Introduction
Breast cancer (BC) is a common malignancy among
women. According to recent cancer statistics, ~2.1 million women
worldwide were diagnosed with BC and ~627,000 individuals died from
the condition in 2018 (1). The
prognosis of patients with BC has substantially improved in recent
decades owing to the rapid advancement in diagnostic methods and
individualized treatments. Nevertheless, this disease poses a
severe threat to female health (2,3). Routine
diagnosis and treatment are rarely effective in certain patients
due to the intratumor heterogeneity of BC. Therefore, the
identification of novel and more reliable molecular biomarkers for
the prediction of outcomes and targeted treatments for BC is
crucial.
As part of the kinetochore complex, ZW10 interacting
kinetochore protein (ZWINT) is required for the mitotic spindle
checkpoint (4). The majority of
studies have proposed that ZWINT acts as a structural protein,
which is part of the inner kinetochore scaffold and recruits zeste
white 10, which is also a centromere protein, to the kinetochore
(5,6). Upregulation of ZWINT has been observed
in numerous types of cancer, including ovarian and hepatocellular
cancer, as well as glioblastoma, and is indicative of a worse
prognosis (7–9). ZWINT depletion has been demonstrated to
attenuate cell proliferation in 293-T and BC MCF-7 cells via
Terf/tripartite motif containing 17 (TRIM17) signaling (10). In addition, abnormal expression of
ZWINT has been reported to be associated with chromosomal
instability and poor clinical outcomes in certain types of cancer
(11). Furthermore, downregulated
expression of ZWINT, mediated by the inhibition of
cyclooxygenase-2, has been reported to impede the proliferation of
prostate cancer cells via prostaglandin E receptor-1 signaling
(12). These results suggested that
ZWINT may serve an essential role in cancer progression and
development.
Since the expression and clinical implication of
ZWINT have not been comprehensively investigated in human BC, the
current study examined the significance of ZWINT expression in
human BC via bioinformatics analysis. The aim of the present study
was to evaluate the expression profile, roles and accuracy of ZWINT
as a biomarker for the long-term prognostic prediction of BC
through pooling and analyzing currently available data.
Materials and methods
Tissue specimens and
immunohistochemistry (IHC)
BC and adjacent normal breast tissue specimens were
collected from 62 patients who had been treated at the Tongji
Hospital, Tongji Medical College of Huazhong University of Science
and Technology (Wuhan, China) between September 2016 and June 2018.
The patients that participated in the study had not received
chemotherapy, radiotherapy, targeted therapy or other treatment
prior to surgery. All patients in this study were women and were
aged between 28 and 77 years (median age, 47 years). Written
informed consent was obtained from all patients and the
experimental protocol was approved by the Ethics Committee of
Tongji Hospital, Tongji Medical College of Huazhong University of
Science and Technology. The collected specimens were immediately
fixed with 10% neutral formaldehyde for 12 h at room temperature.
The fixed specimens were paraffin-embedded and cut into 4 µm thick
sections. IHC was conducted using Polink-2 plus® Polymer
HRP Detection system (cat. no. PV-0023; BIOSS) according to the
manufacturer's instructions. Sections were incubated with the
primary antibody against ZWINT (cat. no. bs-7852R; BIOSS;1:200) at
4°C overnight, and with horseradish peroxidase-conjugated
anti-rabbit immunoglobulin G (cat.no. bs-0295D-HRP; BIOSS; 1:500)
at 37°C for 20 min. The specimens were examined using an Olympus
BX-51 microscope (Olympus, Corporation; magnification, ×200) by two
experienced pathologists. Results were scored according to the
staining intensity and the percentage of positive cells on a 3- and
4-point scale. Staining intensity was graded as follows: 0,
negative; 1, weak; 2, moderate; or 3, strong. The ratio of stained
positive cells was scored as follows: 0, no staining; 1, ≤25%; 2,
26–50%; 3, 51–75%; and 4, >75%. The total score was calculated
as the sum of the intensity and percentage scores. Additionally,
for semi-quantitative analysis, a score ≤4 was considered to
indicate low ZWINT expression and a score >4 high ZWINT
expression.
Gene Expression Profiling Interactive
Analysis (GEPIA)-based cancer data analysis
GEPIA (http://gepia.cancer-pku.cn/) analysis based on The
Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression,
which contains a large number of RNA sequencing data for cancer and
normal tissues, was conducted to examine the mRNA expression levels
of ZWINT in BC and adjacent normal tissues (13).
Oncomine database analysis
Data regarding the transcription levels of ZWINT in
BC were retrieved from the Oncomine database (https://www.oncomine.org/), a cancer research database
that contains a large number of datasets and samples, on the basis
of microarray and high-throughput sequencing. A fold change >2,
P<1×10−4 and genes ranking in the top 10%, with
simultaneous data restricted to mRNA were set as significance
cut-off levels. Four datasets [TCGA Breast (14), Curtis Breast (15), Ma Breast (16) and Richardson Breast (17)] were used to analyze the differential
expression of ZWINT in multiple pathological types of BC and normal
breast tissue.
Expression levels of ZWINT and
clinical outcomes of BC
mRNA expression of ZWINT in different types of
molecular and clinicopathological BC, and its association with
prognosis were evaluated by employing Breast Cancer Gene-Expression
Miner (version 4.1; bc-GenExMiner) which is a mining tool and a web
source of published BC genomic data (18,19). A
P-value indicating statistically significant differences among
groups (age, nodal status, ER, PR, Her-2, SBR grade, triple
negative status and basal-like status) was generated using Welch's
test followed by the Dunnett-Tukey-Kramer's test. Moreover,
Kaplan-Meier Plotter (https://kmplot.com/), which is an online database that
contains data from 5,143 patients with BC, 2,437 patients with lung
cancer, 1,816 patients with ovarian cancer, 1,065 patients with
gastric cancer and 364 patients with liver cancer combined with
relapse-free survival (RFS) and overall survival (OS) data
(20), was used to assess the
relationship between the transcription levels of ZWINT and OS or
RFS among patients with BC. Specifically, clinical samples were
divided into high expression and low expression groups in terms of
the median expression value of ZWINT. Subsequently, the log rank
P-value and hazard ratio (HR) with 95% confidence interval (CI)
were calculated. In addition, the impact of ZWINT on metastatic
relapse-free survival (MRFS) and the risk of metastatic relapse
(MR) of BC patients were evaluated in all cohorts in bc-GenExMiner
pooled by means of univariate Cox regression model, and were
illustrated with a Kaplan-Meier curve and a forest plot,
respectively.
Mutation and copy number variation
(CNV) analysis of ZWINT
Mutations of ZWINT were evaluated using the
Catalogue of Somatic Mutations in Cancer (COSMIC) database, which
is a comprehensive resource of somatic mutations identified in
numerous types of human cancer (21,22). The
distribution of various mutations was included and presented in the
form of a pie chart. In addition, the cBio Cancer Genomics Portal
(cBioPortal) (http://cbioportal.org) database was
used to examine the alteration frequency of ZWINT mutations and
CNVs in BC (23,24). The following data were used in
cBioPortal: i) Breast Cancer [MSK, Cancer Cell 2018 (25)] ii) Breast Cancer [METABRIC (26)] iii): Breast [BCCRC 2012 (27)] iv): Breast [Broad 2012 (28)] v): Breast [Sanger (29)] vi) Breast [TCGA (30)] vii): Breast [BCCRC Xenograft
(31)]; viii) BRCA [INSERM 2016
(32)].
Gene set enrichment analysis
(GSEA)
The functional role of ZWINT was determined by
performing GSEA. GSEA acts as a computational tool to evaluate
whether a pre-defined set of genes demonstrates statistically
significant, consistent differences between two biological states,
including phenotypes (33). In the
present study, the original mRNA sequencing data of BC were
downloaded from TCGA and the samples were sorted in terms of ZWINT
expression levels. A hundred samples were divided into high or low
categories (50 samples in each category) based on the ZWINT
expression levels, in order to annotate phenotype and the
‘Hallmarks’ gene set from the Molecular Signatures Database (MSigDB
h.all.v6.2.symbols.gmt) was used for enrichment analysis. False
discovery rate <0.25 and nominal P-value <0.05 were set as
cut-off criteria.
Co-expression and correlation
analysis
The ‘coexpression analysis’ module of Oncomine
database was used to investigate the co-expressed genes of ZWINT in
BC. The feature of ‘gene correlation analysis’ of the cBioPortal
and bc-GenExMiner was used to verify the co-expression of ZWINT and
CDK1 in BC, and linear (Pearson) and/or nonparametric (Spearman)
correlation coefficients were calculated to assess the correlation
intensity between these two genes. P<0.05 was considered to be
statistically significant.
Statistical analysis
IHC data were analyzed using SPSS software (version
21.0; IBM Corp.). A Wilcoxon matched-pairs signed rank test was
conducted to compare BC tissues and adjacent normal tissues, and
data are presented as the median ± interquartile range. Fisher's
exact test was used to analyze the association between ZWINT
expression and clinicopathological characteristics. Moreover, data
from the bc-GenExMiner database were used to analyze the mRNA
expression of ZWINT in terms of clinicopathological characteristics
with Welch's test and the Dunnett-Tukey-Kramer's test. The MRFS and
MR of BC patients were estimated by the univariate Cox regression.
P<0.05 was considered to indicate a statistically significant
difference.
Results
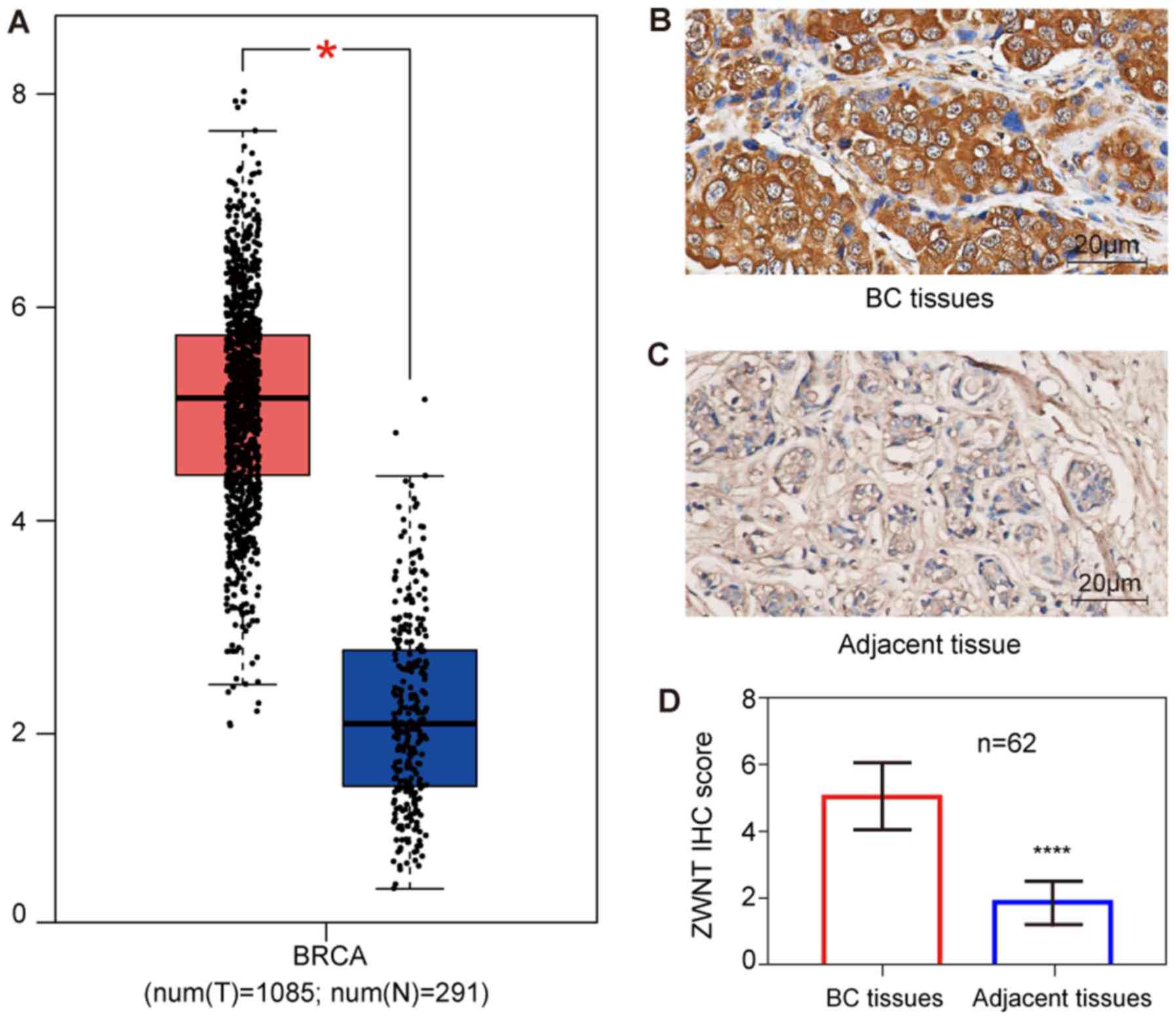
ZWINT expression in human BC. The mRNA expression
profile of ZWINT in BC was studied by searching GEPIA and the
Oncomine database. Elevated ZWINT expression was identified in BC
tissues compared with normal tissues (Fig. 1A). Furthermore, the results
demonstrated that ZWINT mRNA was significantly overexpressed in
mucinous, medullary, invasive lobular and invasive ductal breast
carcinoma compared with the normal breast tissues (Table I). In addition, IHC analysis
confirmed that ZWINT expression was higher in BC tissues compared
with adjacent normal breast tissues (Fig. 1B-D).
 | Table I.ZW10 interacting kinetochore protein
expression in different subtypes of breast carcinoma. |
Table I.
ZW10 interacting kinetochore protein
expression in different subtypes of breast carcinoma.
| Subtype | Cancer | Normal | P-value | Fold change | Rank (%) | Source of raw data
(ref) |
|---|
| Invasive Breast
Carcinoma | 76 | 61 |
3.19×10−35 | 5.133 | 1 |
|
| Invasive Lobular
Breast Carcinoma | 36 | 61 |
1.47×10−18 | 4.373 | 1 | TCGA (14) |
| Invasive Ductal
Breast Carcinoma | 389 | 61 |
2.73×10−44 | 5.989 | 1 |
|
| Invasive Ductal
Breast Carcinoma | 1556 | 144 |
3.04×10−121 | 2.809 | 1 |
|
| Invasive Breast
Carcinoma | 21 | 144 |
1.09×10−8 | 2.313 | 1 |
|
| Invasive Lobular
Breast Carcinoma | 148 | 144 |
3.80×10−49 | 2.253 | 1 |
|
| Invasive Ductal and
Invasive Lobular | 90 | 144 |
4.24×10−35 | 2.564 | 1 | Curtis et al
(15) |
| Breast
Carcinoma |
|
|
|
|
|
|
| Medullary Breast
Carcinoma | 32 | 144 |
5.66×10−15 | 2.789 | 1 |
|
| Mucinous Breast
Carcinoma | 46 | 144 |
3.96×10−17 | 2.208 | 2 |
|
| Breast
Carcinoma | 14 | 144 |
8.10×10−6 | 2.547 | 3 |
|
| Tubular Breast
Carcinoma | 67 | 144 |
2.45×10−23 | 2.087 | 3 |
|
| Invasive Ductal
Breast Carcinoma | 9 | 14 |
6.50×10−5 | 3.553 | 2 | Ma et al
(16) |
| Ductal Breast
Carcinoma In Situ | 9 | 14 |
3.74×10−5 | 3.787 | 2 |
|
| Ductal Breast
Carcinoma | 40 | 7 |
9.56×10−6 | 7.593 | 6 | Richardson et
al (17) |
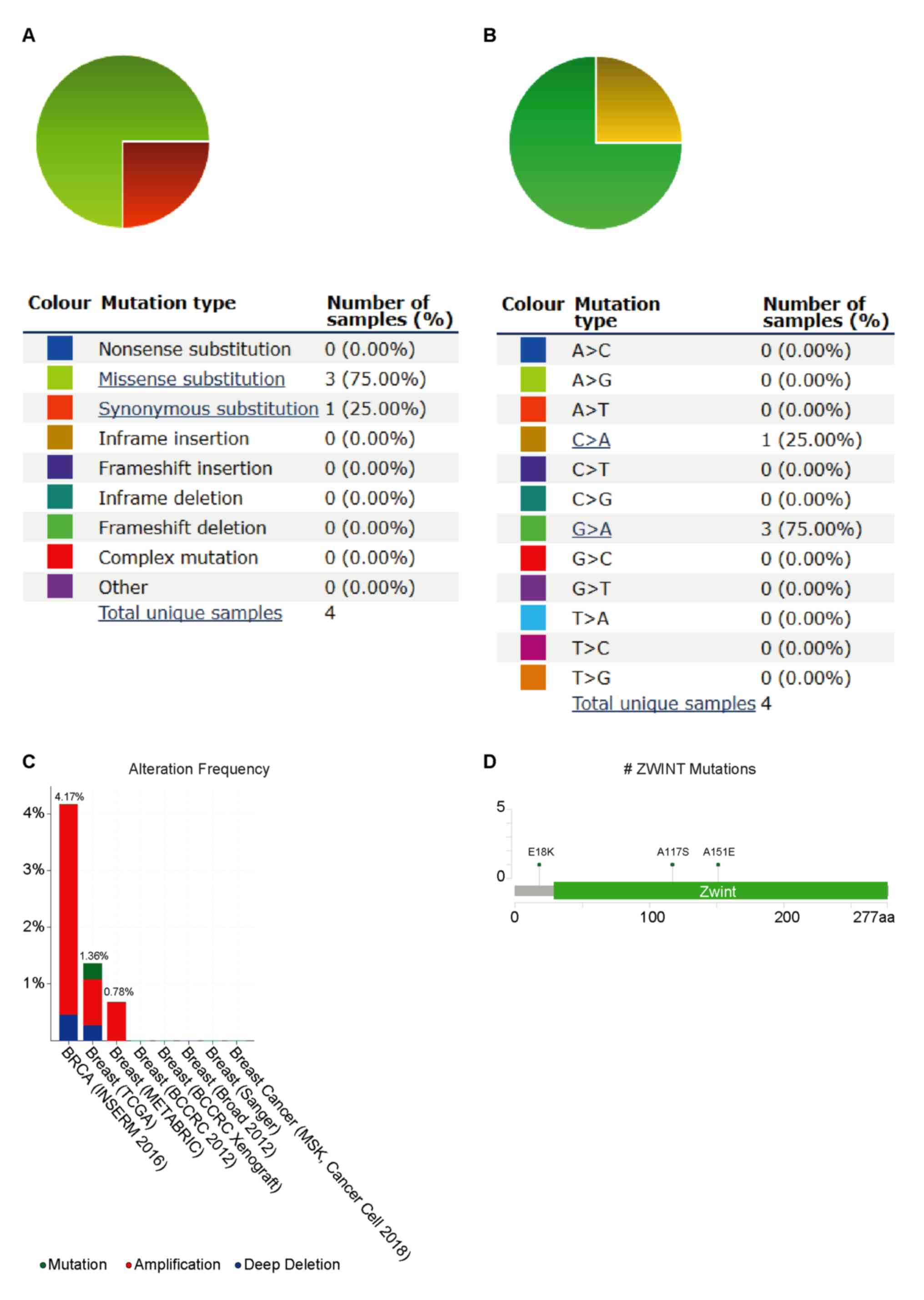
ZWINT mutations in human BC
COSMIC database analysis identified two types of
ZWINT mutations in BC, missense and synonymous substitutions
(Fig. 2A). For substitution
mutations, the data revealed that C>A and G>A mutations
accounted for 25 and 75% of the ZWINT coding strand, respectively
(Fig. 2B). cBioPortal was applied to
assess the genomic alteration frequency of ZWINT in BC and the
results revealed that ZWINT genomic alteration exceeded 5%,
including amplification, mutation and deep deletion (Fig. 2C). A total of three locations of
ZWINT mutation, including E18K, A117S and A151E, were found in
cBioPortal database (Fig. 2D).
Association between ZWINT expression
and different clinicopathological indicators
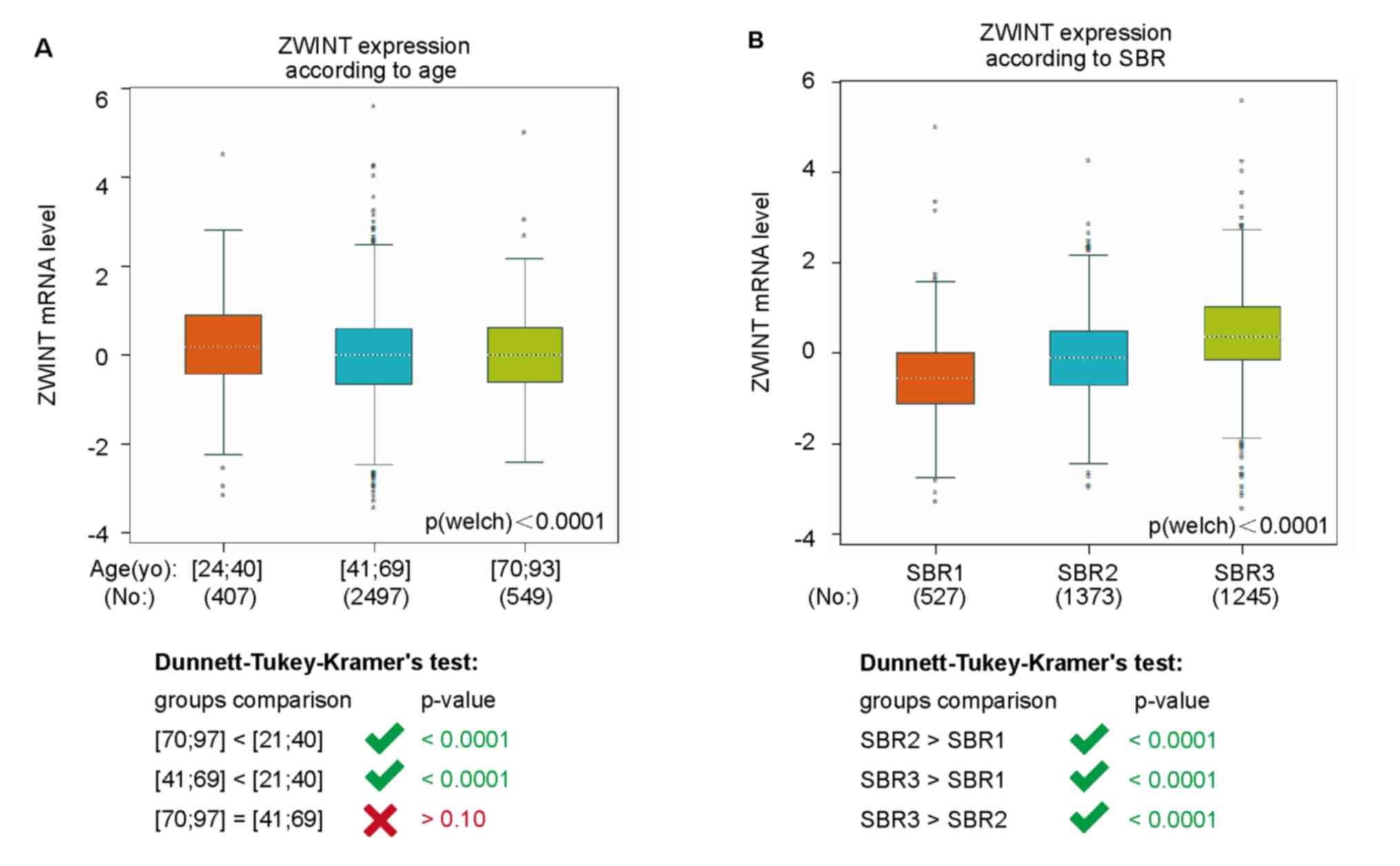
In bc-GenExMiner, the mRNA expression of ZWINT among
groups of patients with different clinicopathological parameters
was assessed using Welch's test followed by the
Dunnett-Tukey-Kramer's test. The mRNA expression levels of ZWINT
were significantly increased in patients aged 24–40, compared with
those aged 41–69 (P<0.0001) and 70–93 (P<0.01). Nevertheless,
no statistical difference was observed between patients aged 41–69
and aged 70–93 (P>0.1; Fig. 3A).
Additionally, no significant differences were found between
patients with positive nodal status and those with negative status
(P=0.7048; Table II). Furthermore,
in terms of classical molecular types of BC, patients with estrogen
receptor (ER)− or progesterone receptor (PR)−
(P<0.0001 for both) and human epidermal growth factor receptor 2
(HER2)+ (P=0.0216) status had higher ZWINT expression
levels than their respective opposite status (Table II). Triple-negative BC (TNBC), an
aggressive type of BC, has been identified to lack expression of
PR, ER and HER2 (34). The
expression of ZWINT was found to be significantly upregulated in
patients with TNBC (P<0.0001; Table
II). Additionally, patients with basal-like features had
significantly increased ZWINT expression compared with patients
without basal-like traits (P<0.0001; Table II). Moreover, in terms of Scarff
Bloom and Richardson (SBR) grades, patients with a more advanced
SBR grade exhibited higher ZWINT expression (SBR3 > SBR1,
P<0.0001; SBR2 > SBR1, P<0.0001; SBR3 > SBR2,
P<0.0001; Fig. 3B). In addition
to these results, IHC indicated significant differences in the
protein expression of ZWINT between various age groups, PR and HER2
expression groups, and TNBC status groups (all P<0.05), whereas
there was no statistically significant difference between groups
with different node status or ER expression status (both P>0.05;
Table III).
 | Table II.Clinicopathological characteristics
and the mRNA expression levels of ZWINT in patients with BC
according to the bc-GenExMiner analysis.a |
Table II.
Clinicopathological characteristics
and the mRNA expression levels of ZWINT in patients with BC
according to the bc-GenExMiner analysis.a
| Variables | Number | mRNA
expression | P-value |
|---|
| Nodal status |
|
| 0.7048 |
|
Negative | 2426 | – |
|
|
Positive | 1480 | – |
|
| ER (IHC) |
|
| <0.0001 |
|
Negative | 1388 | Increased |
|
|
Positive | 3675 | – |
|
| PR (IHC) |
|
| <0.0001 |
|
Negative | 804 | Increased |
|
|
Positive | 1249 | – |
|
| HER2 (IHC) |
|
| 0.0216 |
|
Negative | 1409 | – |
|
|
Positive | 201 | Increased |
|
| Triple-negative
status |
|
| <0.0001 |
| No | 3831 | – |
|
|
Yes | 374 | Increased |
|
| Basal-like
status |
|
| <0.0001 |
|
None-basal-like | 3904 | – |
|
|
Basal-like | 1048 | Increased |
|
 | Table III.ZWINT expression in subgroups as
detected by immunohistochemistry. |
Table III.
ZWINT expression in subgroups as
detected by immunohistochemistry.
|
| ZWINT
expression |
|---|
|
|
|
|---|
| Variables | Number (n=62) | Low | High | P-value |
|---|
| Age (years) |
|
|
| 0.0063 |
|
≤40 | 38 | 8 | 30 |
|
|
>40 | 24 | 14 | 10 |
|
| Nodal status |
|
|
| 0.2884 |
|
Negative | 37 | 11 | 26 |
|
|
Positive | 25 | 11 | 14 |
|
| ER |
|
|
| 0.2891 |
|
Negative | 32 | 9 | 23 |
|
|
Positive | 30 | 13 | 17 |
|
| PR |
|
|
| 0.0008 |
|
Negative | 38 | 7 | 31 |
|
|
Positive | 24 | 15 | 9 |
|
| HER2 |
|
|
| 0.0277 |
|
Negative | 40 | 10 | 30 |
|
|
Positive | 22 | 12 | 10 |
|
| Triple-negative
status |
|
|
| 0.0018 |
| No | 44 | 20 | 24 |
|
|
Yes | 18 | 2 | 16 |
|
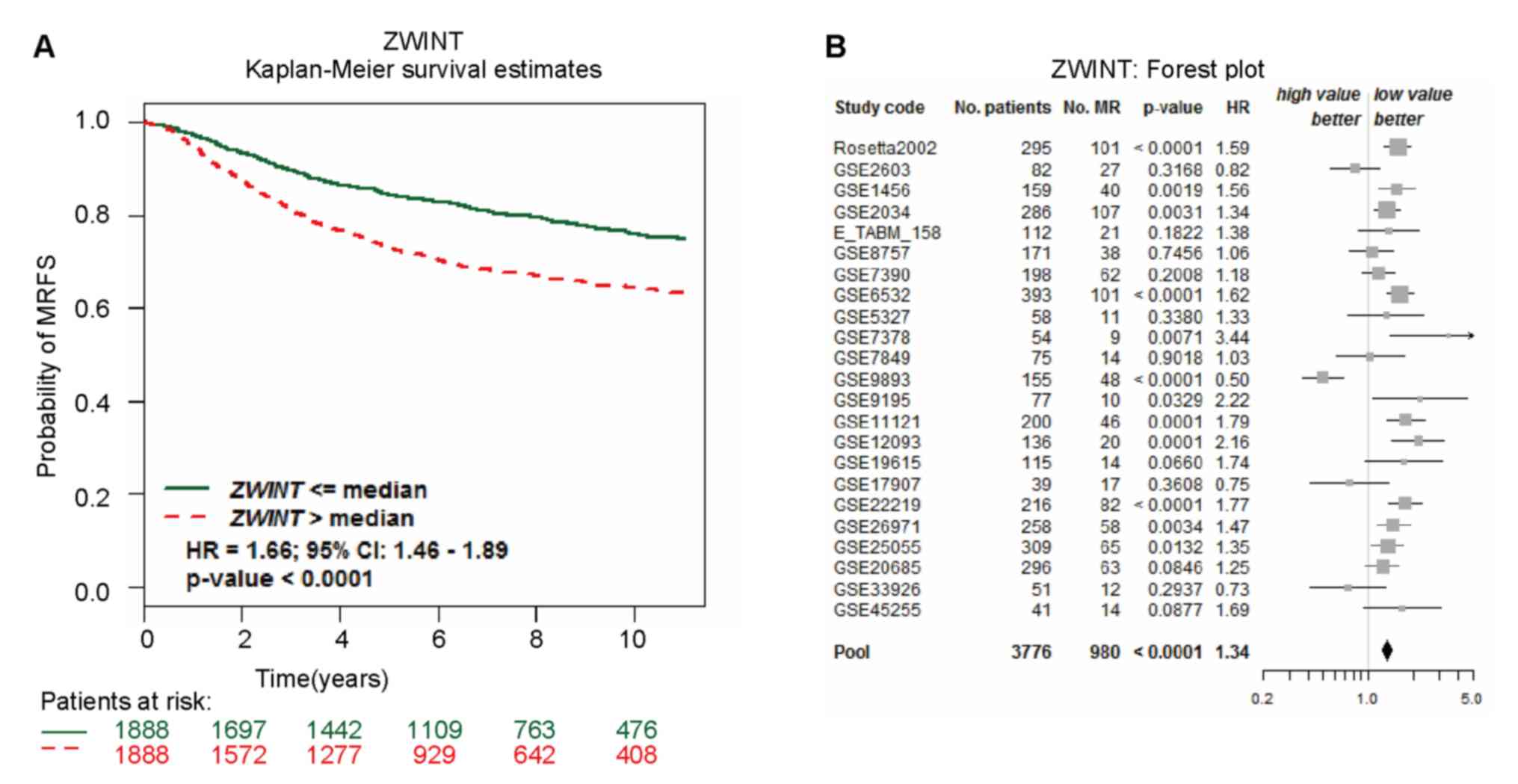
ZWINT expression and the prognosis of
patients with BC
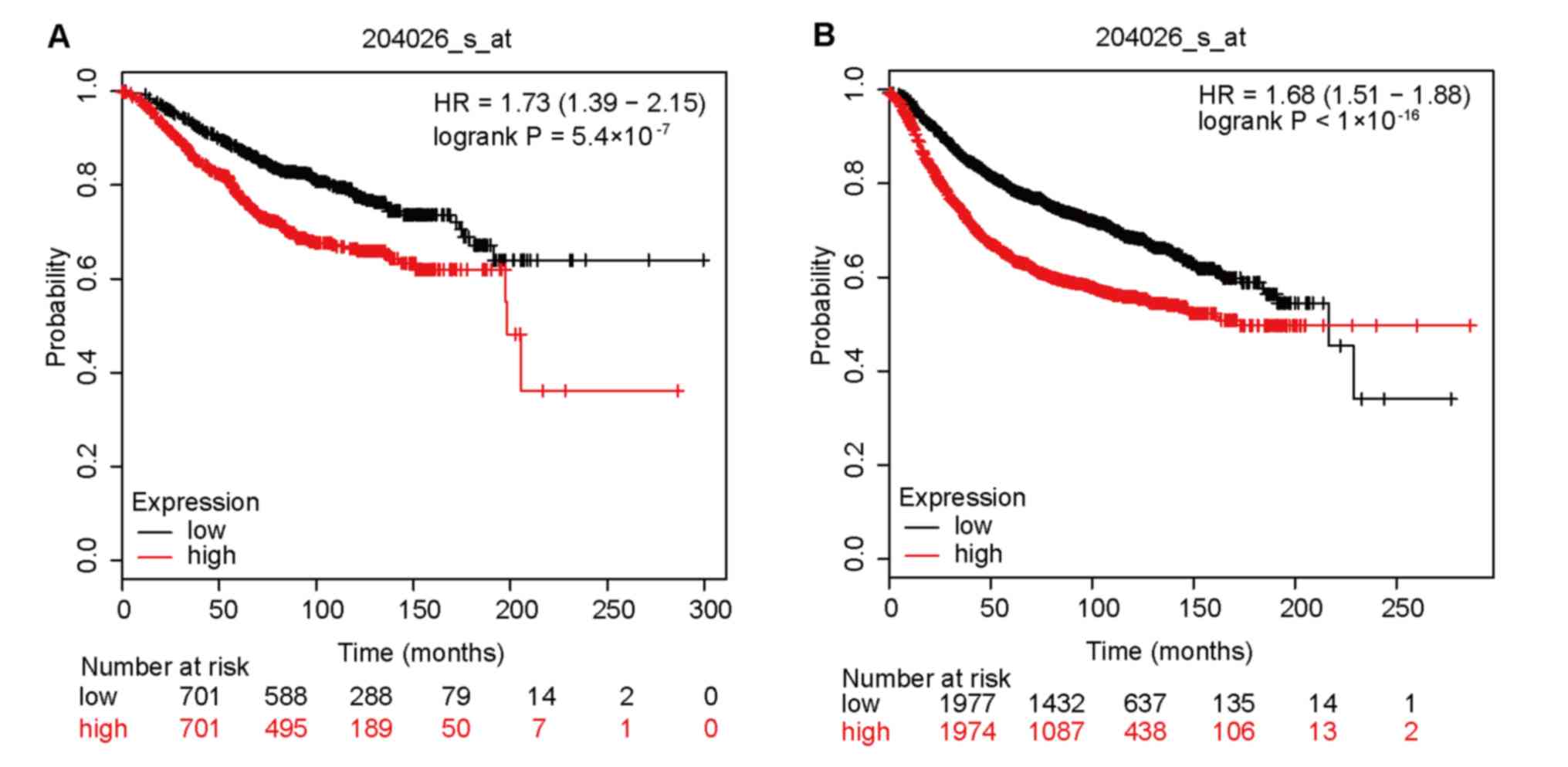
According to Kaplan-Meier Plotter analysis, higher
ZWINT expression was associated with a shorter OS (HR=1.73; 95%
CI=1.39–2.15; P=5.4×10−7) and a shorter RFS (HR=1.68;
95% CI=1.51–1.88; P<1×10−16) (Fig. 4). Gene prognostic analysis was
conducted with bc-GenExMiner, which involved the integration of the
accessible annotated genomic resources in order to explore the
association between ZWINT expression and metastatic relapse-free
survival (MRFS). As presented in Fig.
5A, increased ZWINT expression levels were related to shorter
MRFS (HR=1.66; 95% CI=1.46–1.89; P<0.0001). Moreover, a higher
risk of metastatic relapse was observed in patients with elevated
ZWINT expression (HR=1.34; 95% CI=1.26–1.43; P<0.0001; Fig. 5B).
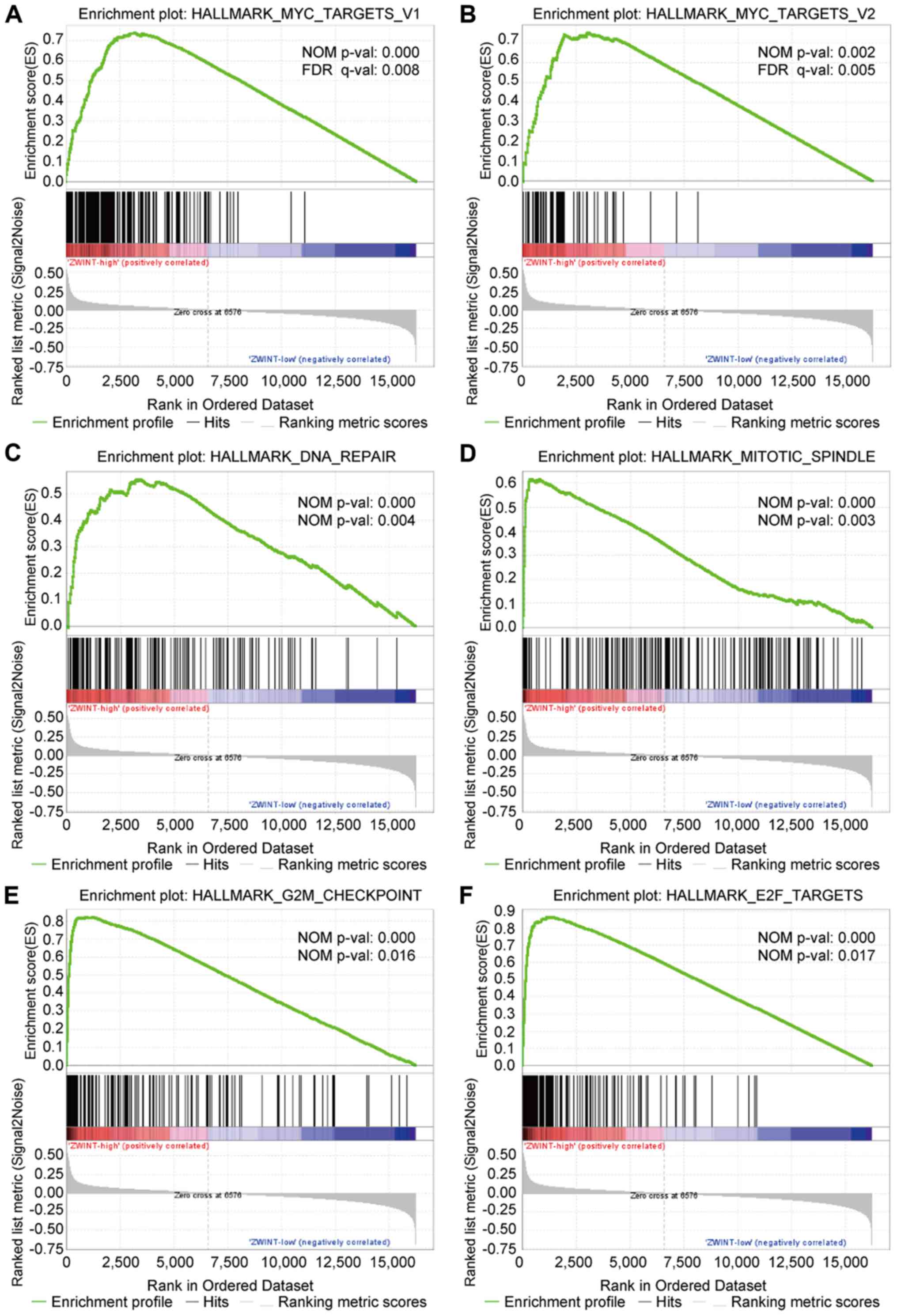
Molecular mechanisms and co-expression
of ZWINT in BC
Increased ZWINT expression was found to be related
to worse prognosis in patients with BC. Nonetheless, the underlying
molecular mechanisms remain to be explored. In order to evaluate
whether the expression levels of ZWINT were related to known gene
signatures, GSEA was performed with BC RNA-sequencing data from
TCGA database. GSEA revealed that 11 hallmark pathways were
upregulated in the high ZWINT expression group, indicating that
ZWINT expression was positively associated with these pathways
(Table IV). Most of these pathways
are implicated in cellular metabolism, biosynthesis and cell damage
repair processes, including Myc targets V1/2, DNA repair, mitotic
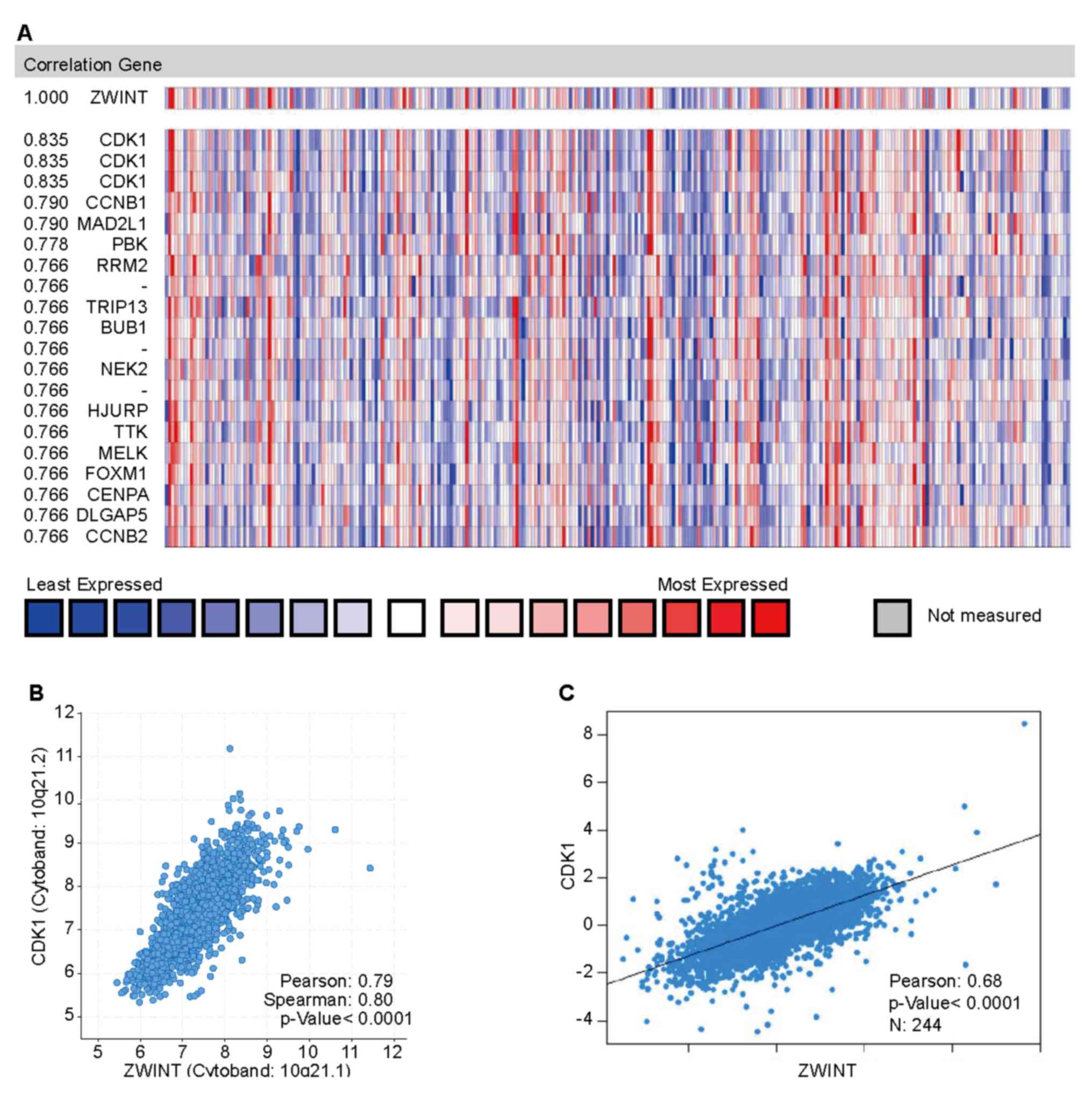
spindle, G2M checkpoint and E2F targets (Fig. 6). In addition, co-expression analysis
was performed using the Oncomine database to further study the
regulation of ZWINT. In total, 124 co-expression genes were
filtered out in a cohort of 289 patients with BC (Fig. 7A) (35). The gene with the greatest correlation
score, cyclin-dependent kinase 1 (CDK1), is considered to serve an
important role in the regulation of the cell cycle process and has
been confirmed to be abnormally upregulated in several types of
cancer by numerous studies (36–40). A
positive correlation was confirmed between ZWINT and CDK1 mRNA
expression in the METABRIC database by employing cBioPortal
(Spearman's correlation=0.80; Pearson's correlation=0.79; Fig. 7B). Data mining in bc-GenExMiner
further verified the positive correlation between ZWINT and CDK1
mRNA expression (Fig. 7C). These
results indicated that ZWINT could be involved in biosynthesis,
cell cycle and damage repair processes, and may be related to CDK1
signaling pathways.
 | Table IV.Association between ZW10 interacting
kinetochore protein expression and predefined gene signatures, as
determined by Gene Set Enrichment Analysis. |
Table IV.
Association between ZW10 interacting
kinetochore protein expression and predefined gene signatures, as
determined by Gene Set Enrichment Analysis.
| Name | Size | ES | NES | NOM P-value | FDR q-value |
|---|
|
HALLMARK_MYC_TARGETS_V1 | 198 | 0.740607 | 2.01761 | 0 | 0.007663 |
|
HALLMARK_DNA_REPAIR | 140 | 0.55583 | 2.001272 | 0 | 0.004239 |
|
HALLMARK_MITOTIC_SPINDLE | 198 | 0.616344 | 1.98359 | 0 | 0.003048 |
|
HALLMARK_MTORC1_SIGNALING | 196 | 0.63402 | 1.968746 | 0.001957 | 0.003571 |
|
HALLMARK_MYC_TARGETS_V2 | 58 | 0.755216 | 1.92142 | 0.001965 | 0.005123 |
|
HALLMARK_UNFOLDED_PROTEIN_RESPONSE | 108 | 0.492682 | 1.888622 | 0.009653 | 0.00677 |
|
HALLMARK_SPERMATOGENESIS | 73 | 0.607111 | 1.791163 | 0.00813 | 0.015301 |
|
HALLMARK_G2M_CHECKPOINT | 196 | 0.822451 | 1.774046 | 0 | 0.016042 |
|
HALLMARK_E2F_TARGETS | 199 | 0.865591 | 1.763097 | 0 | 0.01676 |
|
HALLMARK_OXIDATIVE_PHOSPHORYLATION | 198 | 0.550228 | 1.748484 | 0.052731 | 0.017751 |
|
HALLMARK_GLYCOLYSIS | 181 | 0.430354 | 1.544676 | 0.040619 | 0.083129 |
Discussion
Previous studies have examined the expression and
roles of ZWINT in multiple types of cancer (7–9,11,12).
Endo et al (10) demonstrated
that the interaction of ZWINT with Terf/TRIM17 resulted in its
degradation and the consequent regulation of cell proliferation in
BC MCF-7 and 293-T cells. Miller et al (41) identified a total of 62 genes,
including ZWINT, whose expression levels were markedly changed in
BC after short-term therapy with letrozole, indicating that ZWINT
was potentially involved in estrogen regulation. However, the
expression profile of ZWINT and its effect on BC prognosis remain
unexplored.
In the present study, the expression profile of
ZWINT in BC was initially evaluated by IHC and GEPIA. Βoth the mRNA
and protein expression levels of ZWINT were found to be elevated in
BC tissues, suggesting that it may serve an oncogenic role in BC.
The Oncomine database search revealed that the transcriptional
expression of ZWINT was significantly upregulated in mucinous,
medullary, invasive lobular and invasive ductal breast carcinoma.
Furthermore, increased ZWINT expression was found to be
significantly associated with HER2 positivity, a basal-like subtype
of breast carcinoma, TNBC and increased histological grades
according to SBR grading. Moreover, significant associations were
observed between increased ZWINT mRNA expression and negative
expression of ER/PR as well as younger age of the patients.
Previous studies have demonstrated that BC in young women (BCYW)
has distinctive clinicopathological characteristics compared with
BC in post-menopausal women (42,43).
Specifically, BCYW is often of a higher grade, is more aggressive,
lacks endocrine receptor expression and has a higher proportion of
HER2+ and triple-negative histology (44–46). The
present study demonstrated that ZWINT mRNA expression was higher in
BCYW than in BC in older patients, suggesting that ZWINT may serve
an important role in the progression of BC, particularly in BCYW.
Furthermore, IHC detection revealed a close association between
ZWINT expression and the age of patients, PR and HER2 status, and
TNBC in BC. Nevertheless, the results were partly in disagreement
with the findings of the bc-GenExMiner. For instance, no
association was found between ZWINT expression and ER status in the
study cohort, which may be due to the relatively small sample size,
and ethnic or geographical differences. Kaplan Meier-plotter
analysis revealed that high ZWINT expression was related to
unfavorable OS and RFS. Additionally, data mining with
bc-GenExMiner demonstrated that high ZWINT expression was strongly
associated with increased risk of metastatic relapse. These
findings suggested that ZWINT may serve as a biomarker for the
prognosis of BC.
To explore the role of ZWINT in BC, GSEA was
performed. This analysis demonstrated that higher expression of
ZWINT was closely associated with cell cycle-related pathways,
including Myc targets V1/2, DNA repair, mitotic spindle, G2M
checkpoint and E2F targets. It is generally believed that aberrant
cellular proliferation is a hallmark of malignancies and tumor
cells tend to exhibit altered expression of genes that directly
modulate their cell cycle (47).
This process occurs either via mutations in the upstream signaling
pathways or as a consequence of genetic impairments in genes
encoding cell cycle-related proteins (48). As a proto-oncogene, Myc encodes a
nuclear phosphoprotein and is involved in cell cycle progression,
apoptosis and cellular transformation (49). Myc is rigorously controlled in normal
cells, whereas it is aberrantly expressed in the majority of types
of human cancer (50). Notably, the
internal interaction between Myc and the E2F family of
transcription factors in BC has been reported by several studies.
For instance, Fujiwara et al (51) generated a hybrid mouse model and
demonstrated that Myc-induced breast tumor latency was
significantly decreased in E2F1 knockout mice and increased in E2F2
and E2F3 knockout mice. A follow-up study, however, revealed that
lack of E2F1 or E2F3 significantly postponed tumor onset in both
ErbB2- and Myc-triggered breast tumorigenesis, whereas lack of E2F2
promoted breast tumorigenesis induced by Myc overexpression
(52). The present results
demonstrated that in addition to recognized involvement in the
mitotic spindle checkpoint, ZWINT may participate in the signaling
pathways of Myc and E2F in BC. Nevertheless, given the complexity
of the cell cycle, the precise role of ZWINT in BC could not be
determined. To further investigate the molecular mechanism of ZWINT
in BC, co-expression and correlation analyses were performed. It
was demonstrated that ZWINT may be closely associated with the CDK1
signaling pathways in BC. Importantly, CDK1 has been observed to
serve an essential role in tumor initiation and progression in
different types of carcinomas (40,53,54).
Ablation of the CDK1 gene induced resistance to NRAS-G12V-induced
tumorigenesis in liver cells (55).
Moreover, CDK1 inhibition in combination with mitogen-activated
protein kinase kinase inhibition synergistically increased the
apoptotic rate of colorectal cancer cells and decreased clonogenic
survival as compared with monotreatment (56). These results suggested that ZWINT may
promote tumor formation and progression by modulating CDK1
expression in BC. Further studies are warranted to determine the
relationship between ZWINT and CDK1 in BC.
In summary, the present study identified that ZWINT
was commonly upregulated in BC and may aid prognostic prediction in
patients with BC. ZWINT may serve as an independent prognostic
biomarker for BC; however, further research is required to
substantiate the findings of the present study.
Acknowledgments
Not applicable.
Funding
This project was supported by the Hubei Provincial
Natural Science Foundation of China (grant no. 2015000250) and the
Clinical Research Physician Program of Tongji Medical College, HUST
(grant no. 2017zqnlxr01).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
HL was completed the bioinformatic research and was
a major contributor of manuscript preparation. WZ designed the
project and made the study plan. YD was responsible for the
analysis of the clinical characteristics of breast cancer
specimens. GW and MD performed the immunohistochemistry and
analyzed the results. ZY and XL contributed to the writing and
revision of the manuscript, and provided valuable input on the
design of the experiment and the interpretation of the results. All
authors read and approved the final version of the manuscript.
Ethics approval and consent to
participate
The experimental protocol of the present study was
approved by the Ethics Committee of Tongji Hospital. Written
informed consent was obtained from all patients.
Patient consent for publication
The experimental protocol of this study was approved
by the Ethics Committee of Tongji Hospital, Tongji Medical College
of Huazhong University of Science and Technology. All patients
provided signed informed consent.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Greenlee H, DuPont-Reyes MJ, Balneaves LG,
Carlson LE, Cohen MR, Deng G, Johnson JA, Mumber M, Seely D, Zick
SM, et al: Clinical practice guidelines on the evidence-based use
of integrative therapies during and after breast cancer treatment.
CA Cancer J Clin. 67:194–232. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Runowicz CD, Leach CR, Henry NL, Henry KS,
Mackey HT, Cowens-Alvarado RL, Cannady RS, Pratt-Chapman ML, Edge
SB, Jacobs LA, et al: American cancer society/American society of
clinical oncology breast cancer survivorship care guideline. J Clin
Oncol. 34:611–635. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Starr DA, Saffery R, Li Z, Simpson AE,
Choo KH, Yen TJ and Goldberg ML: HZwint-1, a novel human
kinetochore component that interacts with HZW10. J Cell Sci.
113:1939–1950. 2000.PubMed/NCBI
|
|
5
|
Vos LJ, Famulski JK and Chan GK: hZwint-1
bridges the inner and outer kinetochore: Identification of the
kinetochore localization domain and the hZw10-interaction domain.
Biochem J. 436:157–168. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kops GJ, Kim Y, Weaver BA, Mao Y, McLeod
I, Yates JR III, Tagaya M and Cleveland DW: ZW10 links mitotic
checkpoint signaling to the structural kinetochore. J Cell Biol.
169:49–60. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tang J, He D, Yang P, He J and Zhang Y:
Genome-wide expression profiling of glioblastoma using a large
combined cohort. Sci Rep. 8:151042018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu Z, Zhou Y, Cao Y, Dinh TL, Wan J and
Zhao M: Identification of candidate biomarkers and analysis of
prognostic values in ovarian cancer by integrated bioinformatics
analysis. Med Oncol. 33:1302016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ying H, Xu Z, Chen M, Zhou S, Liang X and
Cai X: Overexpression of Zwint predicts poor prognosis and promotes
the proliferation of hepatocellular carcinoma by regulating
cell-cycle-related proteins. Onco Targets Ther. 11:689–702. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Endo H, Ikeda K, Urano T, Horie-Inoue K
and Inoue S: Terf/TRIM17 stimulates degradation of kinetochore
protein ZWINT and regulates cell proliferation. J Biochem.
151:139–144. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pérez de Castro I, de Cárcer G and
Malumbres M: A census of mitotic cancer genes: New insights into
tumor cell biology and cancer therapy. Carcinogenesis. 28:899–912.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bieniek J, Childress C, Swatski MD and
Yang W: COX-2 inhibitors arrest prostate cancer cell cycle
progression by down-regulation of kinetochore/centromere proteins.
Prostate. 74:999–1011. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cancer Genome Atlas Network, .
Comprehensive molecular portraits of human breast tumours. Nature.
490:61–70. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Curtis C, Shah SP, Chin SF, Turashvili G,
Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et
al: The genomic and transcriptomic architecture of 2,000 breast
tumours reveals novel subgroups. Nature. 486:346–352. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ma XJ, Dahiya S, Richardson E, Erlander M
and Sgroi DC: Gene expression profiling of the tumor
microenvironment during breast cancer progression. Breast Cancer
Res. 11:R72009. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Richardson AL, Wang ZC, De Nicolo A, Lu X,
Brown M, Miron A, Liao X, Iglehart JD, Livingston DM and Ganesan S:
X chromosomal abnormalities in basal-like human breast cancer.
Cancer Cell. 9:121–132. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jézéquel P, Campone M, Gouraud W,
Guérin-Charbonnel C, Leux C, Ricolleau G and Campion L:
bc-GenExMiner: An easy-to-use online platform for gene prognostic
analyses in breast cancer. Breast Cancer Res Treat. 131:765–775.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jézéquel P, Frénel JS, Campion L,
Guérin-Charbonnel C, Gouraud W, Ricolleau G and Campone M:
bc-GenExMiner 3.0: New mining module computes breast cancer gene
expression correlation analyses. Database (Oxford).
2013:bas0602013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Forbes SA, Beare D, Bindal N, Bamford S,
Ward S, Cole CG, Jia M, Kok C, Boutselakis H, De T, et al: COSMIC:
High-resolution cancer genetics using the catalogue of somatic
mutations in cancer. Curr Protoc Hum Genet. 91:10.11.1–10.11.37.
2016. View
Article : Google Scholar
|
|
22
|
Forbes SA, Beare D, Boutselakis H, Bamford
S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al:
COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids
Res. 45:D777–D783. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Razavi P, Chang MT, Xu G, Bandlamudi C,
Ross DS, Vasan N, Cai Y, Bielski CM, Donoghue MTA, Jonsson P, et
al: The genomic landscape of endocrine-resistant advanced breast
cancers. Cancer Cell. 34:427.e6–438.e6. 2018. View Article : Google Scholar
|
|
26
|
Pereira B, Chin SF, Rueda OM, Vollan HK,
Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ,
et al: The somatic mutation profiles of 2,433 breast cancers
refines their genomic and transcriptomic landscapes. Nat Commun.
7:114792016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shah SP, Roth A, Goya R, Oloumi A, Ha G,
Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al: The clonal
and mutational evolution spectrum of primary triple-negative breast
cancers. Nature. 486:395–399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Banerji S, Cibulskis K, Rangel-Escareno C,
Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY,
Sougnez C, Zou L, et al: Sequence analysis of mutations and
translocations across breast cancer subtypes. Nature. 486:405–409.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Stephens PJ, Tarpey PS, Davies H, Van Loo
P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell
GR, et al: The landscape of cancer genes and mutational processes
in breast cancer. Nature. 486:400–404. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ciriello G, Gatza ML, Beck AH, Wilkerson
MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, et
al: Comprehensive molecular portraits of invasive lobular breast
cancer. Cell. 163:506–519. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Eirew P, Steif A, Khattra J, Ha G, Yap D,
Farahani H, Gelmon K, Chia S, Mar C, Wan A, et al: Dynamics of
genomic clones in breast cancer patient xenografts at single-cell
resolution. Nature. 518:422–426. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lefebvre C, Bachelot T, Filleron T,
Pedrero M, Campone M, Soria JC, Massard C, Lévy C, Arnedos M,
Lacroix-Triki M, et al: Mutational profile of metastatic breast
cancers: A retrospective analysis. PLoS Med. 13:e10022012016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ivshina AV, George J, Senko O, Mow B,
Putti TC, Smeds J, Lindahl T, Pawitan Y, Hall P, Nordgren H, et al:
Genetic reclassification of histologic grade delineates new
clinical subtypes of breast cancer. Cancer Res. 66:10292–10301.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Malumbres M: Cyclin-dependent kinases.
Genome Biol. 15:1222014. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ravindran Menon D, Luo Y, Arcaroli JJ, Liu
S, KrishnanKutty LN, Osborne DG, Li Y, Samson JM, Bagby S, Tan AC,
et al: CDK1 interacts with Sox2 and promotes tumor initiation in
human melanoma. Cancer Res. 78:6561–6574. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wei D, Parsels LA, Karnak D, Davis MA,
Parsels JD, Marsh AC, Zhao L, Maybaum J, Lawrence TS, Sun Y and
Morgan MA: Inhibition of protein phosphatase 2A radiosensitizes
pancreatic cancers by modulating CDC25C/CDK1 and homologous
recombination repair. Clin Cancer Res. 19:4422–4432. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Whalley HJ, Porter AP, Diamantopoulou Z,
White GR, Castañeda-Saucedo E and Malliri A: Cdk1 phosphorylates
the Rac activator Tiam1 to activate centrosomal Pak and promote
mitotic spindle formation. Nat Commun. 6:74372015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu CX, Wang XQ, Chok SH, Man K, Tsang SHY,
Chan ACY, Ma KW, Xia W and Cheung TT: Blocking CDK1/PDK1/β-Catenin
signaling by CDK1 inhibitor RO3306 increased the efficacy of
sorafenib treatment by targeting cancer stem cells in a preclinical
model of hepatocellular carcinoma. Theranostics. 8:3737–3750. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Miller WR, Larionov AA, Renshaw L,
Anderson TJ, White S, Murray J, Murray E, Hampton G, Walker JR, Ho
S, et al: Changes in breast cancer transcriptional profiles after
treatment with the aromatase inhibitor, letrozole. Pharmacogenet
Genomics. 17:813–826. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cancello G, Maisonneuve P, Rotmensz N,
Viale G, Mastropasqua MG, Pruneri G, Veronesi P, Torrisi R,
Montagna E, Luini A, et al: Prognosis and adjuvant treatment
effects in selected breast cancer subtypes of very young women
(<35 years) with operable breast cancer. Ann Oncol.
21:1974–1981. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Narod SA: Breast cancer in young women.
Nat Rev Clin Oncol. 9:460–470. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Azim HA Jr, Michiels S, Bedard PL, Singhal
SK, Criscitiello C, Ignatiadis M, Haibe-Kains B, Piccart MJ,
Sotiriou C and Loi S: Elucidating prognosis and biology of breast
cancer arising in young women using gene expression profiling. Clin
Cancer Res. 18:1341–1351. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Han W and Kang SY; Korean Breast Cancer
Society, : Relationship between age at diagnosis and outcome of
premenopausal breast cancer: Age less than 35 years is a reasonable
cut-off for defining young age-onset breast cancer. Breast Cancer
Res Treat. 119:193–200. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Keegan TH, DeRouen MC, Press DJ, Kurian AW
and Clarke CA: Occurrence of breast cancer subtypes in adolescent
and young adult women. Breast Cancer Res. 14:R552012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sherr CJ: Cancer cell cycles. Science.
274:1672–1677. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Otto T and Sicinski P: Cell cycle proteins
as promising targets in cancer therapy. Nat Rev Cancer. 17:93–115.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bretones G, Delgado MD and León J: Myc and
cell cycle control. Biochim Biophys Acta. 1849:506–516. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wahlström T and Henriksson MA: Impact of
MYC in regulation of tumor cell metabolism. Biochim Biophys Acta.
1849:563–569. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fujiwara K, Yuwanita I, Hollern DP and
Andrechek ER: Prediction and genetic demonstration of a role for
activator E2Fs in Myc-induced tumors. Cancer Res. 71:1924–1932.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wu L, de Bruin A, Wang H, Simmons T,
Cleghorn W, Goldenberg LE, Sites E, Sandy A, Trimboli A, Fernandez
SA, et al: Selective roles of E2Fs for ErbB2- and Myc-mediated
mammary tumorigenesis. Oncogene. 34:119–128. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu X, Gao Y, Ye H, Gerrin S, Ma F, Wu Y,
Zhang T, Russo J, Cai C, Yuan X, et al: Positive feedback loop
mediated by protein phosphatase 1α mobilization of P-TEFb and basal
CDK1 drives androgen receptor in prostate cancer. Nucleic Acids
Res. 45:3738–3751. 2017.PubMed/NCBI
|
|
54
|
Saatci Ö, Borgoni S, Akbulut Ö, Durmuş S,
Raza U, Eyüpoğlu E, Alkan C, Akyol A, Kütük Ö, Wiemann S and Şahin
Ö: Targeting PLK1 overcomes T-DM1 resistance via CDK1-dependent
phosphorylation and inactivation of Bcl-2/xL in HER2-positive
breast cancer. Oncogene. 37:2251–2269. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Diril MK, Ratnacaram CK, Padmakumar VC, Du
T, Wasser M, Coppola V, Tessarollo L and Kaldis P: Cyclin-dependent
kinase 1 (Cdk1) is essential for cell division and suppression of
DNA re-replication but not for liver regeneration. Proc Natl Acad
Sci USA. 109:3826–3831. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang P, Kawakami H, Liu W, Zeng X,
Strebhardt K, Tao K, Huang S and Sinicrope FA: Targeting CDK1 and
MEK/ERK overcomes apoptotic resistance in BRAF-mutant human
colorectal cancer. Mol Cancer Res. 16:378–389. 2018. View Article : Google Scholar : PubMed/NCBI
|