Introduction
Breast cancer is the second leading cause of
cancer-related mortality, following lung cancer, in women globally,
with almost one third of cases resulting in mortality (1). It was reported that the expected
numbers of new breast cancer cases in 2012 was 230,480, which is
expected to account for 30% of all new cancer cases among women
(2). Understanding the underlying
molecular mechanisms of tumor suppressor genes and then effectively
incorporating this knowledge into a clinical environment has long
been a focus of cancer research and translational medicine.
Maspin is a non-inhibitory member of the serine
protease inhibitor super family; previous studies have demonstrated
that maspin may be useful in clinical practice (3,4). Maspin
is typically silenced or expressed at a decreased level in breast
cancer cells, and has long been considered a type II tumor
suppressor that regulates cell adhesion and invasion (5). The MCF-7 cell line originated from
human breast cancer cells and has retained a number of
characteristics of differentiated mammary epithelial cells
(6).
microRNAs (miRNAs) are highly conserved noncoding
RNA molecules that are approximately 17–25 nucleotides in length.
They control gene expression at the post-transcriptional level via
interacting with a specific target mRNA (7–9). miRNAs
are regulatory molecules recognized to be aberrantly expressed in
cancer and contribute to various aspects of tumor biology,
including the proliferation and invasive abilities of tumors
(10). In addition, miRNAs may
provide insights into the molecular pathogenesis of breast cancer
(11). Although maspin is considered
to be a tumor suppressor gene, the underlying molecular mechanisms
of maspin-induced inhibition of breast cancer cell proliferation
and invasion remains unknown.
In the present study, maspin expression was induced
in MCF-7 cells using cloning techniques. Subsequently, the
differential expression of miRNA was determined between the maspin
and mock groups using gene chip analysis. Furthermore, the effect
of maspin on the corresponding target genes of miR-21 gene was
investigated. The present study aimed to identify how maspin
affected the proliferation and differentiation of MCF-7 cells.
Materials and methods
Cell culture
MCF-7 cells were purchased from the American Type
Culture Collection (Manassas, VA, USA). TRIzol® was
purchased from Invitrogen (Thermo Fisher Scientific, Inc., Waltham,
MA, USA) and cell culture was conducted according to the method
described previously (12). MCF-7
cells were cultured in Dulbecco's modified Eagle's medium
supplemented with 10% fetal bovine serum (both from Gibco; Thermo
Fisher Scientific, Inc.) at 37°C in an atmosphere containing 5%
CO2.
Plasmid transfection
Total mRNA was extracted from MCF-7 cells using
TRIzol with NanoDrop™ 2000 (Thermo Fisher Scientific, Inc.).
Following the quantification, 500 µg total RNA was
reverse-transcribed with a cDNA Reverse Transcription kit (Beijing
TransGen Biotech Co., Ltd., Beijing, China), according the
manufacturer's protocols. The experiment used 50 µl PCR reaction
volume and 500 ng cDNA template with Trans-start Tip Green qPCR
SuperMix (Beijing TransGen Biotech Co., Ltd.), and the method was
performed under the following conditions: Denaturing for 30 sec at
94°C; annealing for 30 sec from 65°C to 60°C, and decreasing at
0.5°C each cycle, and extending for 3 min at 72°C for each 10
cycles; annealing for 30 sec at 60°C, and extending for 3 min at
72°C for 25 cycles, and a final extension at 72°C for 5 min.
Additionally the following primers were used in PCR: Forward,
5′-GGAATTCCCCGCAATGGATGCCCTGCAACTAG-3′ and reverse,
5′-CCCTCGAGACTTAAGGAGAACAGAATTTGCCAA-3′. The forward primers
contain an EcoRI restriction enzyme site, and the reverse primers
contain a XhoI restriction enzyme site. Human maspin cDNA was
inserted into a pEF expression vector (Promega Corporation,
Madison, WI, USA) using restriction enzyme digestion with
EcoRI and XbaI, and the pcDNA3.1-maspin contained a
cDNA encoding amino acids 1–375 of human maspin. The vector
pcDNA3.1 alone was used as a negative control. Transient
transfection of cells with plasmids was performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol, and 20
µg DNA was diluted in 200 µl Lipofectamine® 2000 reagent
in 60 mm well. Each experiment was conducted immediately following
transfection.
Cell viability assay
For the quantitative determination of the
proliferation of MCF-7 cells, a Cell Counting Kit-8 (CCK-8; Nanjing
Jiancheng Bioengineering Institute, Nanjing, China) assay was
performed. Following the transfection of maspin pcDNA into MCF-7
cells for 24 h, the cells were washed, counted and seeded at a
density of 4×105 cells/ml/well in 96-well plates. CCK-8
solution was added and the cells were incubated for an additional 4
h at 37°C in a 5% CO2 incubator. Cell proliferation was
determined using a spectrophotometer at an absorbance of 450 nm.
The experiments were repeated ≥5 times/group.
In vitro invasion assay
The maspin-MCF-7 cells were seeded at a density of
1×104 cells/well in Dulbecco's modified Eagle's medium
with 10% fetal bovine serum (both from Gibco; Thermo Fisher
Scientific, Inc.) in Matrigel-coated invasion chambers in a 24-well
plate (BD Biosciences, Franklin Lakes, NJ, USA) and Matrigel
solution was polymerized in Transwell inserts for 45 min at 37°C.
Cells (5×105) were plated in the top chamber in
Dulbecco's modified Eagle's medium without serum, and Dulbecco's
modified Eagle's medium supplemented with 10% fetal bovine serum
(Gibco; Thermo Fisher Scientific, Inc.) was placed into the lower
chamber. Following this, cells were incubated for an additional 4 h
at 37°C in an atmosphere containing 5% CO2, and then for
24 h at 37°C, the cells on the upper membrane surface were removed
with a cotton swab. Cells that invaded to the lower side of the
membrane were stained with 0.1% crystal violet at room temperature
for 20 min and fixed with 4% paraformaldehyde at room temperature
for 20 min, and then stained by 0.1% crystal violet for another 20
min at 37°C. The mean number of invasive cells were counted under a
×100 fluorescence microscope using four separate fields of view for
each well. The experiments were repeated ≥3 times.
miRNA microarray
miRNA microarray analysis was performed as
previously described (13). Total
RNA was isolated from cells with TRIzol reagent and quantified with
NanoDrop 2000 (Thermo Fisher Scientific, Inc.). Following the
quantification, 500 µg total RNA was reverse-transcribed with a
cDNA Reverse Transcription kit (Beijing TransGen Biotech Co.,
Ltd.), and miRNA was isolated using the mirVana™ miRNA isolation
kit (Ambion; Thermo Fisher Scientific, Inc.). cRNA was synthesized
by using the 3′IVT Express kit (Affymetrix; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. The
fragmented labeled cRNA was applied using the MicroRNA 2.0 Array
(Affymetrix; Thermo Fisher Scientific, Inc.), and hybridized at
45°C for 18 h using the GeneChip Hybridization Oven 640
(Affymetrix; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocols. The arrays were scanned using a GeneChip
Scanner 3000 (Affymetrix; Thermo Fisher Scientific, Inc.). The
expression levels of each miRNA in the array were compared and
normalized with the mean using Partek GS 6.5 (Partek Inc., St.
Louis, MO, USA). Average-linkage hierarchical clustering of the
data was applied by using the Cluster (2.20; http://rana.lbl.gov/EisenSoftware.htm), and for
cluster analysis, log transformed data were centered to the mean
values of each gene expression (14)
and the results were displayed using TreeView graphical (1.6.6)
analysis (15).
Quantitative PCR (qPCR) for mRNA
Total RNA was isolated from MCF-7 cells with TRIzol
reagent, according to the manufacturer's protocol, using 1,000 µl
TRIzol and incubated for 5 min at room temperature. Subsequently,
200 µl chloroform was added, vortexed for 15 sec, and incubated for
2–3 min at room temperature. This was followed by centrifugation at
12,000 × g at 4°C for 15 min. The upper watery phase was removed
and 1.5 times its volume (100%) ethanol was added. Additionally,
700 µl of this mixture were placed in RNeasy Mini spin column in 2
ml collection tube and centrifuged at 8,000 × g at room temperature
for 15 sec. After the mixture had completely passed the column, 700
µl of buffer RWT (Qiagen GmbH, Hilden, Germany) was added to each
column, and again centrifuged at 8,000 × g at room temperature for
15 sec. Then, 500 µl buffer RPE was added to the column and
centrifuged at 8,000 × g at room temperature for 15 sec. The
previous process was repeated for 2 min at 8,000 × g. The column
was transferred to a new 1.5 ml collection tube and 50 µl
RNase-free water was pipetted directly onto the column and
centrifuged for 1 min at 8,000 × g to elute RNA. The extracted mRNA
then stored at −80°C until use. qPCR was performed with an Applied
Biosystems StepOne™ real-time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.), and the fast
SYBR®-Green Master mix (Thermo Fisher Scientific, Inc.)
was used for qPCR, according to the manufacturer's protocol. The
method of quantification for PCR was calculated with the
2−ΔΔCq method (16).
Phosphatase and tensin homolog (PTEN), programmed cell death 4
(PDCD4) and B-cell lymphoma-2 (Bcl-2) gene expression was detected
and the primers used are listed in Table
I. Data are presented as the relative expression levels
compared with β-actin.
 | Table I.Primer sequences used in the reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Primer sequences used in the reverse
transcription-quantitative polymerase chain reaction.
| Gene | Primer sequences |
|---|
| Maspin | F:
5′-TATCCCTGTTGCCGGTTCA-3′ |
|
| R:
5′-AGATGGGAGAAGGAATGTCAC-3′ |
| U6 | F:
5′-CTCGCTTCGGCAGCACA-3′ |
|
| R:
5′-AACGCTTCACGAATTTGCGT-3′ |
| β-actin | F:
5′-TCCTCCTGAGCGCAAGTACTC-3′ |
|
| R:
5′-CTGCTTGCTGATCCACATCTG-3′ |
| PTEN | F:
5′-TCGACTACTTGCTTTGTAGA-3′ |
|
| R:
5′-TTTACAGCCCCGATTGGGCT-3′ |
| PDCD4 | F:
5′-TCTCAAATGCCCTTTCATCC-3′ |
|
| R:
5′-TGGATTAACTGTGCCAACCA-3′ |
| Bcl-2 | F:
5′-GCTTTTCCTCTGGGAAGGAT-3′ |
|
| R:
5′-CCTCCGTTATCCTGGATCCA-3′ |
| HOXD10 | F:
5′-ATGTACATGCCACCACCTAGC-3′ |
|
| R:
5′-TTGCTGTGTAACAGGTTGCTCTA-3′ |
| MDR1 | F:
5′-TCACCAAGCGGCTCCGATACAT-3′ |
|
| R:
5′-CCCGGCTGTTGTCTCCATAGGC-3′ |
Reverse transcription (RT-)qPCR for
miRNA
Total RNA was isolated from cells by using TRIzol.
cDNA was synthesized using the miRNA Reverse Transcription kit 20
µl RT reactions, incubated for 60 min at 37°C, followed by 5 min at
95°C, according to the manufacturer's instructions (Applied
Biosystems; Thermo Fisher Scientific, Inc.). A total of 20 ng cDNA
was used as a template in a total volume of 20 µl reaction with the
following conditions: Denaturation at 95°C for 15 min followed by
40 cycles of 94°C for 15 sec, 55°C for 30 sec, and 72°C for 34 sec,
in which fluorescence was acquired and detected via Real-time PCR
system (Qiagen GmbH). After the PCR cycles, melting curve analyses
were performed to validate the specific generation of the expected
PCR product small nuclear RNA U6 was used as an endogenous control.
Forward and Reverse target sequences are shown in Table I.
A SYBR green miRNA assay was used to quantify the
relative expression levels of miR-10b, miR-21 and miR-451,
according to the manufacturer's protocol. The primers of miR-10b
were 5′-TACCCTGTAGAACCGAATTTG-3′ (forward) and
5′-AACTGGTGTCGTGGAGTCGGC-3′ (reverse), and the primers of miR-21
were 5′-GCCGCTAGCTTATCAGACT-3′ (forward) and
5′-AGTGCAGGGTCCGAGGTA-3′ (reverse). The fold-change between the
selected genes and U6 transcripts for miRNA was calculated with the
2−ΔΔCq method (16). And
all reactions were run in triplicate.
Western blot analysis
The method was performed as previously described
(17). The antibodies used in the
present study were all purchased from Abcam (Cambridge, UK). The
membranes were incubated with primary antibodies against maspin
(dilution, 1:1,500; catalog no. ab182785), PDCD4 (dilution,
1:1,000; catalog no. ab45124), HOXD10 (dilution, 1:350; catalog no.
ab76897), multi-drug resistance gene (dilution, 1:1,500; MDR1;
catalog no. ab170904), PTEN (dilution, 1:500; catalog no. ab31392),
Bcl-2 (dilution, 1:150; catalog no. ab117115) and β-actin
(dilution, 1:1,000; catalog no. ab8226) at 4°C overnight. Briefly,
normal or treated cells were harvested on ice with ice-cold lysis
buffer containing 10 mM Tris pH 7.4, 100 mM NaCl, 1 mM EDTA, 1 mM
EGTA, 1 mM NaF, 20 mM Na4P2O7, 1%
Triton X-100, 10% glycerol, 0.1% SDS and 0.5% deoxycholate, and
protease and phosphatase inhibitor cocktails. Samples (30 µg
protein) were loaded to a 10% SDS-PAGE gel, fractioned through
electrophoresis, and transferred onto nitrocellulose membranes. The
membranes were blocked with 5% BSA and wash the membrane in three
washes of TBS with 0.05% Tween-20 (TBST), 5 min each, then probed
with appropriate primary and horseradish peroxidase
(HRP)-conjugated rabbit secondary antibodies (dilution, 1:3,000;
catalog no. ab150077; Abcam) for 1 h at room temperature. The
membranes were washed three times with TBST, 5 min each. The blots
were detected using ECL western blotting detection reagents
(catalog no. RPN2109; GE Healthcare, Chicago, IL, USA). The images
were captured and analyzed using the UVP gel documentation system
(UVP, LLC, Phoenix, AZ, USA).
Statistical analysis
Data are presented as the mean ± standard deviation.
Mean values were compared and P-values were determined using the
two-tailed Student's t-test or the Pearson's χ2 test.
P<0.05 was considered to indicate a statistically significant
difference. All statistical calculations were performed using
GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA)
software and SPSS (version 17.0 for Windows; SPSS Inc., Chicago,
IL, USA).
Results
Maspin inhibits the proliferation and
invasion of MCF-7 cells
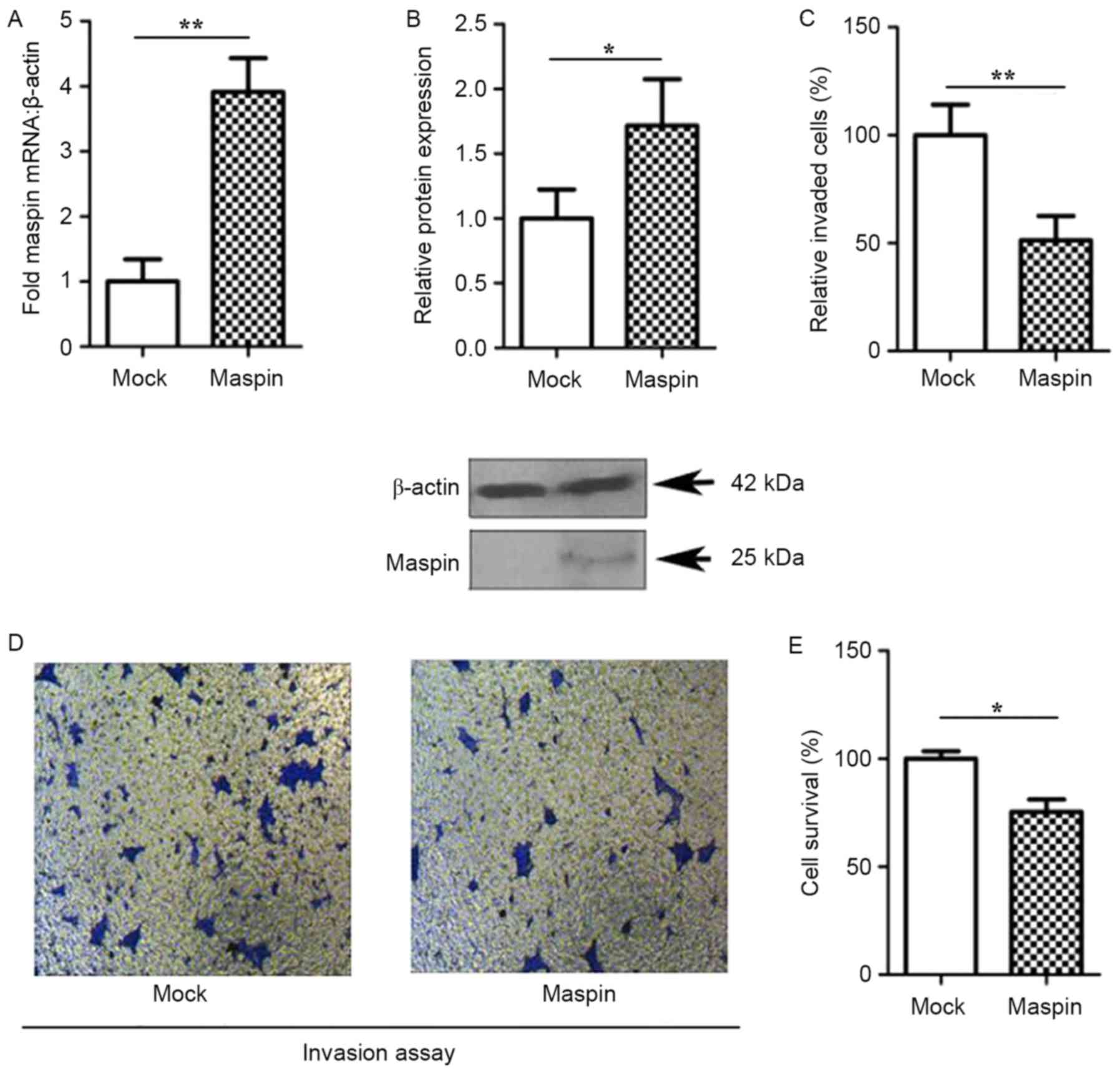
To assess whether maspin expression was required for
the proliferation and invasive properties of MCF-7 cells, human
maspin cDNA (1.3 kb full length) was inserted into a pEF expression
vector for restriction enzyme digestion with EcoRI and
XbaI. Maspin clones were examined using RT-qPCR and western
blot analysis. As presented in Fig.
1, the gene and protein expression of maspin were upregulated
in maspin cells compared with the mock cells (P<0.05);
therefore, maspin was successfully cloned into the MCF-7 cells
(Fig. 1A and B). The expression of
maspin in MCF-7 resulted in a significant reduction in the invasion
(Fig. 1C and D; P<0.05) and
proliferation (Fig. 1E; P<0.05)
of MCK-7 cells, when compared with the mock cells.
Maspin downregulates miR-10b, miR-21
and miR-451 expression in MCF-7 cells
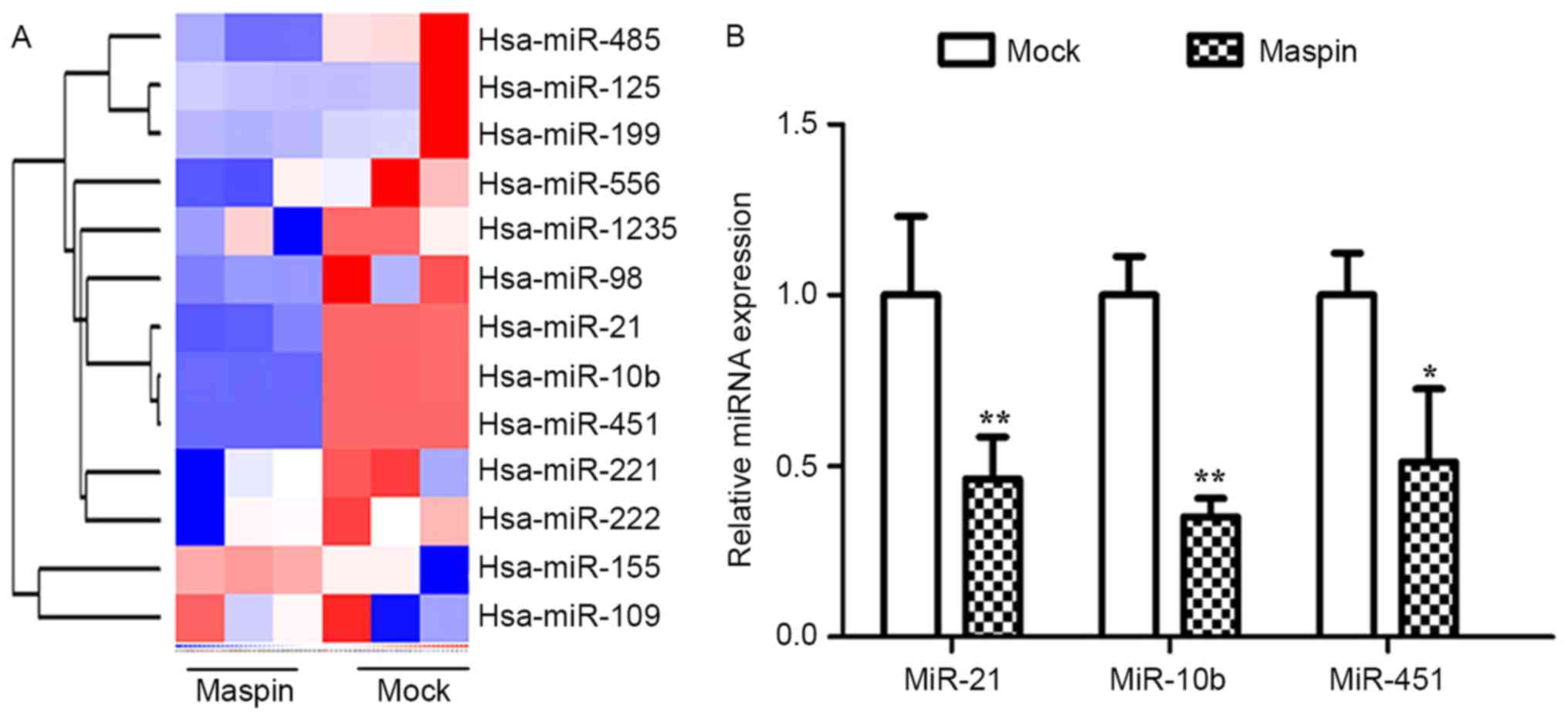
Using miRNA array analysis, it was identified that
maspin altered the expression of a number of microRNAs in MCF-7
cells, including miR-21, miR-10b and miR-451 (Fig. 2A). The aberrant expression of miR-21,
miR-10b and miR-451 in association with tumorigenesis, tumor growth
and tumor metastasis has been identified in distinct types of
malignancy; therefore, in the present study, the expression of
these miRNAs in MCF-7 cells, that had been stably or transiently
transfected with maspin, were analyzed. Using qPCR, with β-actin as
an endogenous control, it was revealed that miR-21, miR-10b and
miR-451 were significantly downregulated between 2- and 2.4-fold by
maspin (Fig. 2B; P<0.05).
Maspin increases the protein
expression of miR-21, miR-10b and miR-451 target genes in MCF-7
cells
In order to explore whether maspin affected the
proliferation and differentiation of MCF-7 cells, the effect of
maspin on the target genes of miR-21, miR-10b and miR-451 was
investigated. Compared with the mock group, the expression levels
of the target genes of miR-21 in maspin pcDNA group were
significantly different (P<0.05). As presented in Fig. 3, maspin increased the mRNA expression
of PTEN, PDCD4 and Bcl-2 by between 1.5- and 2.0-fold (Fig. 3A). Fig. 3B
and C demonstrate that maspin significantly increased the
expression of HOXD10 (P<0.01) and MDR1 (P<0.05).
Additionally, the protein expression levels of PTEN, PDCD4 and
Bcl-2 in the mock and maspin pcDNA groups (Fig. 3D-F). The relative expression is
depicted in Fig. 3G-I.
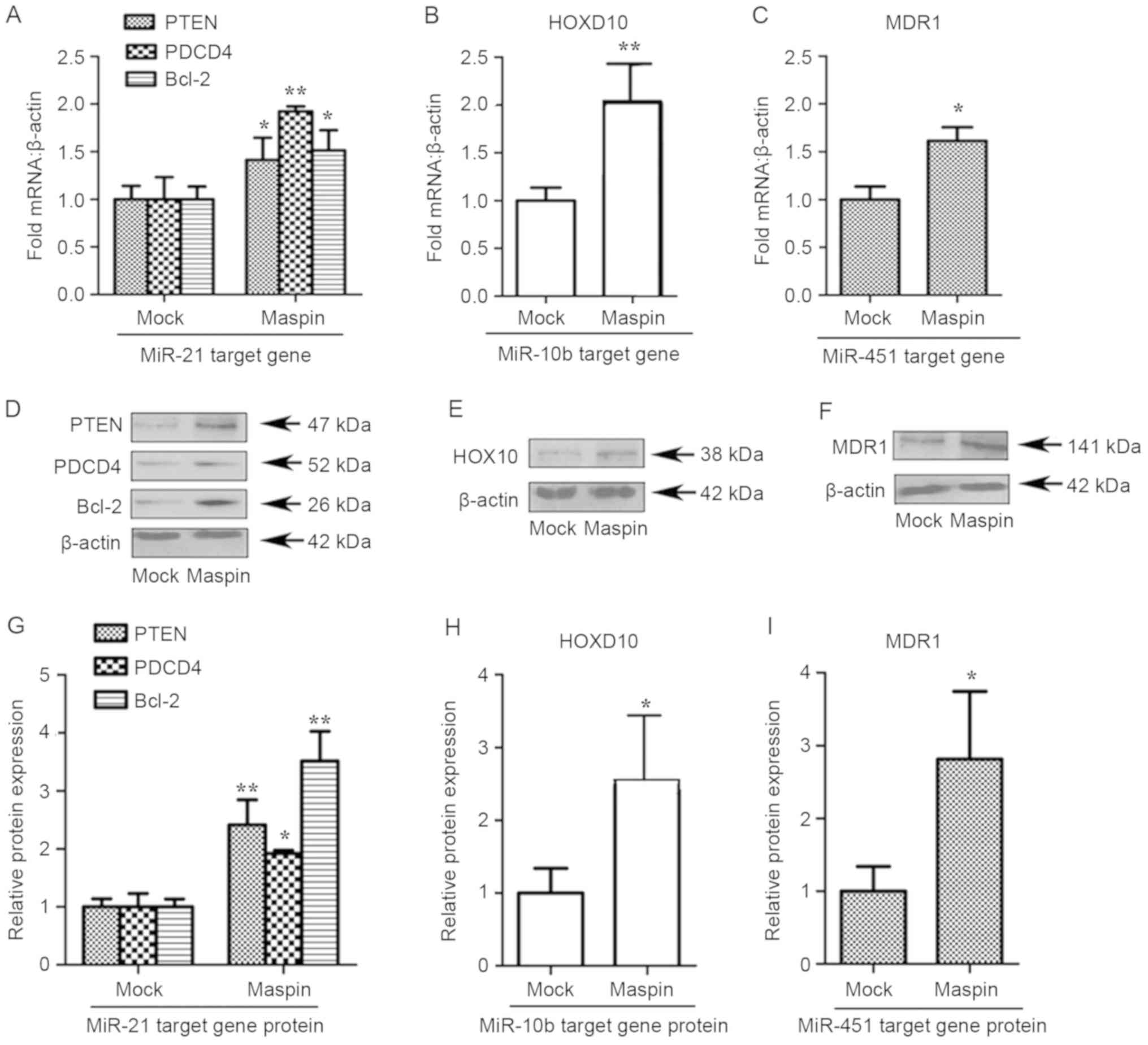 | Figure 3.Maspin increases the expression of
miR-21, miR-10b and miR-451. MCF-7 cells were transfected with mock
or maspin pcDNA as indicated. The specificity of maspin to
influence the mRNA of endogenous miR-21, miR-10b and miR-451 target
gene was examined using RT-qPCR or western blotting in parallel
with the mock group as a negative control. (A) RT-qPCR analysis of
(A) miR-21, (B) miR-10b and (C) miR-451 target genes expression in
MCF-7 cells transfected with maspin, compared with mock. (D)
Western blot analysis of miR-21 target genes in maspin cells,
compared with mock cells. (E) Western blot analysis of miR-10b
target genes in maspin cells, compared with mock cells. (F) Western
blot analysis of miR-451 target genes in maspin cells, compared
with mock cells. The fold of relative protein expression of (G)
miR-21 target genes, (H) miR-10b target genes and (I) miR-451
target genes. Data are presented as the mean ± standard deviation
from a single experiment in duplicate, representative of ≥3
separate experiments. β-actin was the loading control. *P<0.05
vs. mock; **P<0.01 vs. mock. miR, microRNA; PTEN, phosphatase
and tensin homolog; PDCD4, programmed cell death 4; Bcl-2, B-cell
lymphoma-2; MDR1, multi-drug resistance gene 1; HOXD10, homeobox
D10. |
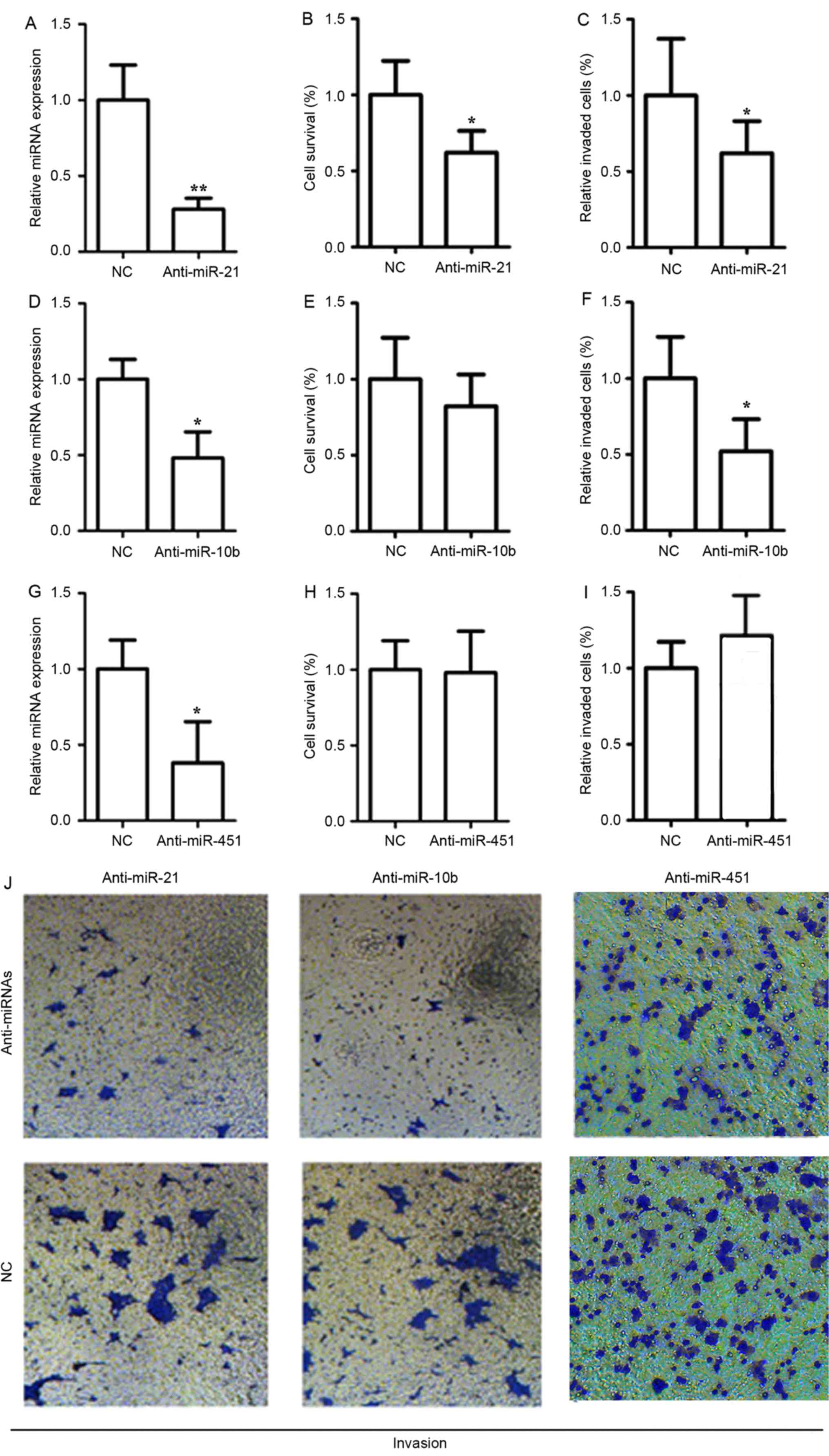
miR-21 promotes the proliferation and
invasion of MCF-7 cells
To determine whether the aforementioned miRNAs may
function as oncogenes, the effect of suppressing miR-21, miR-10b
and miR451 on cell proliferation and invasion was investigated. An
anti-miR-21 inhibitor was used to inhibit the expression of miR-21.
The anti-miR-21 inhibitor is a sequence-specific and chemically
modified oligonucleotide to specifically target and knockdown
miR-21 molecule. TaqMan real-time PCR revealed that anti-miR-21
significantly reduced miR-21 level (Fig.
4A; P<0.05), indicating that anti-miR-21 is efficiently
introduced into the cells and knock down miR-21. A significant
decrease (P<0.05) was observed in the proliferation and invasion
of MCF-7 cells, following transfection of anti-miR-21 inhibitor
(Fig. 4B and C). Additionally, the
expression of miR-10b was down-regulated in MCF-7 cells following
the use of an antisense anti-miR-10b inhibitor, which resulted in a
2.6-fold decrease in the gene expression levels of miR-10b
(Fig. 4D; P<0.05). The results of
the present study suggest that the transfected antisense
anti-miR-10b inhibitor achieved >50% inhibition of miR-10b.
Fig. 4E and F demonstrate that
miR-10b function is required for invasiveness in MCF-7 cells
(P<0.05), but not for the proliferation or invasive ability of
these cells (P>0.05). Following transfection with an
anti-miR-451 inhibitor, the expression of miR-451 was significantly
downregulated in MCF-7 cells (Fig.
4G; P<0.05); however, there was no significant difference
observed in the proliferation (Fig.
4H) and invasion (Fig. 4I) of
MCF-7 cells (P>0.05). The transfection with anti-miRNA
inhibitors resulted in a marked reduction in the invasion (Fig. 4J).
Discussion
Maspin is a unique member of the serpin (serine
protease inhibitor) family; the downregulation of maspin has been
associated with the development of breast cancer (18). In the present study, increased maspin
gene expression was associated with decreased invasive capacity and
increased overall survival in MCF-7 cells, which is consistent with
the results of a previous study (19).
miRNAs have been extensively studied in a number of
types of cancer; however, the knowledge of the aberrant expression
and the association between maspin and miRNAs remains unknown. In
the present study, the results revealed an association between
increased maspin expression and the downregulation of miR-21,
miR-10b and miR-451 in MCF-7 breast cancer cells.
miR-21 regulates genes involved in a number of
cellular processes, and has been identified to promote cell
proliferation, invasion and migration by downregulating the
expression of the tumor-suppressor genes PDCD4 and PTEN (20,21). A
previous study revealed that the inhibition of miR-21 may
upregulate the expression of Bcl-2 (22), but this remains controversial
(23). In the present study, the
expression of maspin led to an increase in miR-21 target gene
expression in MCF-7 cells, including of PTEN, PDCD4 and Bcl-2. The
results of the present study were concordant with those of a prior
study (18), demonstrating that
maspin inhibits miR-21. To understand the mechanisms by which
miR-10b induces tumor invasion and metastasis, HOXD10 was of
particular note, as its expression is progressively reduced in
breast tumors of increasing degrees of malignancy (24,25).
Furthermore, miR-10b has been revealed to promote the migration and
invasion of cells via HOXD10 in certain human cancer types
(26). The results of the present
study demonstrated that maspin may inhibit miR-10b and result in
increased levels of HOXD10. Therefore, maspin increased the
expression of HOXD10, and the underlying molecular mechanisms may
be associated with miR-10b. In addition, inducing the expression of
maspin resulted in a decrease in miR451 gene expression; however,
the proliferation and invasive abilities of MCF-7 cells were not
altered by this regulation of miR-451.
The results of the present study identified that
maspin may increase the protein expression of miR-21 target genes.
In the present study, following demonstrating that maspin may
inhibit miR-21, miR-10b and miR451, it was hypothesized that maspin
may affect the properties of MCF-7 cells by decreasing the
expression of miR-21 and increasing its target genes. Cells
transfected with an anti-miR-21 inhibitor resulted in the
inhibition of proliferation and invasion of MCF-7 cells;
conversely, the transfection of cells with an anti-miR-10b
inhibitor prevented invasion, but did not affect the proliferation
of MCF-7 cells. Additionally, transfection of MCF-7 cells with an
anti-miR-451 inhibitor did not alter the proliferation or invasion
of cells. These results were consistent with those of prior studies
that explored the functions of miR-21, miR10b and miR-451 in other
types of tumor cells (27–29).
The results of the present study demonstrated that
maspin inhibits the invasion and proliferation of MCF-7 cells, and
is dependent on the downregulation of miR-21 expression and the
upregulation of target gene expression. Identifying the underlying
molecular mechanisms by which maspin inhibits the invasion and
proliferation of cancer cells will be beneficial for the management
of breast cancer.
Acknowledgements
Not applicable.
Funding
The authors acknowledge funding from National
Natural Science Foundation of China (grant nos. 81460514, 71463030
and 81360447), the Natural Science Foundation of Jiangxi (grant no.
20151BAB205065) and Jiangxi Natural Science Foundation (grant no.
20181BAB205067.
Availability of data and materials
All data generated or analyzed during this study
were included in this published article.
Authors' contributions
SXH conceived and designed the study. WYF, LW, HL,
XW and HZ performed the experiments. WBJ analyzed the results. SXH
wrote, reviewed and edited the manuscript. All authors read and
approved the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Benson JR, Jatoi I, Keisch M, Esteva FJ,
Makris A and Jordan VC: Early breast cancer. Lancet. 373:1463–1479.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Latha K, Zhang W, Cella N, Shi HY and
Zhang M: Maspin mediates increased tumor cell apoptosis upon
induction of the mitochondrial permeability transition. Mol Cell
Biol. 25:1737–1748. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shay JW and Roninson IB: Hallmarks of
senescence in carcinogenesis and cancer therapy. Oncogene.
23:2919–2933. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bodenstine TM, Seftor RE, Khalkhali-Ellis
Z, Seftor EA, Pemberton PA and Hendrix MJ: Maspin: Molecular
mechanisms and therapeutic implications. Cancer Metastasis Rev.
31:529–551. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hill SM and Blask DE: Effects of the
pineal hormone melatonin on the proliferation and morphological
characteristics of human breast cancer cells (MCF-7) in culture.
Cancer Res. 48:6121–6126. 1988.PubMed/NCBI
|
|
7
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lau NC, Lim LP, Weinstein EG and Bartel
DP: An abundant class of tiny RNAs with probable regulatory roles
in Caenorhabditis elegans. Science. 294:858–862. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee RC and Ambros V: An extensive class of
small RNAs in Caenorhabditis elegans. Science. 294:862–864.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tie Y, Liu B, Fu H and Zheng X:
Circulating miRNA and cancer diagnosis. Sci China C Life Sci.
52:1117–1122. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Roth C, Rack B, Müller V, Janni W, Pantel
K and Schwarzenbach H: Circulating microRNAs as blood-based markers
for patients with primary and metastatic breast cancer. Breast
Cancer Res. 12:R902010. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bacus SS, Kiguchi K, Chin D, King CR and
Huberman E: Differentiation of cultured human breast cancer cells
(AU-565 and MCF-7) associated with loss of cell surface HER-2/neu
antigen. Mol Carcinog. 3:350–362. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shi JA, Lu DL, Huang X and Tan W: miR-219
inhibits the proliferation, migration and invasion of
medulloblastoma cells by targeting CD164. Int J Mol Med.
34:237–243. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wilks DS: Chapter 15-Cluster analysis, in
international geophysics. Academic Press. 603–616. 2011.
|
|
15
|
Liu CL, Prapong W, Natkunam Y, Alizadeh A,
Montgomery K, Gilks CB and van de Rijn M: Software tools for
high-throughput analysis and archiving of immunohistochemistry
staining data obtained with tissue microarrays. Am J Pathol.
161:1557–1565. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang S, Ye J, Yu J, Chen L, Zhou L, Wang
H, Li Z and Wang C: The accumulation and efflux of lead partly
depend on ATP-dependent efflux pump-multidrug resistance protein 1
and glutathione in testis Sertoli cells. Toxicol Lett. 226:277–284.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zou Z, Anisowicz A, Hendrix MJ, Thor A,
Neveu M, Sheng S, Rafidi K, Seftor E and Sager R: Maspin, a serpin
with tumor-suppressing activity in human mammary epithelial cells.
Science. 263:526–529. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Berardi R, Morgese F, Onofri A, Mazzanti
P, Pistelli M, Ballatore Z, Savini A, De Lisa M, Caramanti M,
Rinaldi S, et al: Role of maspin in cancer. Clin Transl Med.
2:82013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang S, Li J, Jiang Y, Xu Y and Qin C:
Programmed cell death 4 (PDCD4) suppresses metastastic potential of
human hepatocellular carcinoma cells. J Exp Clin Cancer Res.
28:712009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wickramasinghe NS, Manavalan TT, Dougherty
SM, Riggs KA, Li Y and Klinge CM: Estradiol downregulates miR-21
expression and increases miR-21 target gene expression in MCF-7
breast cancer cells. Nucleic Acids Res. 37:2584–2595. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dong S, Ma W, Hao B, Hu F, Yan L, Yan X,
Wang Y, Chen Z and Wang Z: microRNA-21 promotes cardiac fibrosis
and development of heart failure with preserved left ventricular
ejection fraction by up-regulating Bcl-2. Int J Clin Exp Pathol.
7:565–574. 2014.PubMed/NCBI
|
|
24
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Makiyama K, Hamada J, Takada M, Murakawa
K, Takahashi Y, Tada M, Tamoto E, Shindo G, Matsunaga A, Teramoto
K, et al: Aberrant expression of HOX genes in human invasive breast
carcinoma. Oncol Rep. 13:673–679. 2005.PubMed/NCBI
|
|
26
|
Xiao HB, Li H, Yu G, Xiao W, Hu J, Tang K,
Zeng J, He W, Zeng G, Ye Z and Xu H: MicroRNA-10b promotes
migration and invasion through KLF4 and HOXD10 in human bladder
cancer. Oncol Rep. 31:1832–1838. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu
W, Xiao S and Lu H: miR-21 plays a pivotal role in gastric cancer
pathogenesis and progression. Lab Invest. 88:1358–1366. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hiyoshi Y, Kamohara H, Karashima R, Sato
N, Imamura Y, Nagai Y, Yoshida N, Toyama E, Hayashi N, Watanabe M,
et al: MicroRNA-21 regulates the proliferation and invasion in
esophageal squamous cell carcinoma. Clin Cancer Res. 15:1915–1922.
2009. View Article : Google Scholar : PubMed/NCBI
|