplan-Meier survival analyses were performed to establish a
potential association between differential RNAs and the prognosis
of GC. The lncRNAs HAND2-AS1, PVT1 and ZNF667-AS1 were strongly
associated with the prognosis of GC and could be used as biomarkers
for predicting the prognosis of the disease.
The role of PVT1 in other cancer types has been
previously reported. For instance, it was shown that PVT1 was
highly expressed in patients with glioma at stages III–IV (versus
patients at stages I–II), and the survival time of these patients
with high expression of PVT1 was significantly shorter (41). PVT1 is also involved in the
sensitivity of radiotherapy affecting lung cancer (42). The role of lncRNA HAND2-AS1, which
was also reported in the present study, has been similarly
identified in other cancer types. As an example, HAND2-AS1 appears
to inhibit the proliferation and migration of esophageal squamous
cell tumors (43). HAND2-AS1 can
also impede the growth and metastasis of colon cancer by
upregulating KLF5 (44). In lung
cancer, HAND2-AS1 can also act as a tumor suppressor (45). However, the roles of ZNF667-AS1 and
HAND2-AS1 in GC has not been previously reported. To the best of
our knowledge, the present study is the first to demonstrate that
high expression of ZNF667-AS1 and HAND2-AS1 is negatively
associated with the survival time of patients with GC.
The construction of an lncRNA-miRNA-mRNA ceRNA
network revealed the associations between novel specific lncRNAs
and other RNAs in GC. Moreover, a putative association between
differentially expressed RNAs and the prognosis of patients with GC
was elucidated. The findings of the present study may have
substantial clinical significance; however, some limitations should
be taken into consideration. Firstly, the data were downloaded from
TCGA databases; thus, experimental methods are required to verify
the results. Secondly, the role of HAND2-AS1, PVT1 and ZNF667-AS1
in GC is still unknown; therefore, in vivo and in
vitro experiments are required to answer this question.
In conclusion, the data from the present study will
serve as a basis for the definition and validation of potential
biomarkers for the diagnosis and targeted therapy for GC. Future
functional investigations are required to explore the mechanisms
underlying the roles of these lncRNAs in GC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by the Chinese National
Natural Science Foundation (grant no. 81871995) and The Special
Funding for Doctoral Team of the First Affiliated Hospital of
Zhengzhou University (grant no. 2016-BSTDJJ-11).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
YF and SP conceived and designed the present study,
SP drafted the manuscript and YF revised the initial manuscript. XY
acquired the data. SP, XY, YZ, WM, TL, YY, JJ and QL analyzed and
interpreted the data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sun Z, Wang ZN, Zhu Z, Xu YY, Xu Y, Huang
BJ, Zhu GL and Xu HM: Evaluation of the seventh edition of American
joint committee on cancer TNM staging system for gastric cancer:
Results from a Chinese monoinstitutional study. Ann Surg Oncol.
19:1918–1927. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hombach S and Kretz M: Non-coding RNAs:
Classification, biology and functioning. Adv Exp Med Biol.
937:3–17. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv
XW and Li J: Long noncoding RNAs: Novel insights into hepatocelluar
carcinoma. Cancer Lett. 344:20–27. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li CH and Chen Y: Insight into the role of
long noncoding RNA in cancer development and progression. Int Rev
Cell Mol Biol. 326:33–65. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kino T, Hurt DE, Ichijo T, Nader N and
Chrousos GP: Noncoding RNA gas5 is a growth arrest- and
starvation-associated repressor of the glucocorticoid receptor. Sci
Signal. 3:ra82010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cai Y, Yu X, Hu S and Yu J: A brief review
on the mechanisms of miRNA regulation. Genomics Proteomics
Bioinformatics. 7:147–154. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chan JJ and Tay Y: Noncoding RNA:RNA
regulatory networks in cancer. Int J Mol Sci. 19:E13102018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li H, Yu B, Li J, Su L, Yan M, Zhu Z and
Liu B: Overexpression of lncRNA H19 enhances carcinogenesis and
metastasis of gastric cancer. Oncotarget. 5:2318–2329. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu X, Zhang P, Zhu H, Li S, Chen X and Shi
L: Long noncoding RNA FEZF1-AS1 indicates a poor prognosis of
gastric cancer and promotes tumorigenesis via activation of Wnt
signaling pathway. Biomed Pharmacother. 96:1103–1108. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen QN, Wei CC, Wang ZX and Sun M: Long
non-coding RNAs in anti-cancer drug resistance. Oncotarget.
8:1925–1936. 2017.PubMed/NCBI
|
|
17
|
Goldman M, Craft B, Swatloski T, Cline M,
Morozova O, Diekhans M, Haussler D and Zhu J: The UCSC cancer
genomics browser: Update 2015. Nucleic Acids Res. 43:D812–D817.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Anders S and Huber W: Differential
expression analysis for sequence count data. Genome Biol.
11:R1062010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kimes PK, Liu Y, Neil Hayes D and Marron
JS: Statistical significance for hierarchical clustering.
Biometrics. 73:811–821. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Otterstad JE, Forfang K, Vatne K and
Frøysaker T: Posterior left ventricular aneurysm due to occlusion
of the circumflex coronary artery with recurrent ventricular
tachycardia. Case report. Scand J Thorac Cardiovasc Surg.
16:205–208. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu H, Dong Y, Gao Y, Du Z, Wang Y, Cheng
P, Chen A and Huang H: The fascinating effects of baicalein on
cancer: A review. Int J Mol Sci. 17:E16812016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wei GH and Wang X: lncRNA MEG3 inhibit
proliferation and metastasis of gastric cancer via p53 signaling
pathway. Eur Rev Med Pharmacol Sci. 21:3850–3856. 2017.PubMed/NCBI
|
|
24
|
Wang K, Li X, Song C and Li M: LncRNA
AWPPH promotes the growth of triple-negative breast cancer by
up-regulating frizzled homolog 7 (FZD7). Biosci Rep.
38:BSR201812232018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu Y, Zhang M, Liang L, Li J and Chen YX:
Over-expression of lncRNA DANCR is associated with advanced tumor
progression and poor prognosis in patients with colorectal cancer.
Int J Clin Exp Pathol. 8:11480–11484. 2015.PubMed/NCBI
|
|
26
|
Muers M: RNA: Genome-wide views of long
non-coding RNAs. Nat Rev Genet. 12:7422011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kornienko AE, Guenzl PM, Barlow DP and
Pauler FM: Gene regulation by the act of long non-coding RNA
transcription. BMC Biol. 11:592013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wahlestedt C: Targeting long non-coding
RNA to therapeutically upregulate gene expression. Nat Rev Drug
Discov. 12:433–446. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li T, Mo X, Fu L, Xiao B and Guo J:
Molecular mechanisms of long noncoding RNAs on gastric cancer.
Oncotarget. 7:8601–8612. 2016.PubMed/NCBI
|
|
30
|
Zeng X, Zhang X and Zou Q: Integrative
approaches for predicting microRNA function and prioritizing
disease-related microRNA using biological interaction networks.
Brief Bioinform. 17:193–203. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu Y, Zeng X, He Z and Zou Q: Inferring
microRNA-disease associations by random walk on a heterogeneous
network with multiple data sources. IEEE/ACM Trans Comput Biol
Bioinform. 14:905–915. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sanchez Calle A, Kawamura Y, Yamamoto Y,
Takeshita F and Ochiya T: Emerging roles of long non-coding RNA in
cancer. Cancer Sci. 109:2093–2100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou X, Chen H, Zhu L, Hao B, Zhang W, Hua
J, Gu H, Jin W and Zhang G: Helicobacter pylori infection related
long noncoding RNA (lncRNA) AF147447 inhibits gastric cancer
proliferation and invasion by targeting MUC2 and up-regulating
miR-34c. Oncotarget. 7:82770–82782. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Miao Z, Guo X and Tian L: The long
noncoding RNA NORAD promotes the growth of gastric cancer cells by
sponging miR-608. Gene. 687:116–124. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang S, Xu J, Wang H and Guo H:
Downregulation of long noncoding RNA LINC00460 expression
suppresses tumor growth in vitro and in vivo in gastric cancer.
Cancer Biomark. 24:429–437. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu Y, Guo G, Zhong Z, Sun L, Liao L, Wang
X, Cao Q and Chen H: Long non-coding RNA FLVCR1-AS1 sponges miR-155
to promote the tumorigenesis of gastric cancer by targeting c-Myc.
Am J Transl Res. 11:793–805. 2019.PubMed/NCBI
|
|
37
|
Li J, Xu Q, Wang W and Sun S: MIR100HG: a
credible prognostic biomarker and an oncogenic lncRNA in gastric
cancer. Biosci Rep. 39:BSR201901712019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cen D, Huang H, Yang L, Guo K and Zhang J:
Long noncoding RNA STXBP5-AS1 inhibits cell proliferation,
migration, and invasion through inhibiting the PI3K/AKT signaling
pathway in gastric cancer cells. Onco Targets Ther. 12:1929–1936.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Noorolyai S, Shajari N, Baghbani E,
Sadreddini S and Baradaran B: The relation between PI3K/AKT
signalling pathway and cancer. Gene. 698:120–128. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Joerger AC and Fersht AR: The p53 pathway:
Origins, inactivation in cancer, and emerging therapeutic
approaches. Annu Rev Biochem. 85:375–404. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fang J and Huang J: Clinical significance
of the expression of long non-coding RNA PVT1 in glioma. Cancer
Biomark. 24:509–513. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang D and Hu Y: Long non-coding RNA PVT1
competitively binds microRNA-424-5p to regulate CARM1 in
radiosensitivity of non-small-cell lung cancer. Mol Ther Nucleic
Acids. 16:130–140. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yan Y, Li S, Wang S, Rubegni P, Tognetti
L, Zhang J and Yan L: Long noncoding RNA HAND2-AS1 inhibits cancer
cell proliferation, migration, and invasion in esophagus squamous
cell carcinoma by regulating microRNA-21. J Cell Biochem.
120:9564–9571. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhou J, Lin J, Zhang H, Zhu F and Xie R:
LncRNA HAND2-AS1 sponging miR-1275 suppresses colorectal cancer
progression by upregulating KLF14. Biochem Biophys Res Commun.
503:1848–1853. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Miao F, Chen J, Shi M, Song Y, Chen Z and
Pang L: LncRNA HAND2-AS1 inhibits non-small cell lung cancer
migration, invasion and maintains cell stemness through the
interactions with TGF-β1. Biosci Rep. 39:BSR201815252019.
View Article : Google Scholar : PubMed/NCBI
|