Introduction
Pancreatic cancer still has an extremely poor
prognosis. The number of deaths due to pancreatic cancer has been
increasing worldwide. In fact, it was predicted that the total
deaths from pancreatic cancer will become the second leading cause
of cancer-related deaths in the United States by 2030 (1). In addition, National Cancer Center
Japan (https://ganjoho.jp/reg_stat/statistics/stat/summary.html)
reported that pancreatic cancer became the fourth leading cause of
cancer deaths in Japan in 2016. There are several reasons for the
low survival rate of pancreatic cancer. Among them, the most
pressing problem that has been very hard to resolve is the
difficulty of diagnosing the disease at an early stage. It is very
difficult to detect pancreatic cancer by imaging studies performed
in routine medical examinations, and by the time it is discovered
due to worsening symptoms, such as jaundice or abdominal/back pain,
it would have already progressed to an incurable phase. Another
problem for its early diagnosis is the lack of a useful
biomarker.
Many types of biomarkers for pancreatic cancer in
patient serum, pancreatic juice, or stool have been proposed. Among
these biomarkers, serum carbohydrate antigen (CA) 19-9 is the most
commonly used in the usual clinical setting. However, several
reports have suggested that the positive predictive value (PPV) of
CA19-9 for pancreatic cancer was very low (0.5 to 0.9%) in
asymptomatic patients (2,3). In addition, serum CA19-9 is elevated in
many other types of cancers and benign diseases. As such, the
utility of serum CA19-9 testing as a routine screening tool to find
curable pancreatic cancer is extremely limited. Although pancreatic
cancers are found at an advanced stage in the majority of cases,
Yachida et al (4) have shown
that the progression of pancreatic cancer is relatively slow.
According to their report, it takes at least 10 years for the
initiated tumor cells to become parental non-metastatic founder
cells, and at least 5 more years for the acquisition of metastatic
ability. This suggests that if a potent diagnostic tool that can
detect early stages of the disease was available, we may have a
better chance than previously thought to find and treat pancreatic
cancer before it becomes fatal.
MicroRNA (miRNA) is a small non-coding RNA that
binds to complementary sequences in the 3′ untranslated region of
target messenger RNAs (mRNAs); this leads to the inhibition of
their translation or their degradation, and thereby negatively
regulate gene expression. Recent studies have revealed that many
types of cancer cells produce specific miRNAs (5–7);
therefore, the utility of specific miRNAs as biomarkers for cancers
has been widely investigated. Many studies have shown that specific
alterations in miRNA expression patterns are observable in the
blood (8,9), pancreatic tissues (10,11), or
pancreatic juice (12,13) of pancreatic cancer patients. For
example, Abue et al (9)
reported that miRNA-483-3p levels were elevated in the blood of
pancreatic cancer patients when compared to intraductal papillary
neoplasm patients and healthy controls, and it could be used to
differentiate pancreatic cancer from intraductal papillary
neoplasms with a sensitivity of 43.8%. In addition, Panarelli et
al (10) showed that miR-21,
miR-221, miR-155, miR-100, and miR-181b were overexpressed in
resected pancreatic cancer tissues when compared to benign
lesions.
One advantage of using miRNAs as biomarkers is their
stability. Even though plasma has a high level of RNase activity,
miRNAs are resistant to being degraded by them (14); they are protected from endogenous
RNase activity because cells release the miRNAs by incorporating
them into exosomes (15). Exosomes
are membrane-bound particles of 40 to 100 nm that are released from
many types of cells; they can be taken up by neighboring cells, and
modulate the bioactivities of the recipient cells with their
components, such as lipids, proteins, and nucleic acids (16). Exosomes are enriched with several
hundred folds of mRNAs and miRNAs as compared to the donor cells,
and the miRNAs in urinary exosomes have been widely explored as
biomarkers, especially for cancers of the urinary tract (17–19).
Moreover, some have raised the possibility that urine exosomes can
be derived from diseases in organs other than the urinary tract
(20,21).
Given these facts, in the present study, we
investigated miRNAs specific for pancreatic cancer in urinary
exosomes, and identified a novel biomarker candidate for this
disease.
Materials and methods
Patients
Patients admitted to the Gastroenterology Department
of Mie University Hospital between October 2015 and February 2017
with pancreatic ductal adenocarcinoma (PDAC) or chronic
pancreatitis (CP), including autoimmune pancreatitis, were enrolled
in this study. The final diagnoses of PDAC were confirmed by
endoscopic ultrasound-guided fine-needle aspiration, endoscopic
retrograde cholangiopancreatography, or endoscopic biopsy. The
diagnoses of CP were made by endoscopic ultrasound-guided
fine-needle aspiration and endoscopic retrograde
cholangiopancreatography. The diagnoses of autoimmune pancreatitis
were made from elevated serum IgG4 levels and positive staining for
IgG4 by immunohistochemistry. Healthy control subjects, adjusted by
age and sex, were also enrolled. All patients were newly diagnosed
and treatment-naïve. Patients with a present or past history of any
type of malignant neoplasm were excluded from this study.
This study was approved by the Ethics Committee of
Mie University Hospital (reference no. 2833). Written informed
consent was obtained from each patient included in the study. The
study protocol conformed to the ethical guidelines of the 1975
Declaration of Helsinki, as reflected in the a priori
approval from the institution's human research committee.
Urine and serum sample collection
Urine samples (more than 10 ml) were collected from
the PDAC, CP, and control subjects before highly invasive
examinations, such as endoscopic procedures. The samples were kept
at 4°C, and centrifuged at 3,000 × g for 15 min within 8 h after
collection. The supernatants were divided into 6-ml aliquots and
kept at −30°C until the analyses.
Serum was also collected from each subject before
highly invasive examinations. The samples were kept at −80°C until
the analyses.
Cell lines
The human PDAC cell lines PANC-1 (RCB2095) and MIA
PaCa-2 (RCB2094), human bile duct cell line HuCCT1 (RCB1960), human
hepatocellular carcinoma cell line HuH-7 (RCB1942), human liver
cancer cell line Hep G2 (RCB1886), and human gastric cancer cell
line KATO III (RCB2088) were purchased from RIKEN BRC. The colon
cancer cell line SW480 (ATCC CCL-228) was purchased from ATCC.
Cell cultures
All cell lines were cultured according to the
manufacturers' instructions. Briefly, PANC-1 cells, SW480 cells,
and HuH-7 cells were cultured at 37°C and 5% CO2 in
Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific,
Inc.) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher
Scientific, Inc.) and 1% penicillin-streptomycin (Thermo Fisher
Scientific, Inc.). MIA PaCa-2 cells were maintained in DMEM with
10% FBS, 2.5% horse serum (Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin. HepG2 cells were maintained in modified
Eagle's medium (Thermo Fisher Scientific, Inc.) with 10% FBS and 1%
penicillin-streptomycin. Kato III cells and HuCCT1 cells were
maintained in Roswell Park Memorial Institute medium 1640 (Thermo
Fisher Scientific, Inc.) containing 10% FBS and 1%
penicillin-streptomycin.
After incubation, the cells were washed with
phosphate-buffered saline and incubated for 24 h in FBS-free
medium. Then, the supernatants were centrifuged twice at 3,000 × g
for 15 min and collected and divided into 6-ml aliquots and kept at
−30°C until the analyses.
Isolation of exosomes from urine
samples and culture media
Exosomes were isolated from the urine samples and
culture media using ExoQuick-TC (System Biosciences) according to
the manufacturer's protocol. Briefly, the frozen urine samples or
culture media were thawed and centrifuged at 3,000 × g for 15 min
to eliminate impure substances, such as cells and cell debris.
Next, 5 ml of each supernatant and 1 ml of ExoQuick-Tc exosome
precipitation solution were mixed well by flicking the tubes. The
mixtures were allowed to settle overnight (for at least 12 h) at
4°C and were then centrifuged at 1,500 × g for 30 min. The exosomes
appeared as pellets and were isolated by aspiration of the
supernatants.
Isolation of exosomes from serum
samples
Exosomes were isolated from serum samples by using
ExoQuick (System Biosciences) according to the manufacturer's
instructions. The frozen sera were thawed and centrifuged at 3,500
× g for 20 min to eliminate impure substances, such as cells and
cell debris. Next, 250 µl of each supernatant and 63 µl of Exo
Quick exosome precipitation solution were mixed well by pipetting.
The solutions were then allowed to settle for 30 min on ice and
were then centrifuged at 1,500 × g for 35 min. The exosomes
appeared as pellets and were isolated by aspiration of the
supernatants.
Purification of the miRNA from
exosomes
The miRNAs were purified from the exosomes by using
a miRCURY RNA Isolation kit (Qiagen). Each exosome pellet was mixed
with 350 µl of lysis solution by vortexing for over 15 sec and was
confirmed to have completely dissolved. Subsequently, 200 µl of
99.5% ethanol was added to the mixture and vortexed for over 10
sec. The lysate was applied to a column assembled with a collection
tube and centrifuged at 14,000 × g for 1 min, after which the
flow-through was discarded. Next, 400 µl of wash solution was
applied to the column, and centrifuged at 14,000 × g for 1 min,
after which the flow-through was discarded. This washing procedure
was repeated twice, and the column was dried by centrifugation at
14,000 × g for 2 min. Afterwards, 50 µl of eluent was applied to
the column settled into an elution tube and centrifuged at 200 × g
for 2 min, then 14,000 × g for 1 min. The extraction liquid was
applied to the column again and centrifuged at 200 × g for 2 min,
followed by 14,000 × g for 1 min. The purified samples were stored
at −80°C.
Quality checking of the miRNA
The quality of the extracted miRNA was examined
using an Agilent Bioanalyzer 2100 and an RNA 6000 Small RNA kit
(Agilent Technologies). All of the miRNA samples were confirmed to
be suitable for further analyses.
miRNA expression profiling
The miRNA expression profiles were examined in nine
PDAC patients and seven controls with SurePrint G3 Human miRNA
Microarray, 8×60K Rel. 21.0, consist of 2549 human miRNA probes
(Agilent Technologies) with 60 ng miRNA per sample according to the
manufacturer's protocol. The hybridized chip was scanned using the
G2539A Microarray Scanner (Agilent Technologies) and analyzed using
Gene Spring GX software, version 13.1 (Agilent Technologies). Raw
intensities were not normalized because the levels of miRNA
expression were low.
Detection of miRNA
The levels of miRNA expression were validated in the
culture media, patient sera, and a large number of urine samples
using 3D digital PCR according to the results of the
microarray.
Complementary DNA (cDNA) was synthesized from 6 µl
of miRNA solution by using the miRCURY Universal cDNA Synthesis kit
II (Qiagen) with the following conditions according to the
manufacturer's instructions: 42°C for 60 min, 95°C for 5 min, and
then held at 4°C. Digital PCR was performed using a
QuantStudio® 3D digital PCR (Thermo Fisher Scientific,
Inc.). The reactions were prepared in a final volume of 18 µl,
containing 9 µl of Mastermix QuantStudio® 3D digital PCR
(Thermo Fisher Scientific, Inc.), 0.72 µl of cDNA, 1.8 µl of
primers from the miRCURY LNA PCR Primer Set (Qiagen), and 1.8 µl of
SYBR® Green I dye (Thermo Fisher Scientific, Inc.). The
working solution of SYBR® Green I dye was diluted
(1:1,000) with 100% dimethyl sulfoxide and stored in aliquots at
−20°C. The diluted SYBR® Green I dye was further diluted
(1:20) with Tris-EDTA buffer (pH 8.0). The reaction mix (14.5 µl
out of 18 µl) was loaded onto QuantStudio 3D digital PCR chips by
using a QuantStudio 3D digital PCR chip loader. The amplification
was performed with the following conditions: 96°C for 10 min, 39
cycles of 60°C for 2 min and 98°C for 30 sec, 60°C for 2 min, and
then holding at 10°C. The amplification results were analyzed with
QuantStudio® 3D AnalysisSuite™ Cloud Software (Applied
Biosystems). The software assessed the quality of the chip data and
estimated the concentrations by counting the number of positive
chambers.
Statistical analysis
All statistical tests were analyzed using SPSS
software, version 24.0 (IBM Inc.). The Fisher exact test was
applied to test the categorical data, and the Wilcoxon rank-sum
test or Kruskal-Wallis test with a Dunn-Bonferroni post hoc test
was applied to test the continuous data. The accuracy was
investigated using receiver operating characteristic (ROC) curve
analysis, the area under the curve (AUC), and accuracy measures for
determining a suitable cutoff value. P<0.05 was considered to
indicate a statistically significant difference.
Results
Identification of the
miR-3940-5p/miR-8069 ratio as a novel PDAC biomarker
Firstly, as a preliminary study, we analyzed the
expression of miRNA in urine exosomes by microarray in nine PDAC
and seven control subjects, and the expression profiles were
compared between these groups to find a candidate PDAC biomarker.
Table I shows the characteristics of
the subjects. There was no significant difference in the
characteristics (age and sex) between the PDAC and control groups.
In total, 38 miRNAs were upregulated by more than two-fold in the
PDAC patients when compared to the controls. The five most
upregulated miRNAs in the PDAC group are listed in Table II. Of note, the expression level of
miR-3940-5p was significantly greater in all of the PDAC patients
than in the controls (fold change: 3.047; P=0.036). On the other
hand, miR-8069 was expressed at a similar level in all of the PDAC
patients and controls (data not shown). Based on these results, we
identified the miR-3940-5p/miR-8069 ratio in urine exosomes as a
candidate novel biomarker for PDAC.
 | Table I.Patient characteristics analyzed by
the microarray. |
Table I.
Patient characteristics analyzed by
the microarray.
| Subjects | Age, years | Sex | Size, mm | Tumor location | T classification
(UICC 8th) | Clinical stage
(UICC 8th) |
|---|
| Patients with
pancreatic ductal adenocarcinoma | 65 | M | 18 | Head | T1 | III |
|
| 83 | M | 22 | Tail | T2 | II |
|
| 76 | F | 22 | Head | T4 | III |
|
| 53 | F | 40 | Head | T4 | III |
|
| 73 | F | 30 | Head | T4 | IV |
|
| 84 | M | 16 | Body-tail | T1 | I |
|
| 67 | M | 20 | Head | T2 | I |
|
| 89 | M | 27 | Head | T2 | III |
|
| 59 | F | 46 | Body-tail | T4 | IV |
| Controls | 78 | M |
|
|
|
|
|
| 62 | M |
|
|
|
|
|
| 76 | F |
|
|
|
|
|
| 68 | F |
|
|
|
|
|
| 57 | M |
|
|
|
|
|
| 81 | F |
|
|
|
|
|
| 76 | M |
|
|
|
|
 | Table II.Top five most upregulated microRNAs
in pancreatic ductal adenocarcinoma. |
Table II.
Top five most upregulated microRNAs
in pancreatic ductal adenocarcinoma.
| microRNA | Sequence | Fold change | P-value |
|---|
|
hsa-miR-3940-5p | CAGAGCCCGCCC | 3.047 | 0.036 |
| hsa-miR-6085 | TGTGCTCCCCCAGC | 2.973 | 0.108 |
| hsa-miR-4516 | GCCCCGACCCTTC | 2.662 | 0.120 |
| hsa-miR-4298 | CTGCCTCCTCCTCC | 2.623 | 0.061 |
|
hsa-miR-6749-5p | GCTCCCCCAACCC | 2.527 | 0.184 |
Preliminary validation in cell
lines
As the next step, we attempted to determine whether
the increase in the miR-3940-5p/miR-8069 ratio was specific for
PDAC by using seven different cancer cell lines. Because the
expression levels of miRNA in the exosomes, especially those
excreted into urine, were very low, it was difficult to quantify
them accurately with the conventional quantitative PCR method;
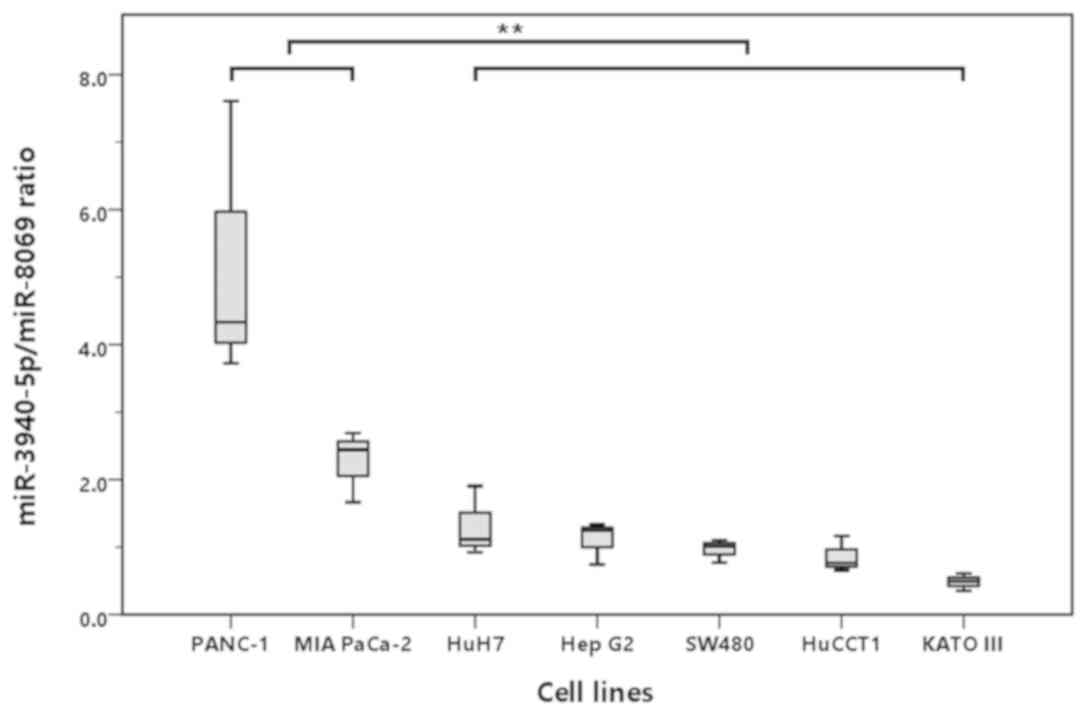
therefore, we measured them by 3D digital PCR. As shown in Fig. 1, the miR-3940-5p/miR-8069 ratio in
exosomes from the culture media was significantly higher in the two
PDAC cell lines (PANC-1 and MIA PaCa-2) than in the non-pancreatic
cancer cell lines (HuH-7, HepG2, SW480, HuCCT1, and Kato III;
P<0.001), suggesting that the increase in the
miR-3940-5p/miR-8069 ratio was specific to PDAC.
Comparison of the expression levels in
serum and urine exosomes
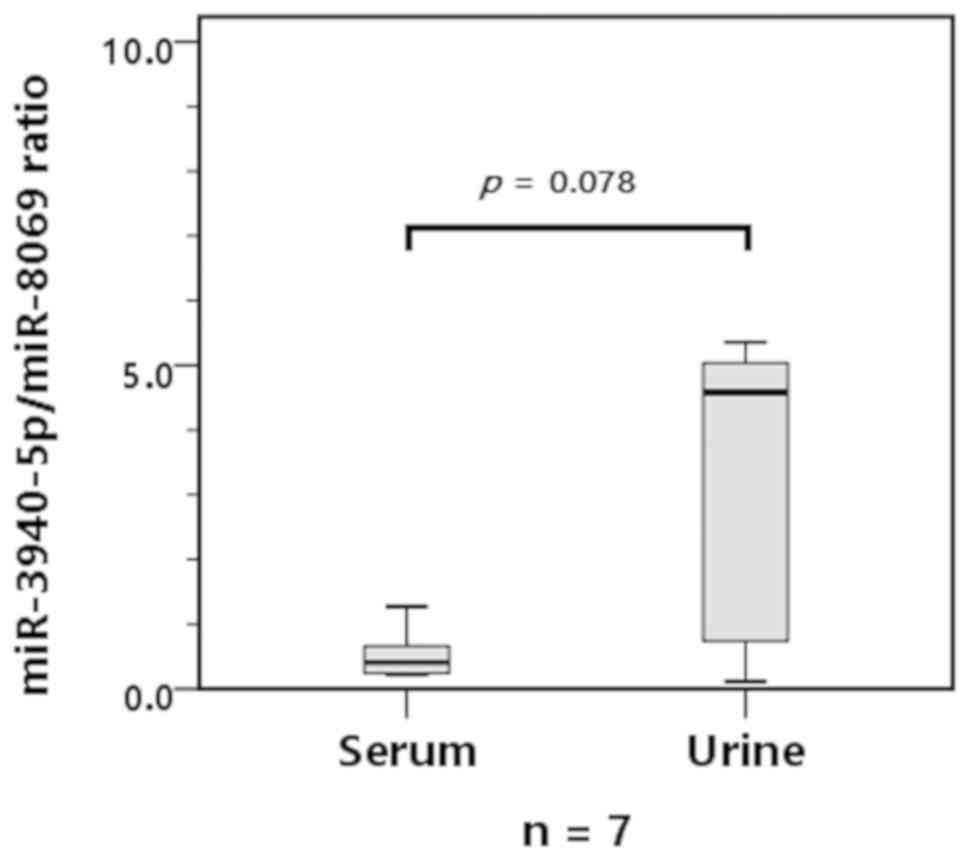
Next, we analyzed the miR-3940-5p/miR-8069 ratio in
exosomes derived from the serum or urine of seven PDAC patients.
Fig. 2 shows that the
miR-3940-5p/miR-8069 ratio was greater in the urine exosomes than
in the serum exosomes, although the difference did not reach a
statistically significant level, perhaps due to the small sample
size. These results suggested that urine samples are more suitable
than serum samples for analyzing the miR-3940-5p/miR-8069 ratio as
a biomarker for PDAC.
Validation in a larger number of
samples
According to the results of the preliminary
analyses, we attempted to further validate the usefulness of the
miR-3940-5p/miR-8069 ratio in urine exosomes as a biomarker for
PDAC in a larger number of samples. We examined a total of 43 PDAC,
12 CP, and 25 control subjects. Table
III shows the characteristics of the patients. The mean tumor
size of the PDAC cases was relatively large at 33.0±16.3 mm.
According to the eighth edition of the International Union Against
Cancer (UICC) classification system, 44.2% of the PDAC patients had
T4 tumors, and 46.5% had clinical stage IV disease. As was
mentioned above, the PDAC group in our study included advanced
cases of PDAC.
 | Table III.Background characteristics of the 43
pancreatic ductal adenocarcinoma cases. |
Table III.
Background characteristics of the 43
pancreatic ductal adenocarcinoma cases.
|
Characteristics | Number/mean ±
SD | Percentage |
|---|
| Age, years | 68.4±10.2 |
|
| Sex (male) | 25 | 58.1 |
| Tumor location |
|
|
|
Head | 26 | 60.5 |
|
Body-tail | 17 | 39.5 |
| Tumor size, mm | 33.0±16.3 |
|
| T classification
(UICC 8th) |
|
|
| T1 | 6 | 14.0 |
| T2 | 13 | 30.2 |
| T3 | 5 | 11.6 |
| T4 | 19 | 44.2 |
| Clinical stage
(UICC 8th) |
|
|
| I | 12 | 27.9 |
| II | 3 | 7.0 |
|
III | 8 | 18.6 |
| IV | 20 | 46.5 |
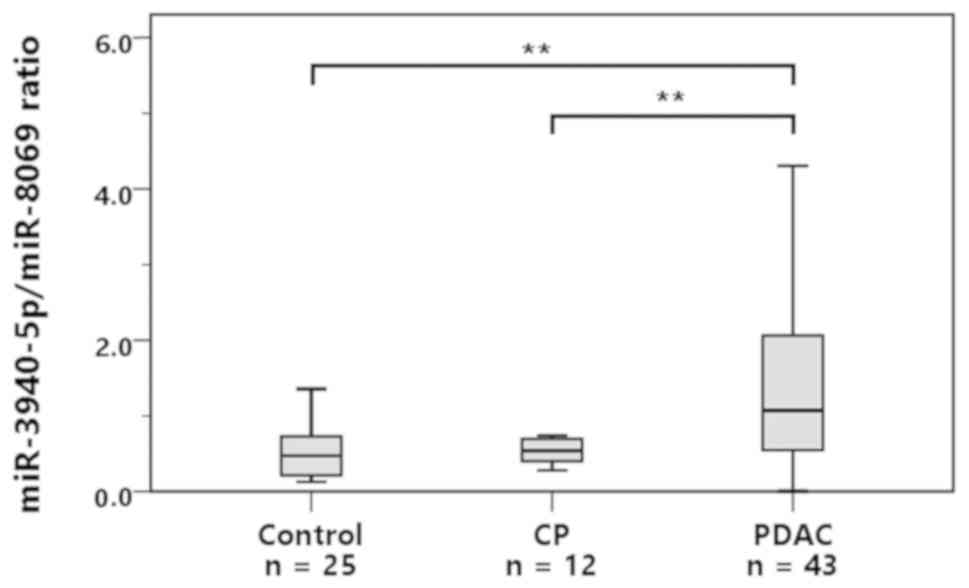
The miR-3940-5p/miR-8069 ratio in the urine exosomes
of each group is shown in Fig. 3.
The median values of the control, CP, and PDAC subjects were 0.47,
0.54, and 1.07, respectively. The miR-3940-5p/miR-8069 ratio in
urine exosomes was significantly higher in the PDAC group than in
the control group (P<0.001) or the CP group (P<0.001).
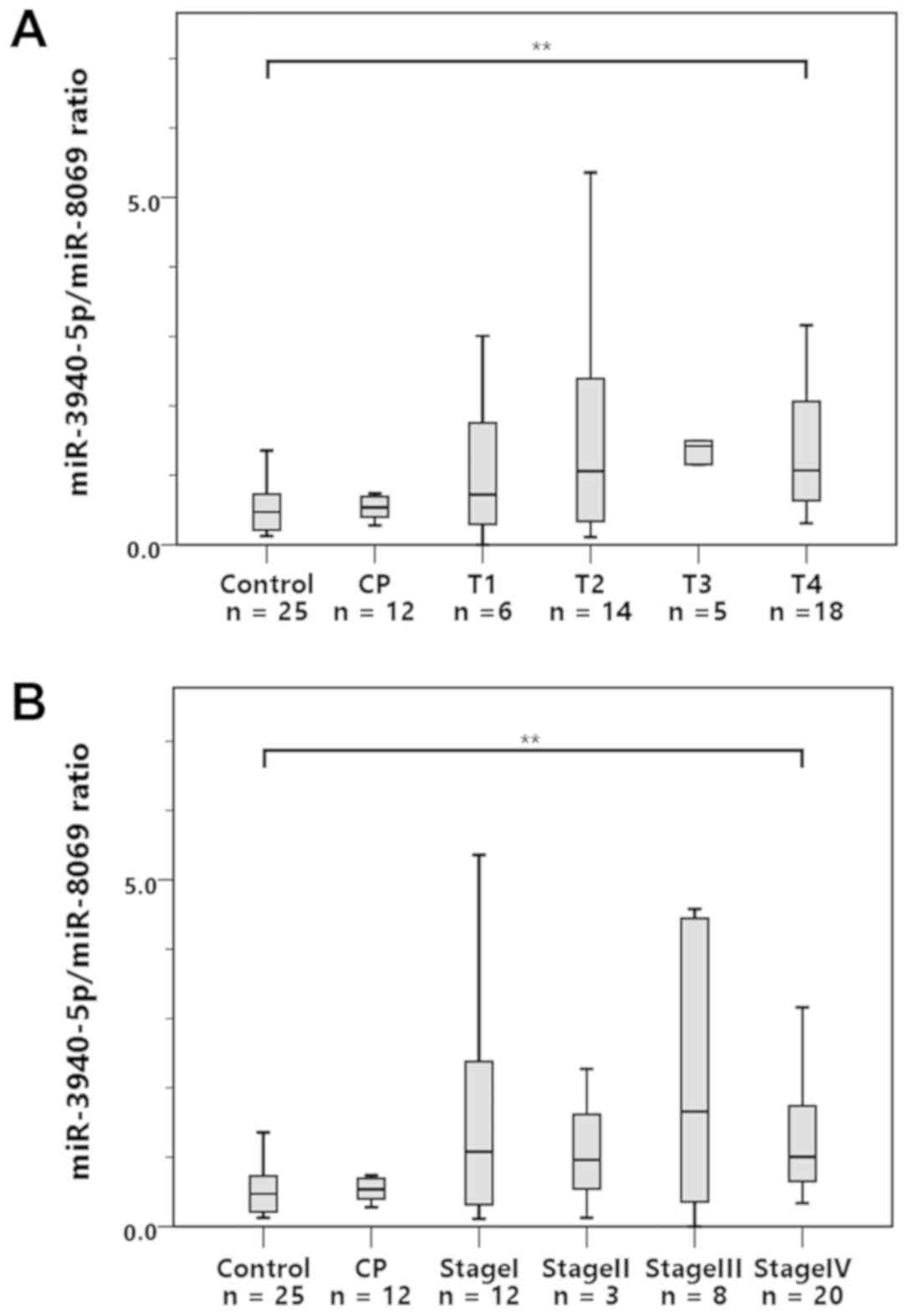
To clarify the stage of PDAC at which the miR-3940-
5p/miR-8069 ratio begins to increase, we analyzed the ratio
according to T classification and clinical stage as defined by the
eighth edition of the UICC classification (Fig. 4A and B). As shown in the graphs, the
miR-3940-5p/miR-8069 ratio in the urine exosomes was significantly
higher in the PDAC patients with T4 tumors or stage IV disease than
in the control subjects. Interestingly, the ratio also tended to be
elevated in T1 or stage I PDAC patients, although the differences
did not reach a statistically significant level, perhaps due to the
small sample sizes. Nevertheless, these results suggest the
possibility that the miR-3940-5p/miR-8069 ratio in urine exosomes
starts to increase from a relatively early stage of PDAC.
Diagnostic value of the
miR-3940-5p/miR-8069 ratio for PDAC
The diagnostic accuracy of using the
miR-3940-5p/miR- 8069 ratio in urine exosomes for differentiating
PDAC from CP and the controls was also evaluated using the receiver
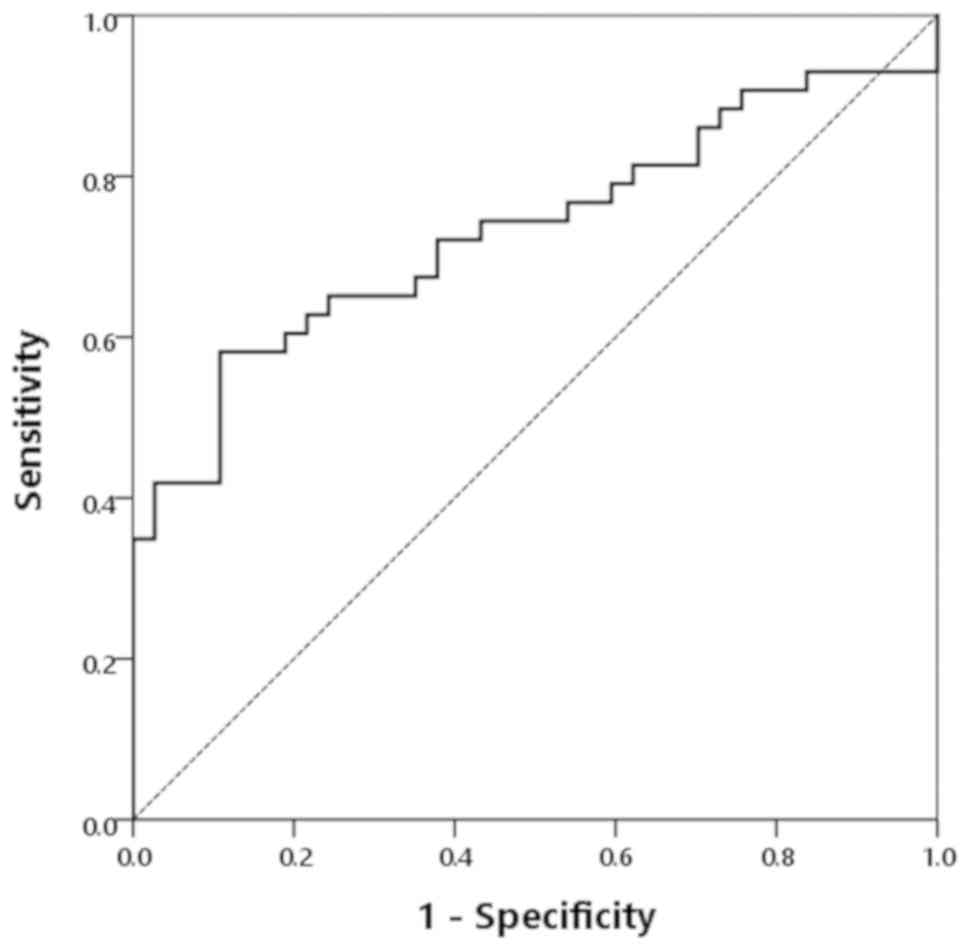
operating characteristic (ROC) curve (Fig. 5). The AUC was 0.732 (95% confidence
interval: 0.621–0.843). The cutoff point established by the ROC
curve was 0.939. By using this cutoff, the sensitivity,
specificity, PPV, and negative predictive value (NPV) for PDAC were
58.1, 89.2, 86.2, and 64.7%, respectively (Table IV).
 | Table IV.Diagnostic ability of the
miR-3940-5p/miR-8069 ratio and CA19-9 for pancreatic ductal
adenocarcinoma. |
Table IV.
Diagnostic ability of the
miR-3940-5p/miR-8069 ratio and CA19-9 for pancreatic ductal
adenocarcinoma.
| Parameters |
miR-3940-5p/miR-8069 | CA19-9 |
miR-3940-5p/miR-8069 + CA19-9 |
|---|
| Sensitivity, % | 58.1 | 79.1 | 93.0a |
| Specificity, % | 89.2 | 81.1 | 70.3b |
| Positive predictive
value, % | 86.2 | 82.9 | 78.4a (100c) |
| Negative predictive
value, % | 64.7 | 76.9 | 89.7b |
Diagnostic value of the miR-3940-5p
ratio with CA19-9
Finally, we examined the utility of the
miR-3940-5p/miR-8069 ratio in urine exosomes in combination with
CA19-9 as biomarkers for differentiating PDAC. In our study
subjects, the sensitivity, specificity, PPV, and NPV of CA19-9
(cutoff: 37 U/ml) were 79.1, 81.1, 82.9, and 76.9%, respectively
(Table IV). After the miR-3940-5p
ratio and CA19-9 were combined, when either of them was positive,
the sensitivity and PPV reached 93.0 and 78.4%, respectively. In
addition, the PPV reached 100% when both were positive. In
contrast, when both were negative, the specificity and NPV were
70.3 and 89.7%, respectively (Table
IV). These results clearly indicated that a combined analysis
of the miR-3940-5p/miR-8069 ratio in urine exosomes and CA19-9 is
more helpful for predicting or ruling out PDAC than when they are
used alone.
Discussion
In the present study, we identified the
miR-3940-5p/miR-8069 ratio in urine exosomes as a novel biomarker
for PDAC. We also found that an elevation of this ratio was
specific for PDAC in cultured cancer cell lines, and the ratio was
greater in the urine exosomes than in the serum exosomes of PDAC
patients. In addition, the diagnostic ability of the
miR-3940-5p/miR-8069 ratio and CA19-9 value for identifying PDAC
was much better when used in combination than when used alone.
Despite the development of several therapeutic
options, the prognosis of PDAC is still poor. One of the biggest
reasons for this is the difficulty of diagnosing the disease at an
early stage. In addition to imaging studies, several biomarkers are
currently used for the diagnosis of PDAC. Among them, CA19-9 is the
most extensively studied biomarker, and it is known to correlate
with prognosis (22); however,
CA19-9 is also occasionally elevated in benign hepatobiliary
diseases, pancreatitis, or other tumors, such as colorectal cancer
and ovarian tumors (23,24), and the PPV in asymptomatic patients
is extremely low (2,3). Therefore, many studies have attempted
to find new diagnostic markers for PDAC in blood or pancreatic
juice, and many studies have been focusing on miRNAs (8–13). In
the present study, we explored miRNAs in urine exosomes, and
identified the miR-3940-5p/miR-8069 ratio as a novel biomarker for
PDAC. There were two major reasons why we focused on urine
exosomes. First, urine is easy to obtain repeatedly from patients
without any invasive procedures; as such, we believe more and more
methodologies using urine samples will be developed hereafter for
the diagnosis of many kinds of diseases. Indeed, recent studies
have suggested the possibility that urine miRNAs can be biomarkers
for several cancers, including PDAC (25,26). The
second reason is that miRNAs are more stable and concentrated in
exosomes than in host cells or serum. The results of preliminary
analyses using microarray data suggested that miR-3940-5p was
upregulated in the urine exosomes of most PDAC patients when
compared to the controls.
The precise mechanism by which exosomes derived from
cells other than urinary tract are excreted into urine is not clear
yet. However, several studies have actually shown that some
disease-specific exosomes can be detected in urine, and they
suggested that the miRNA contained in the exosomes can act as
biomarkers for non-urologic diseases, including cancers (20,21,27). In
addition, Weber et al (28)
showed the differences of miRNA expression profiles in different
human body fluids, and some miRNAs showed higher expression levels
in urine compared to serum. However, the amounts of urinary
exosomes and the miRNAs encapsulated in them are extremely low, so
it has been difficult to obtain and assess a sufficient amount of
them for use as biomarkers. To overcome this problem, Yasui et
al (27) developed a new
nanowire-based methodology to collect miRNAs encapsulated in urine
extracellular vesicles, including exosomes, microvesicles, and
apoptotic bodies. Using this method, they found that many miRNAs in
urine extracellular vesicles were overexpressed or downregulated in
lung, liver, and pancreatic cancers (27). In our present study, we used 3D
digital PCR to accurately quantify the low levels of miRNA for
validation in a large number of samples. Digital PCR, which is a
third generation of PCR, has increased precision and sensitivity
for detecting low amounts of target copies when compared to
conventional quantitative real-time PCR (29,30). As
such, it can be used for the detection and quantification of low
levels of pathogens, rare genetic sequences, and cancer-related
miRNAs (31–33). To more precisely compare the
expression levels of miR-3940-5p in a large number of samples, we
normalized the miR-3940-5p levels to the miR-8069 levels, because
the results of microarray analysis indicated that miR-8069 was
expressed at a similar level in all samples, including PDAC and
control samples. We found that the miR-3940-5p/miR-8069 ratio in
urine exosomes was significantly elevated in PDAC patients when
compared to the controls or CP patients. We also found that this
elevation could be observed at a relatively early stage of
PDAC.
Sun et al (34) reported that the expression of
miR-3940-5p was downregulated in non-small cell lung carcinoma
(NSCLC) tissues when compared to normal lung tissues or
tumor-adjacent tissues. They also found that miR-3940-5p was
specifically reduced in nuclear cell proliferation antigen
Ki-67-positive NSCLC cases (34). In
addition, Ren et al (35)
reported that miR-3940-5p may target cyclin D1 and ubiquitin
specific peptide-28 to inhibit the growth of NSCLC. In the present
study, the miR-3940-5p/miR-8069 ratio was increased in the PDAC
group, and experimental results from studies using cultured cancer
cell lines showed that this increase was specific to PDAC. However,
the biological relevance of its upregulation and the reason for the
discrepancy in results between PDAC and NSCLC remain to be
elucidated. Furthermore, we found that the miR-3940-5p/miR-8069
ratio was greater in urine exosomes than in serum exosomes. The
reason for this remains unclear, however, these results suggests
that although the amount of PDAC-related exosomes that have a high
miR-3940-5p/miR-8069 ratio in serum is extremely low, and a small
proportion of them is excreted into the urine, the concentration of
these exosomes is still higher in urine than in blood.
One of the most important findings of this study is
that the miR-3940-5p/miR-8069 ratio and CA19-9 value as biomarkers
can better predict PDAC when used in combination than when used
alone. In fact, when we applied the cutoff point of 0.939, which
was established from the ROC curve, for the miR-3940-5p/miR-8069
ratio, and the widely clinically used cutoff of 37 U/ml for CA19-9,
the sensitivity and PPV improved to 93.0 and 78.4%, respectively,
when either of them was positive. Moreover, the PPV reached 100%
when they were both positive. Similarly, the NPV also improved to
89.7% when both were negative.
We acknowledge that this study has some limitations.
First, it was carried out with a small number of samples. To
improve the reliability of the findings of this study, more
analyses with a larger number of patients is needed. Second, we did
not clarify the relationship between the miR-3940-5p/miR-8069 ratio
in urine exosomes and the prognosis of PDAC, nor possible changes
in the ratio after therapy for PDAC. Although these points were
beyond the scope of this study, further investigations focusing on
these points are necessary as the next step in examining the
miR-3940-5p/miR-8069 ratio in urine exosomes as a biomarker for
PDAC.
In conclusion, the results of the present study
showed that the miR-3940-5p/miR-8069 ratio in urine exosomes is
elevated in PDAC patients, suggesting that it may be a potent
diagnostic tool for PDAC, especially in combination with
CA19-9.
Acknowledgements
The authors would like to thank Ms. Mina Tenpaku
(Department of Gastroenterology and Hepatology, Mie University
Graduate School of Medicine, Mie, Japan) for her technical
assistance.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KS and MT conceived and designed the study. NY, HI
and MU recruited the subjects, and collected and analyzed the data.
NY, YI, MT and MIk performed experiments. KS, NY and MT wrote the
manuscript. MIt and YT interpreted the data and supervised the
study. MIk, HI, MU, MIt and YT revised the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Mie University Hospital (reference no. 2833). Written
informed consent was obtained from each patient included in the
present study.
Patient consent for publication
All participants provided consent for
publication.
Competing interest
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
PDAC
|
pancreatic ductal adenocarcinoma
|
|
CP
|
chronic pancreatitis
|
|
CA19-9
|
carbohydrate antigen 19-9
|
|
PPV
|
positive predictive value
|
|
NPV
|
negative predictive value
|
|
miRNA
|
microRNA
|
References
|
1
|
Rahib L, Smith BD, Aizenberg R, Rosenzweig
AB, Fleshman JM and Matrisian LM: Projecting cancer incidence and
deaths to 2030: The unexpected burden of thyroid, liver, and
pancreas cancers in the United States. Cancer Res. 74:2913–2921.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kim JE, Lee KT, Lee JK, Paik SW, Rhee JC
and Choi KW: Clinical usefulness of carbohydrate antigen 19-9 as a
screening test for pancreatic cancer in an asymptomatic population.
J Gastroenterol Hepatol. 19:182–186. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chang CY, Huang SP, Chiu HM, Lee YC, Chen
MF and Lin JT: Low efficacy of serum levels of CA 19-9 in
prediction of malignant diseases in asymptomatic population in
Taiwan. Hepatogastroenterology. 53:1–4. 2006.PubMed/NCBI
|
|
4
|
Yachida S, Jones S, Bozic I, Antal T,
Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, et al:
Distant metastasis occurs late during the genetic evolution of
pancreatic cancer. Nature. 467:1114–1117. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Michael MZ, O'Connor SM, van Holst
Pellekaan NG, Young GP and James RJ: Reduced accumulation of
specific microRNAs in colorectal neoplasia. Mol Cancer Res.
1:882–891. 2003.PubMed/NCBI
|
|
6
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Johnson SM, Grosshans H, Shingara J, Byrom
M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D and Slack
FJ: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li A, Omura N, Hong SM, Vincent A, Walter
K, Griffith M, Borges M and Goggins M: Pancreatic cancers
epigenetically silence SIP1 and hypomethylate and overexpress
miR-200a/200b in association with elevated circulating miR-200a and
miR-200b levels. Cancer Res. 70:5226–5237. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Abue M, Yokoyama M, Shibuya R, Tamai K,
Yamaguchi K, Sato I, Tanaka N, Hamada S, Shimosegawa T, Sugamura K
and Satoh K: Circulating miR-483-3p and miR-21 is highly expressed
in plasma of pancreatic cancer. Int J Oncol. 46:539–547. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Panarelli NC, Chen YT, Zhou XK,
Kitabayashi N and Yantiss RK: MicroRNA expression aids the
preoperative diagnosis of pancreatic ductal adenocarcinoma.
Pancreas. 41:685–690. 2012.PubMed/NCBI
|
|
11
|
Hong TH and Park IY: MicroRNA expression
profiling of diagnostic needle aspirates from surgical pancreatic
cancer specimens. Ann Surg Treat Res. 87:290–297. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sadakari Y, Ohtsuka T, Ohuchida K,
Tsutsumi K, Takahata S, Nakamura M, Mizumoto K and Tanaka M:
MicroRNA expression analyses in preoperative pancreatic juice
samples of pancreatic ductal adenocarcinoma. JOP. 11:587–592.
2010.PubMed/NCBI
|
|
13
|
Wang J, Raimondo M, Guha S, Chen J, Diao
L, Dong X, Wallace MB, Killary AM, Frazier ML, Woodward TA, et al:
Circulating microRNAs in pancreatic juice as candidate biomarkers
of pancreatic cancer. J Cancer. 5:696–705. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang J, Li S, Li L, Li M, Guo C, Yao J
and Mi S: Exosome and exosomal microRNA: Trafficking, sorting, and
function. Genomics Proteomics Bioinformatics. 13:17–24. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mlcochova H, Hezova R, Stanik M and Slaby
O: Urine microRNAs as potential noninvasive biomarkers in urologic
cancers. Urol Oncol. 32:41.e1–e9. 2014. View Article : Google Scholar
|
|
18
|
Øverbye A, Skotland T, Koehler CJ, Thiede
B, Seierstad T, Berge V, Sandvig K and Llorente A: Identification
of prostate cancer biomarkers in urinary exosomes. Oncotarget.
6:30357–30376. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Franzen CA, Blackwell RH, Foreman KE, Kuo
PC, Flanigan RC and Gupta GN: Urinary exosomes: The potential for
biomarker utility, intercellular signaling and therapeutics in
urological malignancy. J Urol. 195:1331–1339. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Conde-Vancells J, Rodriguez-Suarez E,
Gonzalez E, Berisa A, Gil D, Embade N, Valle M, Luka Z, Elortza F,
Wagner C, et al: Candidate biomarkers in exosome-like vesicles
purified from rat and mouse urine samples. Proteomics Clin Appl.
4:416–425. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gildea JJ, Carlson JM, Schoeffel CD, Carey
RM and Felder RA: Urinary exosome miRNome analysis and its
applications to salt sensitivity of blood pressure. Clin Biochem.
46:1131–1134. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ballehaninna UK and Chamberlain RS: The
clinical utility of serum CA 19-9 in the diagnosis, prognosis and
management of pancreatic adenocarcinoma: An evidence based
appraisal. J Gastrointest Oncol. 3:105–119. 2012.PubMed/NCBI
|
|
23
|
Stiksma J, Grootendorst DC and van der
Linden PW: CA 19-9 as a marker in addition to CEA to monitor
colorectal cancer. Clin Colorectal Cancer. 13:239–244. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pandey D, Sharma R, Sharma S and Salhan S:
Unusually high serum levels of CA 19-9 in an ovarian tumour:
Malignant or benign? J Clin Diagn Res. 11:QD08–QD10.
2017.PubMed/NCBI
|
|
25
|
Debernardi S, Massat NJ, Radon TP,
Sangaralingam A, Banissi A, Ennis DP, Dowe T, Chelala C, Pereira
SP, Kocher HM, et al: Noninvasive urinary miRNA biomarkers for
early detection of pancreatic adenocarcinoma. Am J Cancer Res.
5:3455–3466. 2015.PubMed/NCBI
|
|
26
|
Kao HW, Pan CY, Lai CH, Wu CW, Fang WL,
Huang KH and Lin WC: Urine miR-21-5p as a potential non-invasive
biomarker for gastric cancer. Oncotarget. 8:56389–56397. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yasui T, Yanagida T, Ito S, Konakade Y,
Takeshita D, Naganawa T, Nagashima K, Shimada T, Kaji N, Nakamura
Y, et al: Unveiling massive numbers of cancer-related
urinary-microRNA candidates via nanowires. Sci Adv. 3:e17011332017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Weber JA, Baxter DH, Zhang S, Huang DY,
Huang KH, Lee MJ, Galas DJ and Wang K: The microRNA spectrum in 12
body fluids. Clin Chem. 56:1733–1741. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Brunetto GS, Massoud R, Leibovitch EC,
Caruso B, Johnson K, Ohayon J, Fenton K, Cortese I and Jacobson S:
Digital droplet PCR (ddPCR) for the precise quantification of human
T-lymphotropic virus 1 proviral loads in peripheral blood and
cerebrospinal fluid of HAM/TSP patients and identification of viral
mutations. J Neurovirol. 20:341–351. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao S, Lin H, Chen S, Yang M, Yan Q, Wen
C, Hao Z, Yan Y, Sun Y, Hu J, et al: Sensitive detection of Porcine
circovirus-2 by droplet digital polymerase chain reaction. J Vet
Diagn Invest. 27:784–788. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pekin D, Skhiri Y, Baret JC, Le Corre D,
Mazutis L, Salem CB, Millot F, El Harrak A, Hutchison JB, Larson
JW, et al: Quantitative and sensitive detection of rare mutations
using droplet-based microfluidics. Lab Chip. 11:2156–2166. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gutiérrez-Aguirre I, Rački N, Dreo T and
Ravnikar M: Droplet digital PCR for absolute quantification of
pathogens. Methods Mol Biol. 1302:331–347. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Conte D, Verri C, Borzi C, Suatoni P,
Pastorino U, Sozzi G and Fortunato O: Novel method to detect
microRNAs using chip-based QuantStudio 3D digital PCR. BMC
Genomics. 16:8492015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun Y, Su B, Zhang P, Xie H, Zheng H, Xu
Y, Du Q, Zeng H, Zhou X, Chen C and Gao W: Expression of miR-150
and miR-3940-5p is reduced in non-small cell lung carcinoma and
correlates with clinicopathological features. Oncol Rep.
29:704–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ren K, Li Y, Lu H, Li Z and Han X:
miR-3940-5p functions as a tumor suppressor in non-small cell lung
cancer cells by targeting cyclin D1 and ubiquitin specific
peptidase-28. Transl Oncol. 10:80–89. 2017. View Article : Google Scholar : PubMed/NCBI
|