Introduction
The antimetabolite agent 5-fluorouracil (5-FU) is
widely used in the treatment of many cancer types, including
gastrointestinal cancers, breast cancers, lung cancers, and cancers
of the aerodigestive tract (1).
5-FU inhibits DNA synthesis and RNA processing,
which in turn affects cell proliferation and survival. 5-FU is
converted to 5-fluorodeoxyuridine monophosphate (FdUMP) in cells.
FdUMP binds to the nucleotide-binding site of thymidylate synthase
(TS), an enzyme that catalyzes the reaction from deoxyuridine
monophosphate (dUMP) to deoxythymidine monophosphate (dTMP), and
inhibits its enzymatic activity. As a result, it causes depletion
or imbalance of the intracellular deoxynucleotide pool (2). FdUMP is also converted into
5-fluorodeoxyuridine triphosphate (FdUTP), which itself is a
substrate for DNA polymerase and is readily misincorporated into
DNA (3). Consequently, 5-FU inhibits
DNA synthesis and repair and results in DNA damage (1). 5-FU can also be converted to
5-fluorouridine triphosphate (FUTP), which misincorporates into RNA
molecules, particularly ribosomal RNA (rRNA), and leads to
inhibition of rRNA processing (4–6). As a
result, 5-FU suppresses cell proliferation.
It has been shown that nucleolar/ribosomal
biogenesis stress, such as inhibition of rRNA synthesis,
processing, or ribosome subunit assembly, activates tumor
suppressor P53 via ribosomal proteins (RPs), including RPL5, RPL11,
and RPL23 (7–13). In response to this stress, the
nucleolus is disrupted, and RPL5, RPL11, and RPL23 are consequently
released from the nucleolus to the nucleoplasm where they bind to
and suppress MDM2, an E3 ubiquitin ligase that ubiquitinates and
targets P53 for proteasome-mediated degradation, and causes P53
stabilization and activation, resulting in cell growth suppression
(7–13). Recently, 5-FU has been reported to
suppress tumor growth through RPL11-mediated activation of P53 in
osteosarcoma (14). However, it is
unknown whether 5-FU inhibits tumor growth through RPL11-mediated
activation of P53 in gastric cancer.
P53 is known to play crucial roles in monitoring
genomic stability and preventing malignant transformation.
Activation of P53 leads to cell cycle arrest, apoptosis, or
senescence, thereby preventing tumorigenesis (15).
It has been reported that 5-FU induced wild-type P53
expression and accumulation, followed by cell growth inhibition in
human gastric, human esophageal adenocarcinoma, and human embryonic
fibroblast cell lines (16,17). In addition, Osaki et al have
reported that 5-FU induced apoptosis in gastric cancer cell lines
carrying the TP53 wild-type gene (18).
Gastric cancer is the fourth most common malignancy
and the second most common cause of death of all malignancies
worldwide (19,20). Despite declining trends globally,
prevention of gastric cancer remains a priority in healthcare.
Therefore, identification of potential novel factors affecting drug
sensitivity of gastric cancer and preventing tumor progression is a
significant clinical challenge.
In the present study, we investigated effects of
RPL11 expression on the sensitivity of gastric cancer against 5-FU
treatment and its underlying mechanism. Our results provide a
relationship between RPL11 expression and susceptibility to 5-FU in
gastric cancer.
Materials and methods
Cell culture and reagents
Four human gastric cancer cell lines were used in
this study: MKN45, NUGC4, MKN7 (all three cell lines from JCRB Cell
Bank), and KE39 (from RIKEN Cell Bank). All cell lines were
cultured in RPMI-1640 medium (Nissui Pharmaceutical Co., Ltd.)
supplemented with 10% fetal bovine serum and 100 U/ml penicillin at
37°C in a humidified atmosphere of 5% CO2. 5-FU was
purchased from FUJIFILM Wako Pure Chemical Corporation.
3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium Bromide
Thiazolyl Blue (MTT) was purchased from Nakalai Tesque.
RNA interference
Cell transfection was performed using Lipofectamine
RNAiMAX Transfection Reagent (Life Technologies; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol for
knockdown experiments. All siRNAs were purchased from FASMAC. siRNA
sequences were as follows: siRPL11#1, 5′-GGUGCGGGAGUAUGAGUUA-3′;
siRPL11#2, 5′-AAGGUGCGGGAGUAUGAGUUA-3′; siControl,
5′-UUCUCCGAACGUGUCACGU-3′. siP53, 5′-CGGCGCACAGAGGAAGAGAAT-3
[Knockdown of siRNA-mediated P53 expression is previously described
(21,22)].
MTT assay
Cells were seeded at 7,000 cells per well in a
96-well plate and transfected with the indicated siRNAs for 24 h.
After transfection, the cells were exposed to different
concentrations of 5-FU for 3 days. Subsequently, the MTT solution
was added to each well, and the cells were cultured for an
additional 4 h. After removing the media, 100 µl of DMSO was added
to each well to dissolve the formazan crystals. The absorbance
values at 570A of each well were measured with a microplate reader
(Sunrise Remote; Tecan Japan Co. Ltd.) and applied to the following
calculation: Relative cell viability=A value of cells treated with
drugs/A value of cells treated with vehicle.
Immunoblot assay
For protein analysis, cells were washed twice with
phosphate-buffered saline (PBS) and lysed with lysis buffer (20 mM
Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM Sodium Vanadate, 1 mM EDTA, 50
mM NaF, 1% Triton X-100) supplemented with Protease Inhibitor
Cocktail (Nakalai Tesque), followed by sonication to reduce
viscosity. Protein concentration of each sample was determined by
the Protein Assay CBB Solution, according to the manufacturer's
protocol (Nakalai Tesque). Lysates containing proteins (20 µg) were
resolved on an SDS-polyacrylamide gel and transferred to an
Immobilon-P membrane (Millipore). The membranes were blocked with
blocking solution (4% BSA in TBST; 50 mM Tris-HCl, pH 7.4, 0.15 M
NaCl, and 0.1% Tween-20) for 1 h at 37°C prior to subsequent
incubation with the following primary antibodies: Anti-P53 antibody
(1:500 dilution; Santa Cruz Biotechnology), anti-P21 antibody
(1:400 dilution; Santa Cruz), anti-RPL11 antibody (1:1,000
dilution; Invitrogen), and anti-Actin (1:3,000 dilution; Bio Matrix
Research) primary antibodies for 1 h at 37°C. Following this, the
membranes were washed three times for 10 min in TBST and incubated
with horseradish peroxidase conjugated anti-rabbit or anti-mouse
IgG secondary antibodies (1:3,000 dilution; Cell Signaling
Technology) for 1 h at 37°C. Immunoreactive bands were visualized
with enhanced chemiluminescence using Clarity Western ECL Substrate
(Bio-Rad). Representative images from repeated experiments are
presented in each figure.
Quantitative real-time PCR
Total RNA from cultured cells was isolated using the
TRIzol reagent (Molecular Research Center) according to the
manufacturer's instructions. RNA (1 µg) was reverse-transcribed
using the ReverTra Ace kit (Toyobo). The mRNA expression levels of
P21, FAS, and RPL11 were determined by real-time
RT-PCR (StepOnePlus Real-Time PCR System; Applied Biosystems) using
GoTaq qPCR Master Mix (Promega) according to the manufacturer's
instructions. Human GAPDH was used for normalization. The
expression of the target gene was quantified using the comparative
cycle threshold method. The primer sequences used were as follows:
RPL11 forward, 5′-GAAAAGGAGAACCCCATGC-3′ and reverse,
5′-CATTTCTCCGGATGCCAA-3′; P21 forward,
5′-CTGGACTGTTTTCTCTCGGCTC-3′ and reverse,
5′-TGTATATTCAGCATTGTGGGAGGA-3′; FAS forward,
5′-TCTGCCATAAGCCCTGT-3′ and reverse, 5′-GTCTGTGTACTCCTTCCCT-3′;
GAPDH forward, 5′-TGCACCACCAACTGCTTAG-3′ and reverse,
5′-GAGGCAGGGATGATGTTC-3′.
Kaplan-Meier plotter
The prognostic significance of the mRNA expression
of RPL11 genes in gastric cancer was evaluated using the
Kaplan-Meier plotter online database, including gene expression
data and clinical data (GSE14210, GSE15459, GSE22377, GSE29272,
GSE51105 and GSE62254). The database automatically divided
RPL11 mRNA expression into high and low expression groups.
The survival curve, log-rank P-value, and hazard ratio with 95%
confidence intervals were calculated and displayed in the results
by the computer. The datasets analyzed during the present study are
available in the Kaplan-Meier plotter online database (http://kmplot.com/analysis/index.php?p=service&cancer=gastric).
Statistical analysis
Data are presented as the mean ± standard deviation.
Differences between multiple groups were determined by one-way
analysis of variance followed by Dunnett's post-hoc test. The
analyses were performed using GraphPad Prism software (version
8.1.1; GraphPad software Inc.). P<0.05 was considered to
indicate a statistically significant difference.
Results
RPL11 is associated with overall
survival of gastric cancer patients treated with 5-FU based
adjuvant therapy
To study the relationship between RPL11
expression and the effect of 5-FU on gastric cancers, we first
performed survival analysis using a Kaplan-Meier plotter
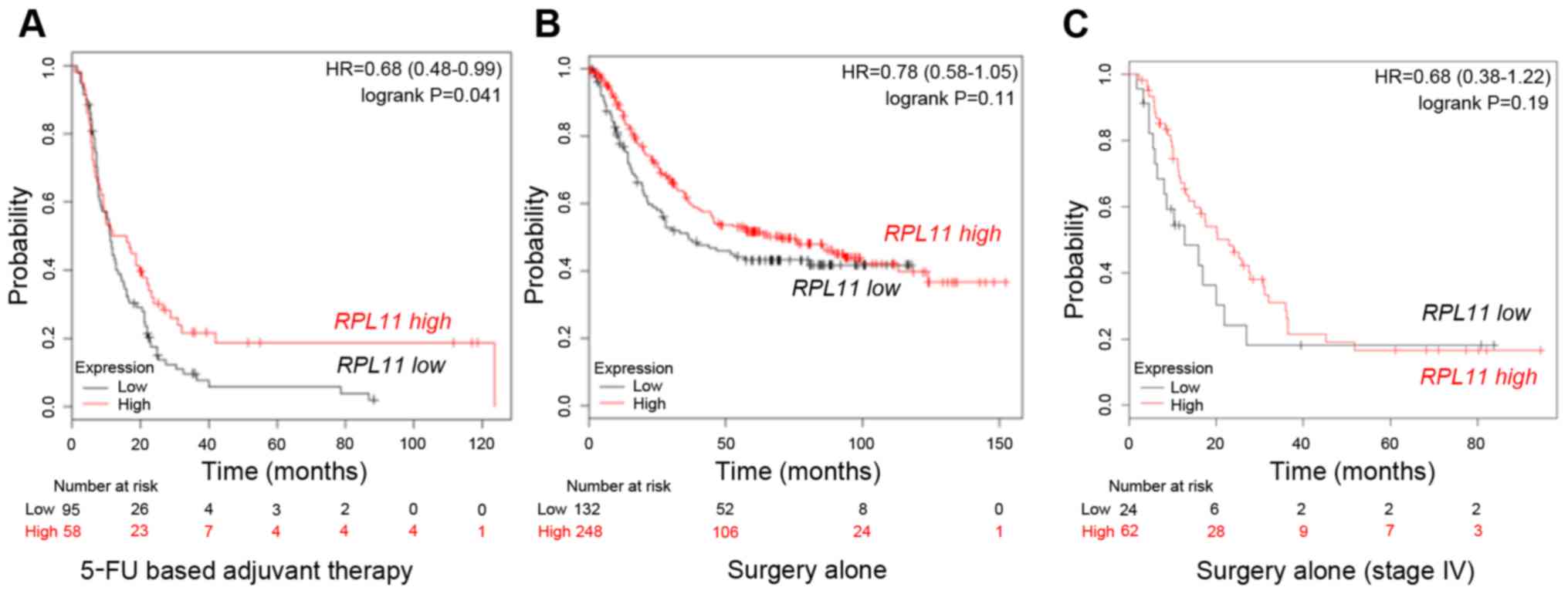
(www.kmplot.com). Fig.
1A indicates that 5-FU based adjuvant therapy-treated gastric
cancer patients in the RPL11-high expression group showed
better prognosis than those in RPL11-low expression group
(P=0.041). In contrast, gastric cancer patients treated by surgery
alone showed no significant difference between the
RPL11-high and RPL11-low expression groups (P=0.11;
Fig. 1B). Similarly, survival
analysis in only stage IV gastric cancer patients with surgery
alone showed no significant difference between the
RPL11-high and RPL11-low expression groups (P=0.19;
Fig. 1C). The results suggest that
RPL11 expression alters the sensitivity of gastric cancer
against 5-FU.
RPL11 inhibits cell proliferation
through regulation of P53 in gastric cancer cell lines
RPL11 is known to regulate the P53 pathway (7). To investigate whether the growth
inhibitory effect of 5-FU in gastric cancer involves regulation of
P53 via RPL11, we performed an MTT assay using gastric cancer cell
lines, MKN45 (wild-type TP53), NUGC4 (wild-type), MKN7
(mutated), and KE39 (mutated) cells. siRPL11-transfected
TP53 wild-type cells, MKN45 and NUGC4 cells, showed more
resistant to 5-FU than their corresponding non-specific control
siRNA-transfected cells. Further, in TP53-mutant cell lines,
MKN7 and KE39 cells, there was no significant difference in growth
suppressive effect of 5-FU between siRPL11-transfected cells and
non-specific control siRNA-transfected cells (Fig. 2A), Further, MKN45 and NUGC4 cells
with siRNA-mediated P53 knockdown showed more resistant to
5-FU than those with scramble siRNA as a control (Fig. 2B); however, double knockdown of
P53 and RPL11 did not show further resistance to 5-FU
in MKN45 and NUGC4 cells in comparison with either P53 or
RPL11 single knockdown (Fig.
2B). These observations suggested that cell growth suppression
by RPL11-mediated P53 activation in 5-FU treated gastric cancer
cell lines is dependent on normal P53 function.
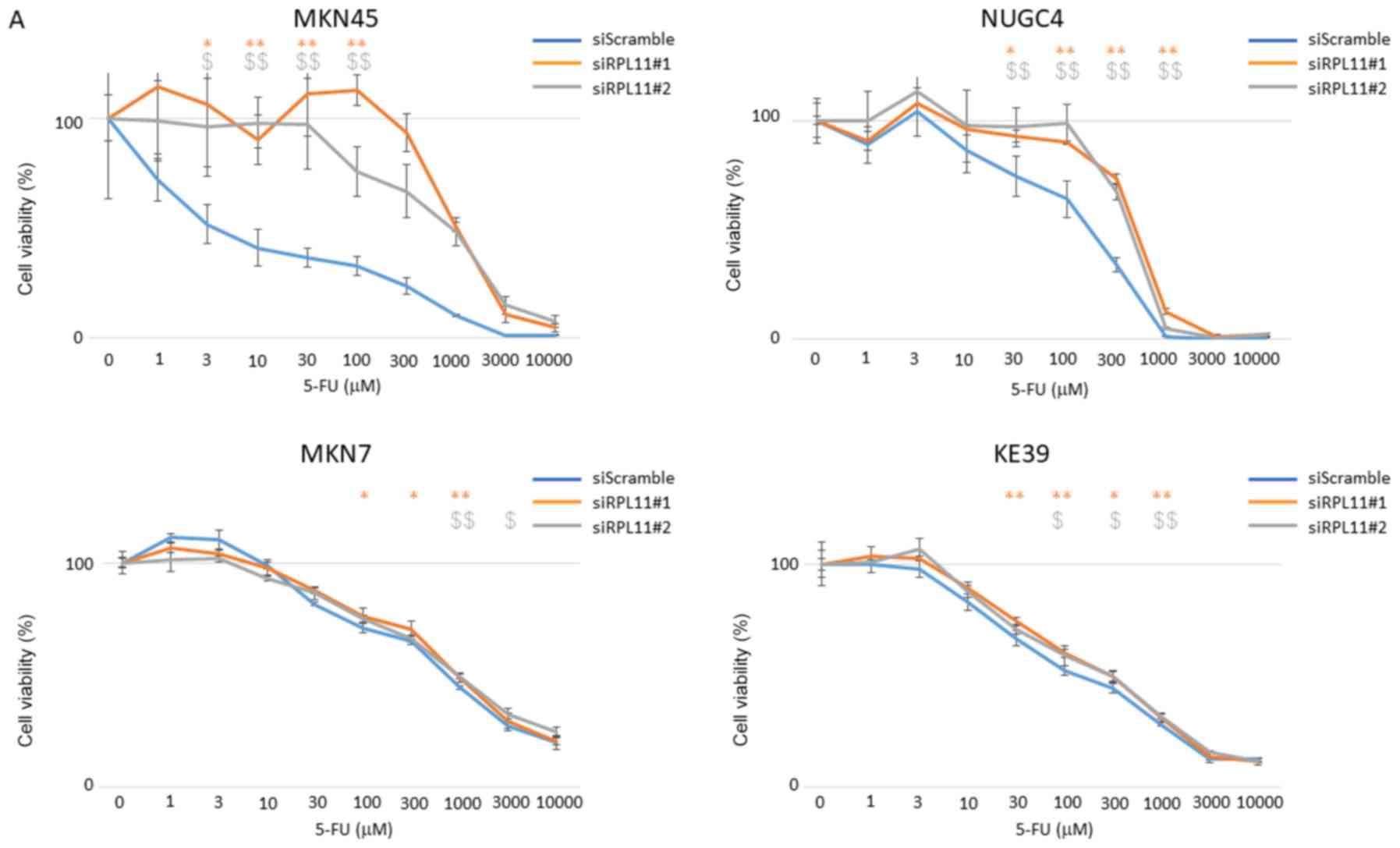 | Figure 2.RPL11 is involved in viability of
TP53 wild-type gastric cancer cells treated with 5-FU. (A)
Viabilities of MKN45, NUGC4, MKN7 and KE39 cells transfected with
siScramble (SC), siRPL11#1, or siRPL11#2 were analyzed by MTT
assay. Cell viabilities were measured upon exposure to the step-up
concentration of 5-FU for 72 h. (B) Viabilities of MKN45, and NUGC4
cells transfected with either siScramble, siRPL11#1, or siP53, both
siRPL11#1 and siP53 were analyzed by MTT assay. Cell viabilities
were measured upon exposure to the step-up concentration of 5-FU
for 72 h. Error bars indicate the standard deviation. *P<0.05
and **P<0.01 vs. siRPL11#1 group; $P<0.05 and
$$P<0.01 vs. RPL11#2 group. #P<0.05 and
##P<0.01 vs. siP53 group, +P<0.05 and
++P<0.01 vs. siRPL11#1 plus siP53 group. RPL11,
ribosomal protein L11; TP53, Tumor protein p53; 5-FU,
5-fluorouracil; si, small interfering RNA. |
5-FU treatment induces RPL11-mediated
P53 activation in gastric cancer cell lines
To investigate whether RPL11 is involved in
regulation of the P53 pathway in 5-FU treated gastric cancer cells,
we examined protein expression levels of P53 and a P53 downstream
target, P21. As a result, 5-FU increased protein expression levels
of P53 and P21 in MKN45 and NUGC4 cells, and they were markedly
reduced by siRNA knockdown of RPL11. Conversely, MKN7 and
KE39 cells did not have any significant effects on P53 and P21
protein expression (Fig. 3A).
Because the extrinsic apoptosis factor FAS is crucial for P53
mediation of apoptosis in 5-FU treated cancer cells (23), 5-FU treatment increased mRNA levels
of P53 target genes, P21 and FAS, which were
determined using the quantitative real-time PCR assay. In MKN45 and
NUGC4 cells, knockdown of RPL11 markedly reduced the
increased mRNA levels of P21 and FAS by 5-FU
treatment. However, in MKN7 and KE39 cells treated with 5-FU, there
was little difference in mRNA levels of P21 and FAS
between siRPL11-transfected cell lines and non-specific control
siRNA-transfected cell lines (Fig.
3B). These results demonstrate that RPL11 expression is
involved in regulating the P53 pathway in 5-FU-treated gastric
cancer cell lines.
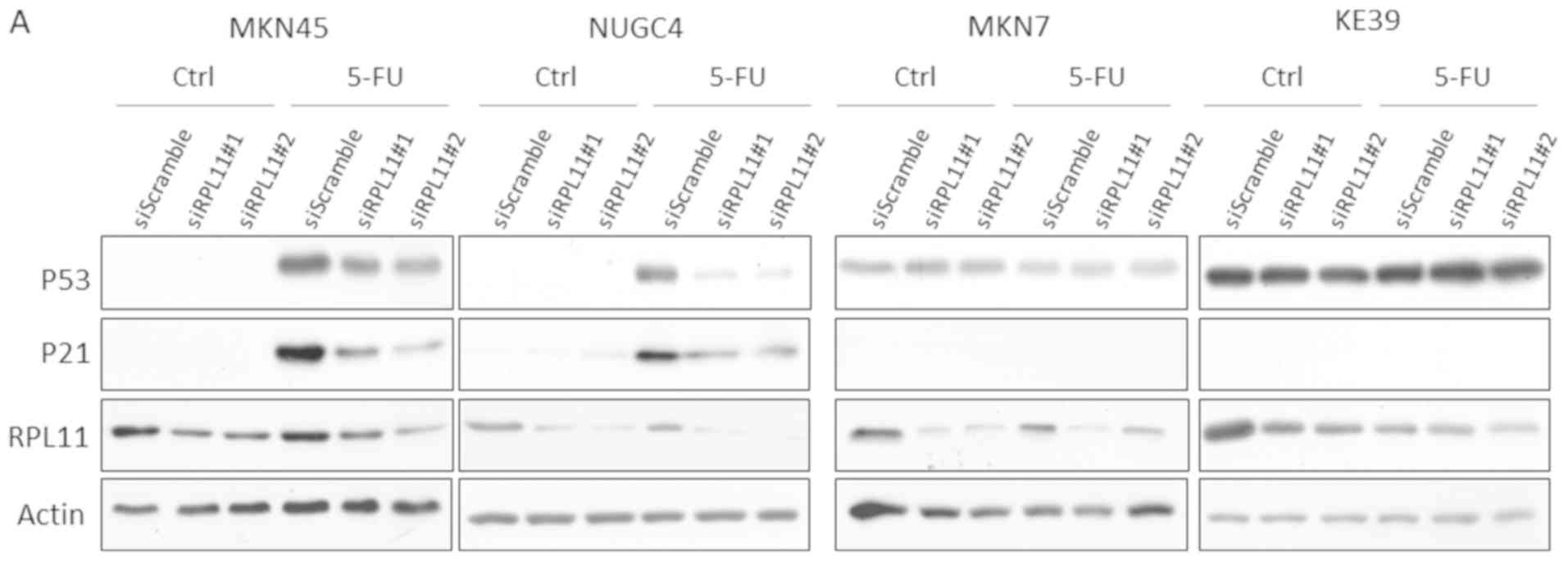 | Figure 3.RPL11 is involved in activation of
the P53 pathway in 5-FU treated gastric cancer cells. (A)
Expression levels of P53, P21 and RPL11 in MKN45, NUGC4, MKN7 and
KE39 cells transfected with siScramble (SC), siRPL11#1, or
siRPL11#2 under mock and 300 µM 5-FU treatment for 24 h were
analyzed by immunoblot assay. (B) Relative P21, FAS, and
RPL11 expression in MKN45, NUGC4, MKN7, and KE39 cells
transfected with siSC, siRPL11#1, or siRPL11#2 under mock and 300
µM 5-FU treatment for 24 h were analyzed by quantitative real-time
PCR. Error bars indicate the standard deviation. *P<0.05 and
**P<0.01 as indicated. ns, not significant; RPL11, ribosomal
protein L11; Ctrl, control; 5-FU, 5-fluorouracil; si, small
interfering RNA; FAS, Fas cell surface death receptor. |
Discussion
The present study investigated whether RPL11
expression was associated with the drug sensitivity of gastric
cancer upon 5-FU treatment. The findings showed that high
RPL11 expression in 5-FU treated gastric cancer patients
have better prognosis than low RPL11 expression in 5-FU
treated gastric cancer patients. In 5-FU-treated gastric cancer
cell lines carrying the TP53 wild-type gene, knockdown of
RPL11 reversed the decreased cell viability through
activation of the P53 pathway. Altogether, these data demonstrate
that RPL11 is closely related to the sensitivity of gastric cancer
against 5-FU and activates the P53 pathway, including P21 and Fas,
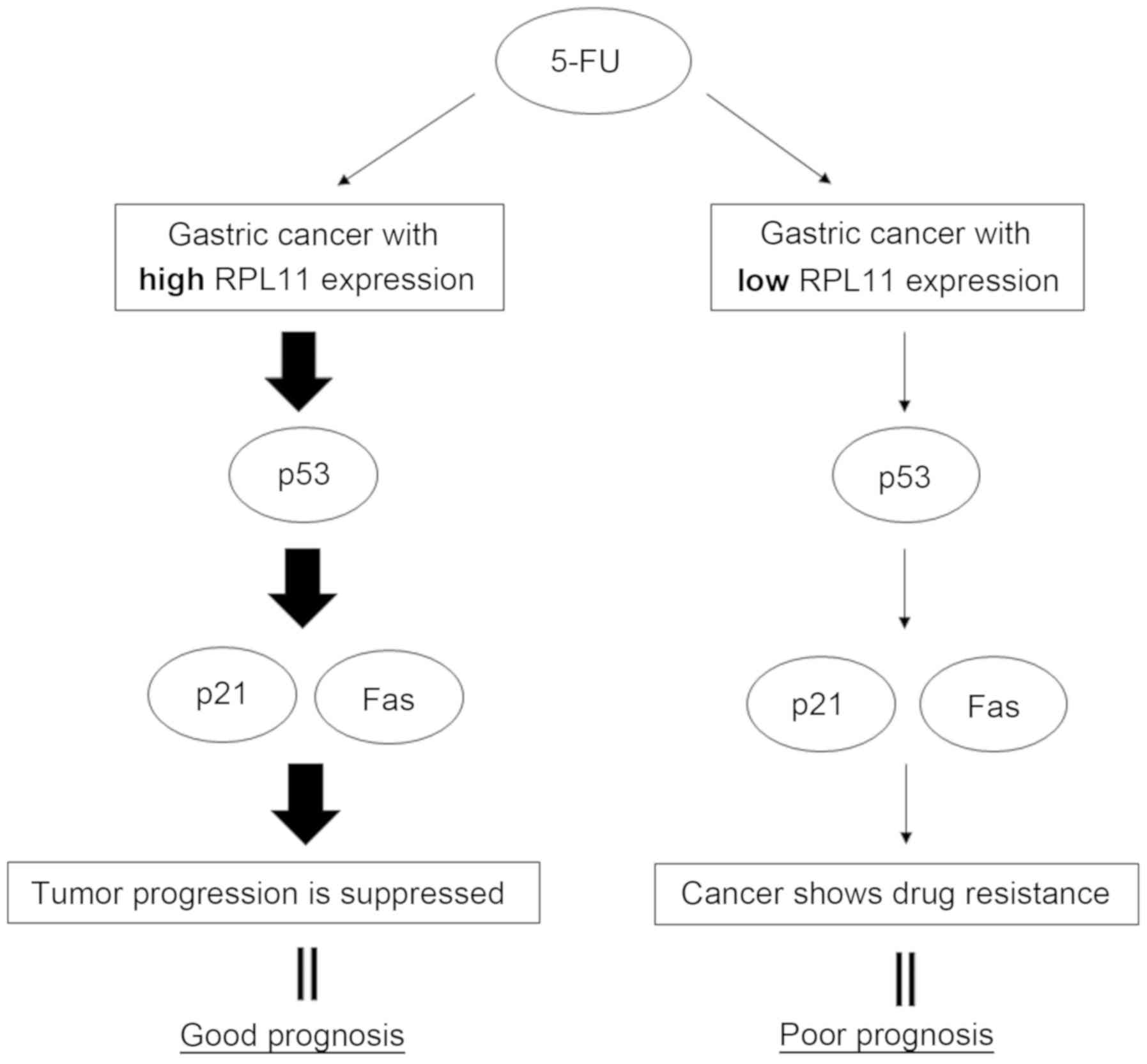
resulting in suppression of tumor progression (Fig. 4).
5-FU is used as an anti-cancer drug for various
cancer types, including gastrointestinal cancers, breast cancers,
lung cancers, and cancers of the aerodigestive tract (1), indicating that 5-FU may exert a tumor
suppression effect via the RPL11-P53 signaling pathway against not
only gastric cancers but also other cancer types. It has been
reported that 5-FU also induces DNA damage-mediated activation of
the P53 pathway (24). In the
present study, RPL11 expression was closely related to P53
pathway-mediated cell growth suppression in 5-FU treated gastric
cancer, but in other cancer types treated with 5-FU. Therefore, DNA
damage may be predominantly related to P53 pathway-mediated cell
growth suppression. Additionally, it has been reported that
5-FU-induced cell growth suppression via inhibition of rRNA
processing involves not only RPL11, but also RPL5 and RPL23 in
osteosarcoma cells (14), suggesting
RPL5 and RPL23 may also be involved in P53 pathway-mediated cell
growth suppression in 5-FU treated gastric cancer. Our data
indicated that there are some differences in RPL11 expression
levels in patients with gastric cancer. RPL11 mRNA expression is
regulated by c-Myc and N-Myc, members of the Myc oncoprotein family
of transcription factors (25,26).
Deferential expression levels and/or activity of Myc in gastric
cancer patients may reflect the differences in RPL11 expression
levels in these patients. Recently, Wang et al reported that
enhancer of zente homologue 2 (EZH2) contributes to 5-FU resistance
in gastric cancer and that high EZH2 expression is
correlated with poor prognosis of gastric cancer patients (27). Although further investigation is
needed, gastric cancer patients with both high RPL11 expression and
low EZH2 expression may show higher sensitivity against 5-FU
treatment than those with either high RPL11 or low EZH2 expression
alone, which may help to more accurately predict 5-FU sensitivity
in gastric cancer. It has been reported that certain ribosomal
proteins, including RPS14, RPL5, RPL22, RPL41, RPS7, RPS15a, RPS24,
RPS27, and RPL15 are involved in neoplastic transformation and/or
cell migration and/or invasion (28). However, there is no report showing
the relationship between ribosomal proteins and drug sensitivity in
human cancer so far. Thus, our present study is the first report
that the ribosomal protein L11 is associated with drug sensitivity
and prognosis in gastric cancer via the P53 pathway.
5-FU is one of the most frequently used first-line
treatments for patients with advanced gastric cancer (29,30).
However, the 5-year survival rate of patients with gastric cancer
remains poor (31). Therefore, there
is an urgent need to identify molecular mechanisms and find novel
therapeutic strategies for 5-FU resistance in gastric cancer.
In the present study, we show that RPL11 expression
is a crucial factor affecting the sensitivity of gastric cancer
against 5-FU, suggesting that RPL11 may be a potential biomarker
for predicting 5-FU sensitivity. Identification of RPL11 as a
biomarker helps gastric cancer patients with 5-FU resistance avoid
unwanted side effects caused by 5-FU treatment. In addition to the
role of RPL11 expression as a biomarker, we found RPL11 is
functionally required to modulate sensitivity to 5-FU. Therefore,
chemotherapy using a drug to elevate RPL11 expression could improve
5-FU resistance in gastric cancer patients with the TP53
wild-type gene. These findings would greatly contribute to a
therapeutic strategy for patients with gastric cancer.
Acknowledgements
The authors would like to thank Ms. Yoshiko Setogawa
and Ms. Hiromi Mitsuo (Department of Molecular Oncology, Kagoshima
University Graduate School of Medical and Dental Sciences) for
their excellent secretarial assistance. We also wish to thank the
Joint Research Laboratory, Kagoshima University Graduate School of
Medical and Dental Sciences, for use of their facilities.
Funding
The present study was supported by JSPS KAKENHI
(grant nos. JP17J05291, JP15K10311, JP15K10338, JP22501047,
JP16K07121, JP17K10871, JP17K07221 and JP18K06732) and the
Molecular Profiling Committee, Grant-in-Aid for Scientific Research
on Innovative Areas ‘Platform of Advanced Animal Model Support’
from The Ministry of Education, Culture, Sports, Science and
Technology, Japan (JSPS KAKENHI grant no. JP 16H06276), Takeda
Medical Foundation, the Shinnihon Foundation of Advanced Medical
Treatment Research, Kodama Memorial Fund for Medical Research, the
Mochida Memorial Foundation for Medical and Pharmaceutical
Research, Foundation for Promotion of Cancer Research in Japan, the
Suzuken Memorial Foundation, the Shimabara Science Promotion
Foundation and Astellas Foundation for Research on Metabolic
Disorders.
Availability of data and materials
The datasets generated and/or analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
KK designed the experiments. TK, AS, and MS
performed the experiments. TK, AS, MS analyzed the experimental
data. TS and KA performed database analysis. KM, MY and YS
performed statistical analysis. TK, TH and TF wrote the paper,
interpreted data and revised it critically for intellectual
content. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
5-FU
|
5-Fluorouracil
|
|
TP53
|
tumor protein p53
|
|
RPL11
|
ribosomal protein L11
|
|
rRNA
|
ribosomal RNA
|
|
MDM2
|
murine double minute 2
|
|
FAS
|
Fas cell surface death receptor
|
References
|
1
|
Longley DB, Harkin DP and Johnston PG:
5-fluorouracil: Mechanisms of action and clinical strategies. Nat
Rev Cancer. 3:330–338. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Houghton JA, Tillman DM and Harwood FG:
Ratio of
2′-deoxyadenosine-5′-triphosphate/thymidine-5′-triphosphate
influences the commitment of human colon carcinoma cells to
thymineless death. Clin Cancer Res. 1:723–730. 1995.PubMed/NCBI
|
|
3
|
Sampath D, Rao VA and Plunkett W:
Mechanisms of apoptosis induction by nucleoside analogs. Oncogene.
22:9063–9074. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wilkinson DS and Pitot HC: Inhibition of
ribosomal ribonucleic acid maturation in Novikoff hepatoma cells by
5-fluorouracil and 5-fluorouridine. J Biol Chem. 248:63–68.
1973.PubMed/NCBI
|
|
5
|
Kanamaru R, Kakuta H, Sato T, Ishioka C
and Wakui A: The inhibitory effects of 5-fluorouracil on the
metabolism of preribosomal and ribosomal RNA in L-1210 cells in
vitro. Cancer Chemother Pharmacol. 17:43–46. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ghoshal K and Jacob ST: Specific
inhibition of pre-ribosomal RNA processing in extracts from the
lymphosarcoma cells treated with 5-fluorouracil. Cancer Res.
54:632–636. 1994.PubMed/NCBI
|
|
7
|
Bhat KP, Itahana K, Jin A and Zhang Y:
Essential role of ribosomal protein L11 in mediating growth
inhibition-induced p53 activation. EMBO J. 23:2402–2412. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dai MS, Zeng SX, Jin Y, Sun XX, David L
and Lu H: Ribosomal protein L23 activates p53 by inhibiting MDM2
function in response to ribosomal perturbation but not to
translation inhibition. Mol Cell Biol. 24:7654–7668. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dai MS and Lu H: Inhibition of
MDM2-mediated p53 ubiquitination and degradation by ribosomal
protein L5. J Biol Chem. 279:44475–44482. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dai MS, Shi D, Jin Y, Sun XX, Zhang Y,
Grossman SR and Lu H: Regulation of the MDM2-p53 pathway by
ribosomal protein L11 involves a post-ubiquitination mechanism. J
Biol Chem. 281:24304–24313. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon
M and Vousden KH: Regulation of HDM2 activity by the ribosomal
protein L11. Cancer Cell. 3:577–587. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jin A, Itahana K, O'Keefe K and Zhang Y:
Inhibition of HDM2 and activation of p53 by ribosomal protein L23.
Mol Cell Biol. 24:7669–7680. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Y, Wolf GW, Bhat K, Jin A, Allio T,
Burkhart WA and Xiong Y: Ribosomal protein L11 negatively regulates
oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress
checkpoint pathway. Mol Cell Biol. 23:8902–8912. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun XX, Dai MS and Lu H: 5-fluorouracil
activation of p53 involves an MDM2-ribosomal protein interaction. J
Biol Chem. 282:8052–8059. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Levine AJ and Oren M: The first 30 years
of p53: Growing ever more complex. Nat Rev Cancer. 9:749–758. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nabeya Y, Loganzo F Jr, Maslak P, Lai L,
de Oliveira AR, Schwartz GK, Blundell ML, Altorki NK, Kelsen DP and
Albino AP: The mutational status of p53 protein in gastric and
esophageal adenocarcinoma cell lines predicts sensitivity to
chemotherapeutic agents. Int J Cancer. 64:37–46. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pickard M, Dive C and Kinsella AR:
Differences in resistance to 5-fluorouracil as a function of cell
cycle delay and not apoptosis. Br J Cancer. 72:1389–1396. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Osaki M, Tatebe S, Goto A, Hayashi H,
Oshimura M and Ito H: 5-Fluorouracil (5-FU) induced apoptosis in
gastric cancer cell lines: Role of the p53 gene. Apoptosis.
2:221–226. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wright NA, Poulsom R, Stamp G, Van Noorden
S, Sarraf C, Elia G, Ahnen D, Jeffery R, Longcroft J, Pike C, et
al: Trefoil peptide gene expression in gastrointestinal epithelial
cells in inflammatory bowel disease. Gastroenterology. 104:12–20.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hong CF, Lin SY, Chou YT and Wu CW:
MicroRNA-7 compromises p53 Protein-dependent apoptosis by
controlling the expression of the chromatin remodeling factor
SMARCD1. J Biol Chem. 291:1877–1889. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang Y, Nakagawa H, Tetreault MP, Billig
J, Victor N, Goyal A, Sepulveda AR and Katz JP: Loss of
transcription factor KLF5 in the context of p53 ablation drives
invasive progression of human squamous cell cancer. Cancer Res.
71:6475–6484. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Petak I, Tillman DM and Houghton JA: p53
dependence of Fas induction and acute apoptosis in response to
5-fluorouracil-leucovorin in human colon carcinoma cell lines. Clin
Cancer Res. 6:4432–4441. 2000.PubMed/NCBI
|
|
24
|
Adamsen BL, Kravik KL and De Angelis PM:
DNA damage signaling in response to 5-fluorouracil in three
colorectal cancer cell lines with different mismatch repair and
TP53 status. Int J Oncol. 39:673–682. 2011.PubMed/NCBI
|
|
25
|
Dai MS, Arnold H, Sun XX, Sears R and Lu
H: Inhibition of c-Myc activity by ribosomal protein L11. EMBO J.
26:3332–3345. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Boon K, Caron HN, Van Asperen R, Valentijn
L, Hermus MC, Van Sluis P, Roobeek I, Weis I, Voûte PA, Schwab M,
et al: N-myc enhances the expression of a large set of genes
functioning in ribosome biogenesis and protein synthesis. EMBO J.
20:1383–1393. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang C, Li X, Zhang J, Ge Z, Chen H and Hu
J: EZH2 contributes to 5-FU resistance in gastric cancer by
epigenetically suppressing FBXO32 expression. Onco Targets Ther.
11:7853–7864. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu X, Xiong X and Sun Y: The role of
ribosomal proteins in the regulation of cell proliferation,
tumorigenesis, and genomic integrity. Sci China Life Sci.
59:656–672. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ajani JA: Evolving chemotherapy for
advanced gastric cancer. Oncologist. 10:49–58. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhe W, Feng ZJ and Fang FG: Effect of
rhizoma curcumate zedoariae and herbra trifolii repentis on
apoptosis of human lung cancer cell line A549. J Capital Univ Med
Sci. 22:304–305. 2001.
|
|
31
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|