Introduction
Colorectal cancer (CRC) is the third most common
cancer type and is a leading cause of cancer-associated mortality
worldwide (1,2). A certain subset of CRC arises from the
sequential accumulation of genetic and epigenetic alterations.
microRNAs (miRNA/miR) have been revealed as candidates in tumor
progression in various cancer types, and changes in their
expression levels are consequently being investigated with the
purpose of identifying clinical biomarkers (3–9). miRNAs
constitute a class of small, non-coding RNA molecules, 18–27
nucleotides in length that function as post-transcriptional
regulators of gene expression, either serving as oncogenes or tumor
suppressor genes (10,11). In CRC, several miRNAs are aberrantly
expressed and target genes downstream of epidermal growth factor
receptor (EGFR) signaling; for example, miR-143 and
−145 target KRAS and BRAF proteins, respectively (12). Nosho et al (13) revealed that high miR-31-5p
(miR-31) expression [miR-31 has the two subtypes;
miR-31 (hsa-mir-31-5p) and miR-31*
(hsa-mir-31-3p)] was significantly associated with
BRAF V600E mutation (P<0.0001) and a poorer prognosis in
a large statistical population of 721 patients with CRC.
Additionally, downregulation of BRAF protein expression, following
transfection of an miR-31 inhibitor into CRC cells was
demonstrated (13). Thus, the
aforementioned evidence indicates that miR-31 may regulate
the activation of BRAF protein in CRC, and may also serve an
important role in the downstream EGFR signaling pathway.
The present study investigated miR-31 that is
significantly associated with advanced CRC with BRAF V600E
mutation, as the presence of BRAF mutations is known to be a
poor prognostic factor in CRC (14–18).
According to the results of the microarray analysis, it was
revealed that miR-31 expression is upregulated in
BRAF-mutant tumors. Therefore, the association between
miR-31 expression levels and BRAF-mutant CRCs was
further investigated using a dataset retrieved from The Cancer
Genome Atlas (TCGA) database. Finally, miR-31 expression
patterns observed in CRC were further supported by investigating
the miR-31 expression level in patients with stage IV
CRC.
Materials and methods
Patients
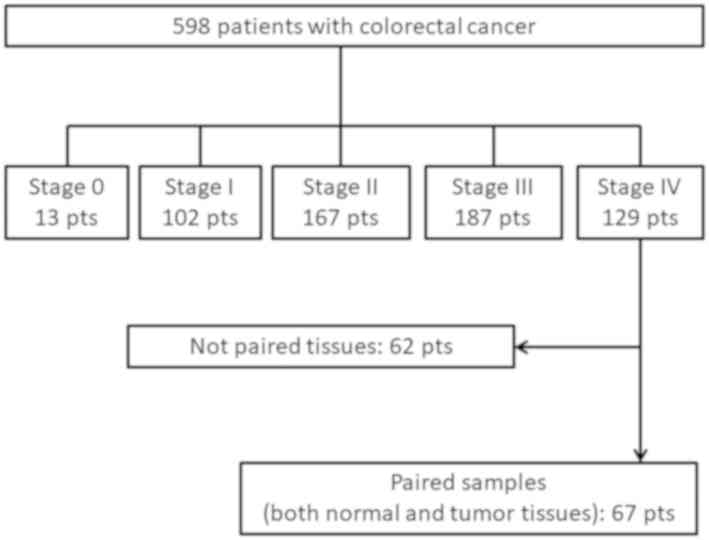
From a cohort of 598 patients with CRC, 129 patients
with stage IV CRC underwent primary tumor resection before other
treatments, such as chemotherapy, radiotherapy or
chemoradiotherapy, at Okayama University Hospital (Okayama, Japan)
between March 2003 and May 2013. Of these, only 67 patients were
evaluated and analyzed in the present study due to availability of
both tumors and the paired normal mucosa (Fig. 1). The tumors and the corresponding
normal mucosa were stored at −80°C following preservation with
RNAlater® (Sigma-Aldrich; Merck KGaA). Clinical,
pathological and survival data were extracted from medical records.
The pathological stage was determined according to the
International Union Against Cancer TNM staging system (seventh
edition) (19). Institutional review
board approval was granted by the Ethics Committee of Okayama
University, and written informed consent was provided from all
patients for the use of their tissues and clinical data.
DNA and RNA extraction
DNA was extracted from 67 fresh-frozen paired tumor
and healthy colonic mucosal tissue specimens, which were obtained
from a site adjacent to the primary tumor, >10 cm from the tumor
border. DNA from the fresh-frozen tissue specimens was extracted
using a QIAamp DNA Mini kit (Qiagen, Inc.) according to the
manufacturer's protocol. Total miRNA was isolated from frozen
tissue specimens preserved by RNAlater® (Sigma-Aldrich;
Merck KGaA) using a miRNeasy Mini kit (Qiagen, Inc.) and quality
and quantity were confirmed using a Qubit fluorometer (Qubit 2.0 or
3.0; Invitrogen; Thermo Fisher Scientific, Inc.).
BRAF and KRAS mutation analyses
A BRAF mutation in codon 600 and KRAS
mutations in codons 12 and 13 were analyzed by direct sequencing
using purified DNA from fresh-frozen tissues of each patient. The
specific primer sequences and PCR conditions have been described
previously (20). The PCR products
were purified using a QIAquick PCR purification kit (Qiagen, Inc.)
according to the manufacturer's protocol and were directly
sequenced on an ABI 310R Genetic Analyzer (Thermo Fisher
Scientific, Inc.).
Microsatellite instability (MSI)
analysis
A multiplex PCR method for the detection of tumors
with MSI was performed to determine the MSI status of all CRC
tissues using four mononucleotide repeat markers (BAT26, NR21, NR27
and CAT25) as described previously (21,22).
Tumors exhibiting MSI in ≥1 mononucleotide repeat marker were
classified as MSI phenotype, whereas those without MSI were
classified as non-MSI phenotype.
Analysis of miRNA expression in paired
primary tumor and normal colonic tissue samples using miRNA
microarray
Total miRNA was isolated from frozen tissue
specimens using a miRNeasy Mini kit (Qiagen, Inc.) and analyzed on
an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.) according
to the manufacturer's protocol. SurePrint G3 Human miRNA 8×60K
Rel.16.0 (Agilent Technologies, Inc.) was used to analyze miRNA
expression in paired primary tumor and normal colonic tissue
samples. The expression level of each probe was calculated as the
sum of 20 spots of raw intensity with the background subtracted.
Target miRNAs that were not detected in any spots were defined as
‘undetected’ and allocated an expression level of ‘0.1’. The data
were normalized to the 90th percentile, and target miRNAs that were
not detected in all the samples were excluded (9).
Preliminary analysis of the
association between miR-31 expression and BRAF mutation using TCGA
database
Freely available datasets regarding miRNA expression
and somatic mutations of colon adenocarcinoma samples were
retrieved from TCGA (23). From TCGA
database (v1.0), a total of 187 CRC samples had data available
regarding miR-31 expression, among which the BRAF
mutation profile was available in 170 CRCs on the GDC Data Portal
(https://portal.gdc.cancer.gov/). Thus,
miR-31 expression in 170 CRC tissues was subsequently
analyzed. Of these, 51 CRCs were categorized as having BRAF
V600E mutation (30%).
Reverse transcription-quantitative
(RT-q)PCR
Total miRNA was isolated from frozen tissue
specimens using a miRNeasy Mini kit (Qiagen, Inc.). The expression
levels of miR-31 (Hs_miR-31_1 miScript Primer Assay; Qiagen,
Inc.) and RNU6B (Hs_RNU6-2_1 miScript Primer Assay; Qiagen,
Inc.) were analyzed using miScript primer assays. The cDNA was
generated by a miScript II RT Kit (Qiagen, Inc.). RT-qPCR was
performed with the gene-specific primers and a miScript
SYBR® Green PCR kit (Qiagen, Inc.) using a LightCycler
480 (Roche Diagnostics), according to the manufacturer's protocol.
Briefly, reactions were incubated in a 96-well plate at 95°C for 15
min, followed by 40 cycles of 95°C for 15 sec, 55°C for 30 sec and
70°C for 30 sec, according to the manufacturer's protocol. The
quantitative value of miRNA in a given sample was calculated by
subtracting that value from the Cq value of RNU6B, which
served as the internal reference gene. Cq values from duplicate
reactions were averaged, and the threshold cycle value (ΔCq) was
calculated by subtracting the Cq of the RNU6B from the Cq of
the target miR-31. The ∆∆Cq value of tumor was calculated by
subtracting the ∆Cq value of the normal counterpart sample
(24). The value of
2−∆∆Cq represents the expression relative quotient (RQ)
of miR-31. Here, the miR-31 expression level (RQ of
miR-31) was presented as the ratio of the miR-31
expression in cancerous tissue vs. adjacent non-cancerous tissue. A
miR-31 expression level <1 indicated that the expression
levels in cancerous tissue were lower compared with that in the
paired normal colonic tissue. Conversely, an miR-31
expression level >1 indicated higher expression in cancerous
tissue compared with that in the paired normal colonic tissue.
Statistical analysis
Statistical analyses were performed using JMP Pro
software (version 14.2; SAS Institute Inc.). The miRNA expression
level is presented as the mean RQ, and was measured three times in
all samples. Categorical variables were compared using a
χ2 test. miR-31 expression levels in
BRAF-mutant CRC tissues from TCGA were compared with those
in BRAF-wild type CRC tissues using Dunnett's Method.
Disease-specific overall survival time (OS) was calculated from the
date of initial treatment to the date of death due to primary colon
cancer or last follow-up for censored patients (the patients were
followed up every month for stage IV cases). Survival data, <120
months, following initial treatment was used. Survival-based
outcomes were analyzed by the Kaplan-Meier method, expressed as
medians and compared with log-rank test. Univariate and
multivariate analyses were performed using the Cox-proportional
hazards model. The hazard ratio (HR) and 95% confidence interval
(CI) were calculated from the model, and the significance of the
parameters was evaluated using Wald's test. All reported P-values
were from two-sided tests, and P<0.05 was considered to indicate
a statistically significant difference.
Results
Identification of miRNAs associated
with CRCs expressing the BRAF V600E mutation
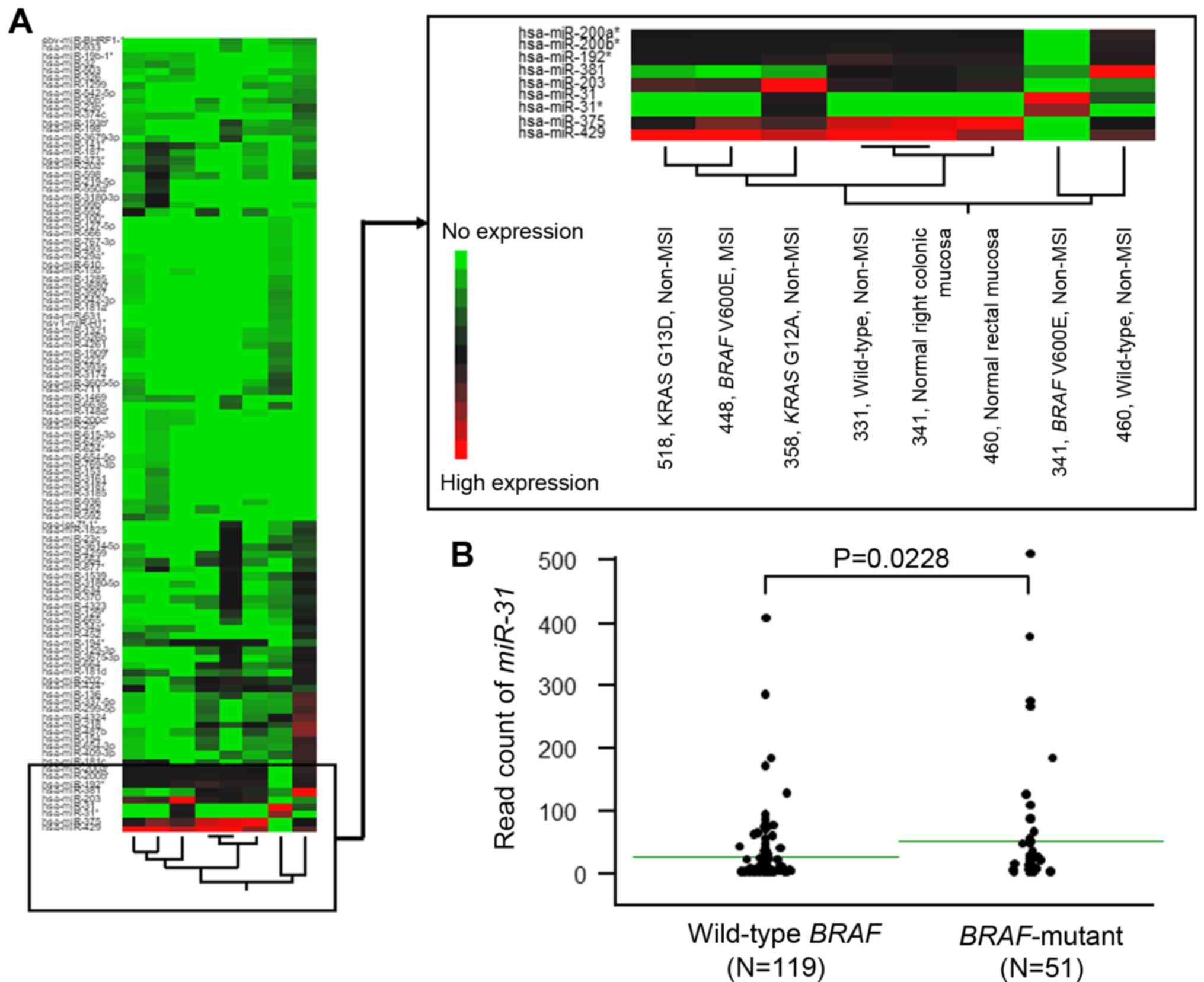
Prior to analyzing the miRNA microarray data, six
independent CRC specimens were evaluated according to their
KRAS/BRAF mutational profiles and MSI status, which resulted
in six subtypes with different genetic and clinical features
(25). No MSI tumors were observed
to exhibit a KRAS-mutation. Therefore, the following
specimens were collected: An MSI with BRAF V600E mutation,
two non-MSI tumors without either KRAS or BRAF
mutation (wild-type), a non-MSI tumor with BRAF V600E
mutation, and two non-MSI tumor with KRAS mutation, each
located in the right or left side of the colon. The two normal
colonic (right colon and rectum) mucosa specimens were used as
controls (Fig. 2A and Table SI). The miRNA array provided the
expression status of a total of 1,368 miRNAs.
To identify miRNAs that were significantly
associated with non-MSI tumors containing the BRAF V600E
mutation, miRNAs exhibiting no expression among the six CRC tissues
and the two corresponding normal colorectal mucosae (803 miRNAs),
and miRNAs that were expressed in all the tissues (286 miRNAs),
were excluded. The cluster analysis revealed that only
miR-31 was significantly upregulated in non-MSI CRC
specimens with the BRAF V600E mutation (sample ID, 341),
while miR-429, −375, −200a, −200b and -192 were
downregulated compared with that in the other CRC phenotypes, such
as CRCs with wild-type BRAF. Generally, molecules that are
significantly and specifically upregulated in tumors are useful for
screening those tumors via liquid biopsy or other screening tools
(26). Therefore, subsequent
analyses focused on miR-31 as a candidate prognostic
biomarker in the present study.
Association between miR-31 expression
and BRAF-mutant CRC in TCGA database
To confirm whether elevated miR-31 expression
is common in BRAF-mutant CRC, miRNA expression and somatic
mutations data from colon adenocarcinoma samples retrieved from
TCGA were analyzed (Fig. 2B). From
the database, a cohort of 170 patients with CRC were available for
analysis of miR-31 expression with respect to BRAF
mutation status. Among the 170 CRCs, the value of miR-31
expression ranged from 0 to 447.6 (median, 6.27). BRAF
mutation was observed in 51 patients with CRC (30%), and all 51
showed higher miR-31 expression (median value, 45.47; 95%
CI, 20.59–70.35) compared to patients with CRC expressing wild-type
BRAF (median value, 21.57; 95% CI, 13.09–30.05;
P=0.0228).
Calculation of miR-31 expression in
stage IV CRC specimens
To examine at what point during CRC progression do
miR-31 expression levels affect clinical outcomes in
advanced CRC, a cohort of 67 patients with stage IV CRCs (out of a
total of 598), had both frozen tumor and adjacent normal mucosa
tissues available for RNA extraction, were selected (Fig. 1). Of the 67 stage IV CRC specimens,
all of them were non-MSI. The median miR-31 expression level
was 3.45 (mean, 142; range, 0.004–6330.531). Therefore, a cut-off
value of 3.5 was used. Cases showing an miR-31 expression
level ≥3.5 were included in the miR-31 high-expression
group, while cases with an miR-31 expression level <3.5
were categorized as low expression. According to this
classification, 33 and 34 cases were defined as miR-31 high
expression and miR-31 low expression, respectively. Table I displays the clinicopathological
features of patients with stage IV CRCs as classified by
miR-31 expression level. Of the 67 stage IV CRCs, only four
had a BRAF mutation and were all included in the
miR-31 high expression group; however, the association was
not significant. Additionally, there were no significant
differences in clinicopathological characteristics between the two
groups.
 | Table I.Clinicopathological features of 67
patients with stage IV CRC stratified by miR-31 expression
level. |
Table I.
Clinicopathological features of 67
patients with stage IV CRC stratified by miR-31 expression
level.
|
|
| miR-31
expression level, n (%) |
|
|---|
|
|
|
|
|
|---|
| Variables | Total, n (%)
(n=67) | Low (<3.5)
(n=34) | High (>3.5)
(n=33) | P-value |
|---|
| Age, years |
|
|
|
|
| Mean
(range) | 64.1 (35–85) | 62.9 (41–85) | 65.3 (35–82) |
|
|
<65 | 28 (41.8) | 17 (50.0) | 11 (33.3) | 0.218 |
|
≥65 | 39 (58.2) | 17 (50.0) | 22 (66.7) |
|
| Gender |
|
|
| 0.621 |
|
Female | 26 (38.8) | 12 (35.3) | 14 (42.4) |
|
|
Male | 41 (61.2) | 22 (64.7) | 19 (57.6) |
|
| Serum CEA level,
ng/µl |
|
|
| 0.262 |
|
<5.0 | 17 (25.4) | 11 (32.4) | 6 (18.2) |
|
|
≥5.0 | 50 (74.6) | 23 (67.6) | 27 (81.8) |
|
| Tumor location |
|
|
| 0.392 |
|
Right | 15 (22.4) | 6 (17.6) | 9 (27.3) |
|
|
Left | 52 (77.6) | 28 (82.4) | 24 (72.7) |
|
| Histology |
|
|
| 0.186 |
| Well or
moderate | 57 (85.1) | 31 (91.2) | 26 (78.8) |
|
| Poor or
mucinous | 10 (14.9) | 3 (8.8) | 7 (21.2) |
|
| No. of distant
metastatic sites |
|
|
| 0.194 |
|
Single | 46 (68.7) | 26 (76.5) | 20 (60.6) |
|
|
Multiple | 21 (31.3) | 8 (23.5) | 13 (39.4) |
|
| T factor |
|
|
| 0.2 |
| 1 | 1 (1.5) | 0 (0.0) | 1 (3.0) |
|
| 2 | 4 (6.0) | 3 (8.8) | 1 (3.0) |
|
| 3 | 38 (56.7) | 22 (64.7) | 16 (48.5) |
|
| 4a | 15 (22.4) | 7 (20.6) | 8 (24.2) |
|
| 4b | 9 (13.4) | 2 (5.9) | 7 (21.2) |
|
| N factor |
|
|
| 0.437 |
| 0 | 13 (19.4) | 9 (26.5) | 4 (13.3) |
|
| 1a | 11 (16.4) | 5 (14.7) | 6 (18.2) |
|
| 1b | 14 (20.9) | 6 (17.6) | 8 (24.2) |
|
| 1c | 1 (1.5) | 1 (2.9) | 0 (0.0) |
|
| 2a | 13 (19.4) | 4 (11.8) | 9 (27.3) |
|
| 2b | 12 (17.9) | 7 (20.6) | 5 (15.2) |
|
| X | 3 (4.5) | 2 (5.9) | 1 (3.0) |
|
| M factor |
|
|
| 0.447 |
| 1a | 44 (65.7) | 24 (54.5) | 20 (45.5) |
|
| 1b | 23 (34.3) | 10 (43.5) | 13 (56.5) |
|
| MSI status |
|
|
| Not calculated |
|
MSI | 0 (0.0) | 0 (0.0) | 0 (0.0) |
|
|
Non-MSI | 67 (100.0) | 34 (100) | 33 (100) |
|
| KRAS/RAF
mutation status |
|
|
| 0.0608 |
|
KRAS mutation | 15 (22.4) | 10 (29.4) | 5 (15.2) |
|
|
BRAF mutation | 4 (6.0) | 0 (0) | 4 (12.1) |
|
|
Wild-type | 48 (71.6) | 24 (70.6) | 24 (72.7) |
|
Clinical outcomes are associated with
miR-31 expression status in patients with stage IV CRCs
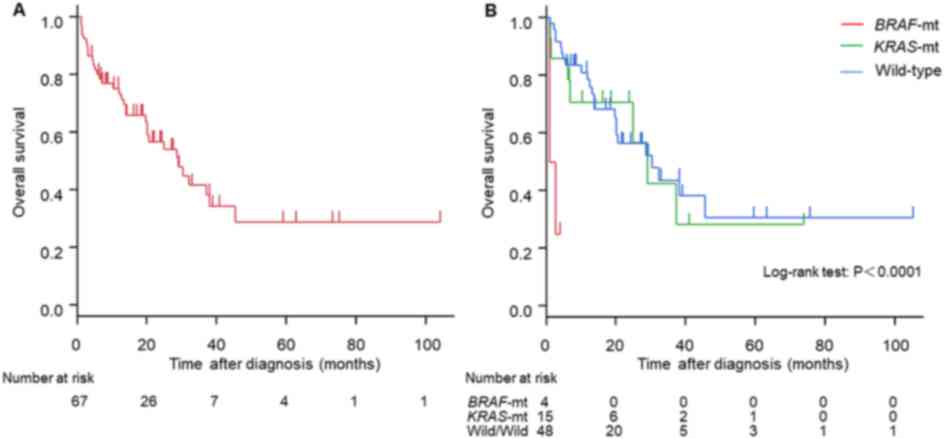
Among the cohort of 67 patients with stage IV CRCs,
the median survival time (MST) was 29 months (95% CI, 19.5–38
months; Fig. 3A). Stratification of
patients according to their BRAF/KRAS mutation status was
then conducted; the MST of patients with BRAF-mutant,
KRAS-mutant and wild-type CRC was 1.8, 29 and 30 months,
respectively (Fig. 3B; P<0.0001).
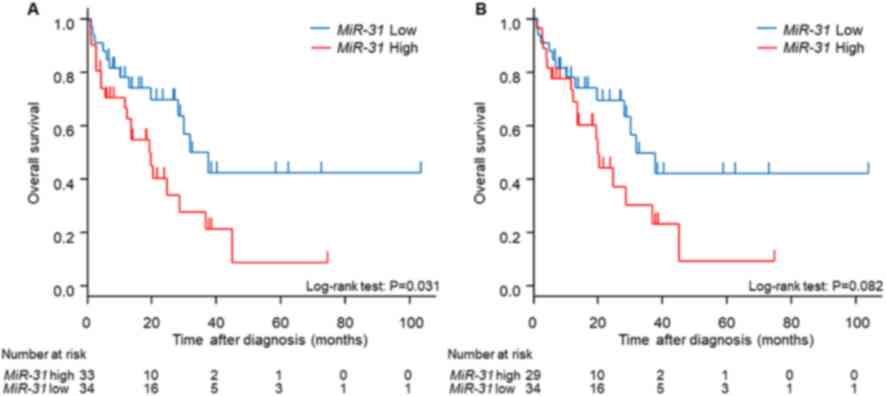
Next, clinical outcomes were analyzed with respect to miR-31
expression status. The MST of the miR-31 high expression
group was 20 months, while that of the miR-31 low expression
group was 38 months (Fig. 4A;
P=0.0314). Of note, the four patients with BRAF-mutant CRCs
(MST, 1.8 months) were all in the miR-31 high-expression
group, and the prognosis of the miR-31 high-expression group
may have been affected by this subgroup of patients. Therefore,
during subsequent prognostic analysis patients with
BRAF-mutant CRC were excluded. The miR-31 high
expression group still maintained a tendency toward poor prognosis
compared with the miR-31 low-expression group, although it
did not reach statistical significance (Fig. 4B).
Finally, univariate and multivariate analyses were
conducted using the Cox-proportional hazards model to determine the
factors that influenced poorer outcomes in patients with stage IV
CRC (Table II). The univariate
analysis revealed that tumors with poor or mucinous histology,
multiple distant metastatic sites, the BRAF V600E mutation
and higher miR-31 expression levels statistically exhibited
a poorer prognosis; however, in the multivariate analysis, only
multiple distant metastatic sites and the presence of the
BRAF V600E mutation, indicated a poorer prognosis.
 | Table II.Cox regression analyses of 67
patients with stage IV colorectal cancer. |
Table II.
Cox regression analyses of 67
patients with stage IV colorectal cancer.
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age, years |
|
|
|
|
| >65
vs. <65 | 1.78
(0.86–3.69) | 0.1197 | 1.41
(0.61–3.26) | 0.4244 |
| Sex |
|
|
|
|
| Female
vs. male | 1.19
(0.59–2.43) | 0.6249 | 1.18
(0.52–2.69) | 0.6891 |
| Serum CEA level,
ng/µl |
|
|
|
|
| >5
vs. <5 | 1.24
(0.55–2.78) | 0.5999 | 1.23
(0.53–2.87) | 0.6243 |
| Tumor location |
|
|
|
|
| Right
vs. left | 1.10
(0.47–2.55) | 0.8252 | 0.34
(0.08–1.42) | 0.1394 |
| Histology |
|
|
|
|
| Poor or
mucinous vs. well or moderate | 3.16
(1.33–7.52) | 0.0094a | 1.83
(0.59–5.67) | 0.2941 |
| No. of distant
metastatic sites |
|
|
|
|
|
Multiple vs. single | 2.95
(1.46–5.96) | 0.0027a | 2.84
(1.31–6.17) | 0.0083a |
| KRAS/BRAF
mutation status |
|
|
|
|
|
BRAF mutation vs.
KRAS mutation | 12.73
(2.68–60.37) | 0.0014a | 13.18
(2.11–82.39) | 0.0058a |
|
KRAS mutation vs.
wild-type | 1.11
(0.47–2.58) | 0.8152 | 2.20
(0.73–6.64) | 0.1611 |
|
BRAF mutation vs.
wild-type | 14.08
(3.37–58.85) | 0.0003b | 29.03
(3.58–235.29) | 0.0016a |
| miR-31
expression level |
|
|
|
|
| High
vs. low | 2.12
(1.05–4.29) | 0.0356a | 1.48
(0.67–3.29) | 0.3358 |
Discussion
In the present study, the potential of miR-31
as a prognostic biomarker for patients with advanced CRC was
demonstrated, particularly in patients with stage IV of the
disease. miR-31 is located on chromosome 9p21.3 and is
reportedly deregulated in various types of human cancer, for
example, esophageal, breast, ovarian and gastric cancers (3,27–30).
Moreover, in CRC, an association has been reported between
upregulation of miR-31 and poor tumor differentiation
(13), oncogenic potential (13,31–34),
deeper invasion (34,35) and advanced disease stages (31,34,35).
In the advanced stages of CRC, the presence of
BRAF mutations is demonstrated to be a poor prognostic
factor (14–18), so specific miRNAs that are
significantly associated with BRAF mutations were
investigated in the present study. The results obtained from a
miRNA array analysis revealed that miR-31 was upregulated in
BRAF-mutant CRC tissues compared with that in other CRC
subtypes, such as KRAS-mutant or wild-type CRCs. The
association between miR-31 and BRAF mutations was
also confirmed in a dataset comprising information on 170 patients
with CRC, retrieved from TCGA database.
In addition to its association with the BRAF
oncogene, miR-31 has been demonstrated to bind to the
3′-untranslated region of special AT-rich sequence-binding protein
2 (SATB2), which is a nuclear matrix-associated transcription
factor and epigenetic regulator, downregulating its expression at
both the mRNA and protein level (36). SATB2 was originally identified as a
highly tissue-specific protein, predominantly expressed in
glandular cells of the lower gastrointestinal tract, and high
expression of SATB2 is an accurate prognostic marker in CRC
(37,38).
Another target of miR-31 is RAS p21
GTPase-activating protein 1 (RASA1), which is a negative regulator
of the RAS-RAF-MEK-ERK signal pathway (39). Overexpression of miR-31
downregulates RASA1 protein but not RASA1 mRNA, suggesting
that miR-31 regulates RASA1 expression via a
post-transcriptional mechanism. RASA1, with its C-terminal GAP
domain, has the ability to stimulate the GTPase activity of normal
RAS p21, which results in the inactivation of RAS. Thus,
overexpression of miR-31 in CRC may repress RASA1 and
consequently upregulate the RAS pathway to promote tumor cell
proliferation. Furthermore, miR-31 promotes
epithelial-mesenchymal transition (EMT) (34,40).
Cottonham et al (34)
reported that CRC cell lines with elevated miR-31 expression
undergo EMT (a factor associated with distant metastasis) in
response to transforming growth factor (TGF)-β, without influencing
the TGF-β pathway. Meng et al (41) reported that miR-31 was
upregulated in lung adenocarcinoma tissues from patients with lymph
node metastases, compared with those without. Additionally, in
vitro functional assays demonstrated that miR-31
increased cell migration, invasiveness and proliferation in an
ERK1/2 signaling-dependent manner (40).
Thus, upregulation of miR-31 may enhance
tumor development, as well as the malignant potential of a tumor,
via inhibition of SATAB2 and RASA1, enhancement of EMT and
upregulation of BRAF expression. Particularly, miR-31
upregulation is commonly observed in BRAF-mutant CRCs, which
may enhance the downstream proteins of RAS-RAF signal cascade.
Regarding molecular targeting therapies, there are
several reports indicating the influence of the expression status
of miR-31 (either −3p alone or both −3p and −5p) on the
sensitivity of metastatic CRC (with wild-type RAS) to
anti-EGFR therapy (42–44). Patients with higher miR-31
expression levels exhibited poorer outcomes during treatment with
anti-EGFR therapy. As miR-31 upregulates BRAF protein levels
irrespective of the RAS/BRAF mutation status and inhibits
RASA1 mRNA (which inactivates activated RAS by stimulating
GTPase) (13,39), it is hypothesized that upregulation
of miR-31 inhibits the efficacy of anti-EGFR therapy, and
acts in a manner similar to oncogenic mutations in the RAS
and RAF genes.
To the best of our knowledge, the most effective
treatment strategy for patients with stage IV CRC and BRAF
mutations, which may be associated with high miR-31
expression levels (13), is
currently unknown. The answer may exist in a recent clinical trial
for unresectable patients with BRAF V600E mutant CRC, called
the BEACON study (45). BRAF
inhibition has been revealed to improve clinical outcomes in
patients with melanoma and non-small cell lung cancer that have a
BRAF V600 mutation; however, in patients with CRC, BRAF
inhibition has only conferred a marginal improvement in the
reduction of tumor burden (46–53).
Notably, in vitro studies demonstrated that in BRAF
V600E-mutant CRC cells, BRAF inhibition results in the rapid
feedback activation of EGFR, permitting sustained MAPK activation
and continued cell proliferation; however, combined inhibition of
BRAF and EGFR resulted in synergistic inhibition of tumor growth in
BRAF V600E mutant CRC xenograft models (54,55).
Subsequent clinical studies of EGFR-targeted therapies, combined
with the BRAF inhibitors vemurafenib or dabrafenib, confirmed that
addition of an EGFR-targeted therapy may improve the activity of
BRAF inhibition in BRAF V600E-mutant CRC (55–57). In
the BEACON study (45), patients
with CRC and the BRAF V600E mutation, who had experienced
treatment failure with one or two prior chemotherapy regimens, were
recruited and treated with three molecular target agents,
encorafenib (a BRAF inhibitor), binimetinib (a MEK inhibitor) and
cetuximab (an EGFR inhibitor). In 29 patients with BRAF
V600E mutant tumors, the overall response rate was 48%, the median
progression-free survival was 8.0 months and the median OS time was
15.3 months (45). This indicates
that the BRAF V600E mutation serves as a facilitator of CRC
progression and therapies are required that inhibit the feedback
activation of RAS-RAF signal cascade.
The present study demonstrated that high
miR-31 expression was associated with BRAF V600E
mutation in patients with stage IV CRC, but also indicated a poor
prognosis, irrespective of BRAF mutation status.
Additionally, an inhibitor to miR-31 may confer a clinical
benefit on patients with CRC with BRAF V600E mutation or
high miR-31 expression levels, perhaps as part of a combination
therapy alongside BRAF inhibitors, MEK inhibitors and EGFR
inhibitors.
In the present study, despite the small population
size, analysis of miRNA microarray data and relevant samples from
TCGA database showed the reproducible result that high
miR-31 expression was associated with a BRAF mutation
and poor outcome in CRC, as demonstrated by a previous study
(13). The results from the current
strongly indicate that the expression level of miR-31 in the
primary cancer site is a promising prognostic factor for CRC
regardless of the RAS/BRAF mutation profile as well as a
predictive factor for anti-EGFR treatment in CRCs with wild-type
RAS/BRAF. Thus, miR-31 may represent a favorable
biomarker and a promising therapeutic target in patients with
CRC.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Mr. Toru Nakai and
Mrs. Tae Yamanishi (Department of Gastroenterological Surgery,
Okayama University Graduate School of Medicine, Dentistry and
Pharmaceutical Sciences, Okayama, Japan) and Mrs. Kikue Tokuda
(Clinical Oncology, Kawasaki Medical School, Okayama, Japan) for
their technical assistance.
Funding
The present study was supported by The Ministry of
Education, Culture, Sports, Science and Technology (MEXT)/Japan
Society of the Promotion of Science (JSPS) KAKENHI (grant nos.
20590572, 25860409, 26462016, 18H03554 and 18K18464).
Availability of data and materials
The datasets generated and analyzed during the
present study are available in the National Cancer Institute
repository (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga).
Authors' contributions
NK performed miRNA analysis and drafted the
manuscript. FT performed TCGA data analysis. AN, YM and HT
performed all DNA extractions and KRAS/BRAF mutation and MSI
analyses. YU collected patient samples, clinicopathological data,
assisted with data interpretation and performed statistical
analyses. TF provided patient samples, clinicopathological data and
designed the study. AT and YY assisted with data interpretation and
revised the manuscript. TN assisted with data interpretation,
designed the project, secured the funding and drafted the
manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The study protocol was approved by The Ethics
Committee of Okayama University Hospital and Kawasaki Medical
School Hospital. All patients provided written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
CI
|
confidence interval
|
|
∆Ct
|
∆ threshold cycle
|
|
EMT
|
epithelial-mesenchymal transition
|
|
HR
|
hazard ratio
|
|
MSI
|
microsatellite instability
|
|
miRNA/miR
|
microRNA
|
|
MST
|
median survival time
|
|
OS
|
overall survival
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
RASA1
|
RAS p21 GTPase-activating protein
1
|
|
RQ
|
expression relative quotient
|
|
SATB2
|
special AT-rich sequence-binding
protein 2
|
|
TCGA
|
The Cancer Genome Atlas
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yu T, Ma P, Wu D, Shu Y and Gao W:
Functions and mechanisms of microRNA-31 in human cancers. Biomed
Pharmacother. 108:1162–1169. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kopcalic K, Petrovic N, Stanojkovic TP,
Stankovic V, Bukumiric Z, Roganovic J, Malisic E and Nikitovic M:
Association between miR-21/146a/155 level changes and acute
genitourinary radiotoxicity in prostate cancer patients: A pilot
study. Pathol Res Pract. 215:626–631. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Braga TV, Evangelista FCG, Gomes LC,
Araujo SSDS, Carvalho MDG and Sabino AP: Evaluation of MiR-15a and
MiR-16-1 as prognostic biomarkers in chronic lymphocytic leukemia.
Biomed Pharmacother. 92:864–869. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Petrovic N: miR-21 might be involved in
breast cancer promotion and invasion rather than in initial events
of breast cancer development. Mol Diagn Ther. 20:97–110. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Toiyama Y, Takahashi M, Hur K, Nagasaka T,
Tanaka K, Inoue Y, Kusunoki M, Boland CR and Goel A: Serum miR-21
as a diagnostic and prognostic biomarker in colorectal cancer. J
Natl Cancer Inst. 105:849–859. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hur K, Toiyama Y, Takahashi M, Balaguer F,
Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR and Goel A:
MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT)
in human colorectal cancer metastasis. Gut. 62:1315–1326. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fuji T, Umeda Y, Nyuya A, Taniguchi F,
Kawai T, Yasui K, Toshima T, Yoshida K, Fujiwara T, Goel A and
Nagasaka T: Detection of circulating microRNAs with Ago2 complexes
to monitor the tumor dynamics of colorectal cancer patients during
chemotherapy. Int J Cancer. 144:2169–2180. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee RC and Ambros V: An extensive class of
small RNAs in caenorhabditis elegans. Science. 294:862–864. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pagliuca A, Valvo C, Fabrizi E, di Martino
S, Biffoni M, Runci D, Forte S, De Maria R and Ricci-Vitiani L:
Analysis of the combined action of miR-143 and miR-145 on oncogenic
pathways in colorectal cancer cells reveals a coordinate program of
gene repression. Oncogene. 32:4806–4813. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nosho K, Igarashi H, Nojima M, Ito M,
Maruyama R, Yoshii S, Naito T, Sukawa Y, Mikami M, Sumioka W, et
al: Association of microRNA-31 with BRAF mutation, colorectal
cancer survival and serrated pathway. Carcinogenesis. 35:776–783.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Samowitz WS, Sweeney C, Herrick J,
Albertsen H, Levin TR, Murtaugh MA, Wolff RK and Slattery ML: Poor
survival associated with the BRAF V600E mutation in
microsatellite-stable colon cancers. Cancer Res. 65:6063–6069.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Saridaki Z, Tzardi M, Sfakianaki M,
Papadaki C, Voutsina A, Kalykaki A, Messaritakis I, Mpananis K,
Mavroudis D, Stathopoulos E, et al: BRAFV600E mutation analysis in
patients with metastatic colorectal cancer (mCRC) in daily clinical
practice: Correlations with clinical characteristics, and its
impact on patients' outcome. PLoS One. 8:e846042013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mori Y, Nagasaka T, Mishima H, Umeda Y,
Inada R, Kishimoto H, Goel A and Fujiwara T: The rare BRAF
VK600-601E mutation as a possible indicator of poor prognosis in
rectal carcinoma-a report of a case. BMC Med Genet. 16:12015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mori Y, Nyuya A, Yasui K, Toshima T, Kawai
T, Taniguchi F, Kimura K, Inada R, Nishizaki M, Haraga J, et al:
Clinical outcomes of women with ovarian metastases of colorectal
cancer treated with oophorectomy with respect to their somatic
mutation profiles. Oncotarget. 9:16477–16488. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Morikawa T, Inada R, Nagasaka T, Mori Y,
Kishimoto H, Kawai T, Umeda Y, Mishima H, Goel A and Fujiwara T:
BRAF V600E mutation is a predictive indicator of upfront
chemotherapy for stage IV colorectal cancer. Oncol Lett.
15:2195–2201. 2018.PubMed/NCBI
|
|
19
|
Edge S, Byrd D, Compton C, Fritz A, Greene
F and Trotti A: AJCC Cancer Staging Manual. https://cancerstaging.org/references-tools/deskreferences/Documents/AJCC%207th%20Ed%20Cancer%20Staging%20Manual.pdfJanuary
7–2013
|
|
20
|
Nagasaka T, Koi M, Kloor M, Gebert J,
Vilkin A, Nishida N, Shin SK, Sasamoto H, Tanaka N, Matsubara N, et
al: Mutations in both KRAS and BRAF may contribute to the
methylator phenotype in colon cancer. Gastroenterology.
134:1950–1960, 60.e1. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Goel A, Nagasaka T, Hamelin R and Boland
CR: An optimized pentaplex PCR for detecting DNA mismatch
repair-deficient colorectal cancers. PLoS One. 5:e93932010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Takehara Y, Nagasaka T, Nyuya A, Haruma T,
Haraga J, Mori Y, Nakamura K, Fujiwara T, Boland CR and Goel A:
Accuracy of four mononucleotide-repeat markers for the
identification of DNA mismatch-repair deficiency in solid tumors. J
Transl Med. 16:52018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cancer Genome Atlas Network: Comprehensive
molecular characterization of human colon and rectal cancer.
Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nagasaka T, Mori Y, Umeda Y and Fujiwara
T: Biomarker for colorectal cancer. Nihon Rinsho. 70:802–808.
2012.(In Japanese). PubMed/NCBI
|
|
26
|
Nikolaou S, Qiu S, Fiorentino F, Rasheed
S, Tekkis P and Kontovounisios C: Systematic review of blood
diagnostic markers in colorectal cancer. Tech Coloproctol.
22:481–498. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang T, Wang Q, Zhao D, Cui Y, Cao B, Guo
L and Lu SH: The oncogenetic role of microRNA-31 as a potential
biomarker in oesophageal squamous cell carcinoma. Clin Sci (Lond).
121:437–447. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Valastyan S, Reinhardt F, Benaich N,
Calogrias D, Szasz AM, Wang ZC, Brock JE, Richardson AL and
Weinberg RA: A pleiotropically acting microRNA, miR-31, inhibits
breast cancer metastasis. Cell. 137:1032–1046. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Creighton CJ, Fountain MD, Yu Z, Nagaraja
AK, Zhu H, Khan M, Olokpa E, Zariff A, Gunaratne PH, Matzuk MM and
Anderson ML: Molecular profiling uncovers a p53-associated role for
microRNA-31 in inhibiting the proliferation of serous ovarian
carcinomas and other cancers. Cancer Res. 70:1906–1915. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang Y, Guo J, Li D, Xiao B, Miao Y,
Jiang Z and Zhuo H: Down-regulation of miR-31 expression in gastric
cancer tissues and its clinical significance. Med Oncol.
27:685–689. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schee K, Boye K, Abrahamsen TW, Fodstad O
and Flatmark K: Clinical relevance of microRNA miR-21, miR-31,
miR-92a, miR-101, miR-106a and miR-145 in colorectal cancer. BMC
Cancer. 12:5052012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cekaite L, Rantala JK, Bruun J, Guriby M,
Agesen TH, Danielsen SA, Lind GE, Nesbakken A, Kallioniemi O, Lothe
RA and Skotheim RI: MiR-9, −31, and −182 deregulation promote
proliferation and tumor cell survival in colon cancer. Neoplasia.
14:868–879. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chang KH, Miller N, Kheirelseid EA,
Lemetre C, Ball GR, Smith MJ, Regan M, McAnena OJ and Kerin MJ:
MicroRNA signature analysis in colorectal cancer: Identification of
expression profiles in stage II tumors associated with aggressive
disease. Int J Colorectal Dis. 26:1415–1422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cottonham CL, Kaneko S and Xu L: miR-21
and miR-31 converge on TIAM1 to regulate migration and invasion of
colon carcinoma cells. J Biol Chem. 285:35293–35302. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang CJ, Zhou ZG, Wang L, Yang L, Zhou B,
Gu J, Chen HY and Sun XF: Clinicopathological significance of
microRNA-31, −143 and −145 expression in colorectal cancer. Dis
Markers. 26:27–34. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mansour MA, Hyodo T, Ito S, Kurita K,
Kokuryo T, Uehara K, Nagino M, Takahashi M, Hamaguchi M and Senga
T: SATB2 suppresses the progression of colorectal cancer cells via
inactivation of MEK5/ERK5 signaling. FEBS J. 282:1394–1405. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Eberhard J, Gaber A, Wangefjord S, Nodin
B, Uhlen M, Ericson Lindquist K and Jirström K: A cohort study of
the prognostic and treatment predictive value of SATB2 expression
in colorectal cancer. Br J Cancer. 106:931–938. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Magnusson K, de Wit M, Brennan DJ, Johnson
LB, McGee SF, Lundberg E, Naicker K, Klinger R, Kampf C, Asplund A,
et al: SATB2 in combination with cytokeratin 20 identifies over 95%
of all colorectal carcinomas. Am J Surg Pathol. 35:937–948. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sun D, Yu F, Ma Y, Zhao R, Chen X, Zhu J,
Zhang CY, Chen J and Zhang J: MicroRNA-31 activates the RAS pathway
and functions as an oncogenic MicroRNA in human colorectal cancer
by repressing RAS p21 GTPase activating protein 1 (RASA1). J Biol
Chem. 288:9508–9518. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang MH, Yu J, Chen N, Wang XY, Liu XY,
Wang S and Ding YQ: Elevated microRNA-31 expression regulates
colorectal cancer progression by repressing its target gene SATB2.
PLoS One. 8:e853532013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Meng W, Ye Z, Cui R, Perry J,
Dedousi-Huebner V, Huebner A, Wang Y, Li B, Volinia S, Nakanishi H,
et al: MicroRNA-31 predicts the presence of lymph node metastases
and survival in patients with lung adenocarcinoma. Clin Cancer Res.
19:5423–5433. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Manceau G, Imbeaud S, Thiebaut R, Liebaert
F, Fontaine K, Rousseau F, Génin B, Le Corre D, Didelot A, Vincent
M, et al: Hsa-miR-31-3p expression is linked to progression-free
survival in patients with KRAS wild-type metastatic colorectal
cancer treated with anti-EGFR therapy. Clin Cancer Res.
20:3338–3347. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mosakhani N, Lahti L, Borze I,
Karjalainen-Lindsberg ML, Sundström J, Ristamäki R, Osterlund P,
Knuutila S and Sarhadi VK: MicroRNA profiling predicts survival in
anti-EGFR treated chemorefractory metastatic colorectal cancer
patients with wild-type KRAS and BRAF. Cancer Genet. 205:545–551.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mlcochova J, Faltejskova-Vychytilova P,
Ferracin M, Zagatti B, Radova L, Svoboda M, Nemecek R, John S, Kiss
I, Vyzula R, et al: MicroRNA expression profiling identifies
miR-31-5p/3p as associated with time to progression in wild-type
RAS metastatic colorectal cancer treated with cetuximab.
Oncotarget. 6:38695–38704. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Van Cutsem E, Huijberts S, Grothey A,
Yaeger R, Cuyle PJ, Elez E, Fakih M, Montagut C, Peeters M, Yoshino
T, et al: Binimetinib, encorafenib, and cetuximab triplet therapy
for patients with BRAF V600E-mutant metastatic colorectal cancer:
Safety lead-in results from the phase III BEACON colorectal cancer
study. J Clin Oncol. 37:1460–1469. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Flaherty KT, Puzanov I, Kim KB, Ribas A,
McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K and
Chapman PB: Inhibition of mutated, activated BRAF in metastatic
melanoma. N Engl J Med. 363:809–819. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wilmott JS, Long GV, Howle JR, Haydu LE,
Sharma RN, Thompson JF, Kefford RF, Hersey P and Scolyer RA:
Selective BRAF inhibitors induce marked T-cell infiltration into
human metastatic melanoma. Clin Cancer Res. 18:1386–1394. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dummer R, Ascierto PA, Gogas HJ, Arance A,
Mandala M, Liszkay G, Garbe C, Schadendorf D, Krajsova I, Gutzmer
R, et al: Overall survival in patients with BRAF-mutant melanoma
receiving encorafenib plus binimetinib versus vemurafenib or
encorafenib (COLUMBUS): A multicentre, open-label, randomised,
phase 3 trial. Lancet Oncol. 19:1315–1327. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Long GV, Stroyakovskiy D, Gogas H,
Levchenko E, de Braud F, Larkin J, Garbe C, Jouary T, Hauschild A,
Grob JJ, et al: Dabrafenib and trametinib versus dabrafenib and
placebo for Val600 BRAF-mutant melanoma: A multicentre,
double-blind, phase 3 randomised controlled trial. Lancet.
386:444–451. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Long GV, Hauschild A, Santinami M,
Atkinson V, Mandala M, Chiarion-Sileni V, Larkin J, Nyakas M,
Dutriaux C, Haydon A, et al: Adjuvant dabrafenib plus trametinib in
stage III BRAF-mutated melanoma. N Engl J Med. 377:1813–1823. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chapman PB, Hauschild A, Robert C, Haanen
JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et
al: Improved survival with vemurafenib in melanoma with BRAF V600E
mutation. N Engl J Med. 364:2507–2516. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Planchard D, Besse B, Groen HJM, Souquet
PJ, Quoix E, Baik CS, Barlesi F, Kim TM, Mazieres J, Novello S, et
al: Dabrafenib plus trametinib in patients with previously treated
BRAF(V600E)-mutant metastatic non-small cell lung cancer: An
open-label, multicentre phase 2 trial. Lancet Oncol. 17:984–993.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hyman DM, Puzanov I, Subbiah V, Faris JE,
Chau I, Blay JY, Wolf J, Raje NS, Diamond EL, Hollebecque A, et al:
Vemurafenib in multiple nonmelanoma cancers with BRAF V600
mutations. N Engl J Med. 373:726–736. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Corcoran RB, Ebi H, Turke AB, Coffee EM,
Nishino M, Cogdill AP, Brown RD, Della Pelle P, Dias-Santagata D,
Hung KE, et al: EGFR-mediated re-activation of MAPK signaling
contributes to insensitivity of BRAF mutant colorectal cancers to
RAF inhibition with vemurafenib. Cancer Discov. 2:227–235. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hong DS, Morris VK, El Osta B, Sorokin AV,
Janku F, Fu S, Overman MJ, Piha-Paul S, Subbiah V, Kee B, et al:
Phase IB study of vemurafenib in combination with irinotecan and
cetuximab in patients with metastatic colorectal cancer with
BRAFV600E mutation. Cancer Discov. 6:1352–1365. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Corcoran RB, Andre T, Atreya CE, Schellens
JHM, Yoshino T, Bendell JC, Hollebecque A, McRee AJ, Siena S,
Middleton G, et al: Combined BRAF, EGFR, and MEK inhibition in
patients with BRAF(V600E)-mutant colorectal cancer. Cancer Discov.
8:428–443. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Connolly K, Brungs D, Szeto E and Epstein
RJ: Anticancer activity of combination targeted therapy using
cetuximab plus vemurafenib for refractory BRAF (V600E)-mutant
metastatic colorectal carcinoma. Curr Oncol. 21:e151–e154. 2014.
View Article : Google Scholar : PubMed/NCBI
|