Introduction
Bladder cancer ranks as the first and second most
common urological malignancies in China and in the USA,
respectively (1,2). The characteristics of bladder cancer
include high recurrence, morbidity and mortality rates (1,2). With
advances in surgical skills, three-year overall survival rates
after radical cystectomy were less than 80% in China in 2015
(1). It is therefore crucial to
determine the underlying mechanism of bladder carcinogenesis and
identify novel molecular targets. To the best of our knowledge,
conventional research on bladder cancer mainly focused on mRNAs;
however, more attention has been paid to numerous non-coding RNAs,
including microRNAs (miRNAs) and long noncoding RNAs (3–5). In
particular, circular RNAs (circRNAs), which represent a novel type
of noncoding RNA with notable regulatory potency (6–9), have
attracted much interest. circRNAs can serve as miRNA sponges and
influence the expression levels of miRNA-targeted transcripts
(10). Furthermore, it was
demonstrated that the circRNA/miRNA axes serve crucial roles in
numerous tumor-associated signaling pathways, such as tight
junction signaling, endocytosis and mTOR signaling pathways
(11,12). Recent studies have determined the
function of certain circRNAs in bladder cancer (13–17). Li
et al (15) found that Cdr1as
was downregulated and sponged multiple miRNAs in bladder cancer,
and demonstrated that it could exert anti-oncogenic functions by
sponging microRNA-135a. In addition, Zhong et al (16) reported that circRNA-MYLK might
function as competing endogenous RNA (ceRNA) for miR-29a, which
could contribute to epithelial-mesenchymal transition and the
development of bladder cancer by activating VEGFA/VEGFR2 and
downstream Ras/ERK signaling pathway. However, the role of all
circRNAs in bladder cancer, and the number of circRNAs acting as
miRNA sponges in bladder carcinoma remain unknown.
The present study aimed to investigate the
expression pattern of circRNAs in four bladder cancer tissues and
matched normal tissues by microarray analysis. The differential
expression levels of representative circRNAs were further
determined in 16 pairs of tissues using reverse
transcription-quantitative polymerase chain reaction (RT-qPCR). The
signaling pathways associated with the dysregulated circRNAs in
bladder cancer tissues were therefore determined using Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway analysis. A
circRNA/miRNA network was then constructed through bioinformatics
methods. The results from the present study indicated that certain
differentially expressed circRNAs may be considered as important
therapeutic targets in bladder cancer.
Materials and methods
Patients and specimens
Four bladder cancer and matched normal samples were
collected from four patients at the First Affiliated Hospital of
Zhengzhou University between January 2016 and November 2016. The
mean age was 62 year-old (range, 50–65 years). These specimens were
used for the circRNA microarray. Additional 16 pairs of tissues
from the Department of Urology of The First Affiliated Hospital of
Zhengzhou University were used to validate circRNA expression by
RT-qPCR. All the samples were collected from patients with advanced
bladder cancer who received radical cystectomy without preoperative
chemotherapy or radiotherapy. Tissue specimens were immediately
stored in liquid nitrogen following surgical resection until
further experiment.
RNA extraction
Total RNA was extracted from frozen specimens using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. RNA concentration was
determined by measuring the OD260 value with a NanoDrop
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc.) and the RNA integrity was assessed using electrophoresis.
Microarray hybridization
Microarray hybridization was performed as previously
described (18). Briefly, total RNAs
were digested with RNase R (Epicentre; Illumina, Inc.) to remove
linear RNAs and enrich circular RNAs. Next, the enriched circular
RNAs were amplified and transcribed into fluorescent cRNA utilizing
a random priming method (Arraystar Super RNA Labeling kit;
Arraystar). The labeled cRNAs were purified by RNeasy Mini kit
(Qiagen GmbH). The concentration and specific activity of the
labeled cRNAs (pmol Cy3/µg cRNA) were measured by NanoDrop ND-1000.
Each labeled cRNA (1 µg) was fragmented by adding 5 µl 10X blocking
agent and 1 µl 25X fragmentation buffer, then heated the mixture at
60°C for 30 min. Finally 25 µl 2X hybridization buffer was added to
dilute the labeled cRNA. Hybridization solution (50 µl) was
dispensed into the gasket slide and assembled to the circRNA
expression microarray slide. The slides were incubated for 17 h at
65°C in an Agilent Hybridization oven. The hybridized arrays were
washed, fixed and scanned using the Agilent Scanner (Agilent
Technologies Inc., part number G2505C). The acquired array images
were analyzed by Agilent Feature Extraction software (version
10.5.1.1; Agilent Technologies Inc.,). Microarray hybridization and
data collection were carried out by Kangchen BioTech Co., Ltd.
Microarray data analysis
Microarray data analysis was performed as previously
described (19). Briefly, circRNAs
expression profiles between bladder cancer and normal control
samples were compared by calculating the fold change for each
circRNA (19). CircRNAs with fold
changes ≥1.5 and P≤0.05 were considered as significantly
differentially expressed.
RT-qPCR
Total RNA was extracted from frozen tissues in
liquid nitrogen using TRIzol (Invitrogen; Thermo Fisher Scientific,
Inc.). The expression levels of the circRNAs were assessed by
RT-qPCR as previously described (19). According to manufacturer's
instructions, M-MLV reverse transcriptase (Invitrogen; Thermo
Fisher Scientific, Inc.) was used for synthesizing cDNA following
RNA extraction. The expression level of the circRNAs was assessed
by RT-qPCR using SYBR Green assay (Arraystar Inc.). Specific
divergent primers (Table SI) were
designed to amplify the circular transcripts. PCR was carried out
in a 10-µl reaction volume, including 2 µl of cDNA, 5 µl 2X Master
mix (Arraystar Inc.), 0.5 µl of forward primer (10 µM), 0.5 µl of
reverse primer (10 µM) and 2 µl of double distilled water. The
reaction was set at 95°C for 10 min for pre-denaturation, followed
by 95°C for 10 sec and at 60°C for 60 sec repeating 40 cycles.
β-actin was used as a reference. Both target and reference were
amplified in triplicate wells. The relative expression levels of
the circRNAs were normalized to the endogenous control β-actin and
expressed as 2−ΔΔCq.
Delineation of circRNA/miRNA
interactions
circRNA/miRNA interactions were predicted using
miRanda (http://www.microrna.org/microrna/home.do) and
TargetScan tools (http://www.targetscan.org). The microRNA response
elements (MREs) on the circRNAs were therefore determined and the
corresponding miRNAs were identified. Cytoscape 3.0 (https://cytoscape.org/) was used to generate the
circRNA/miRNA network diagram.
Statistical analysis
Student's t-tests (two-tailed) was used to compare
the expression levels of circRNAs between bladder cancer and normal
control samples. P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification of differentially
expressed circRNAs in bladder cancer samples
The circRNA microarray was used to investigate the
circRNA profiles in bladder cancer and matched normal samples. A
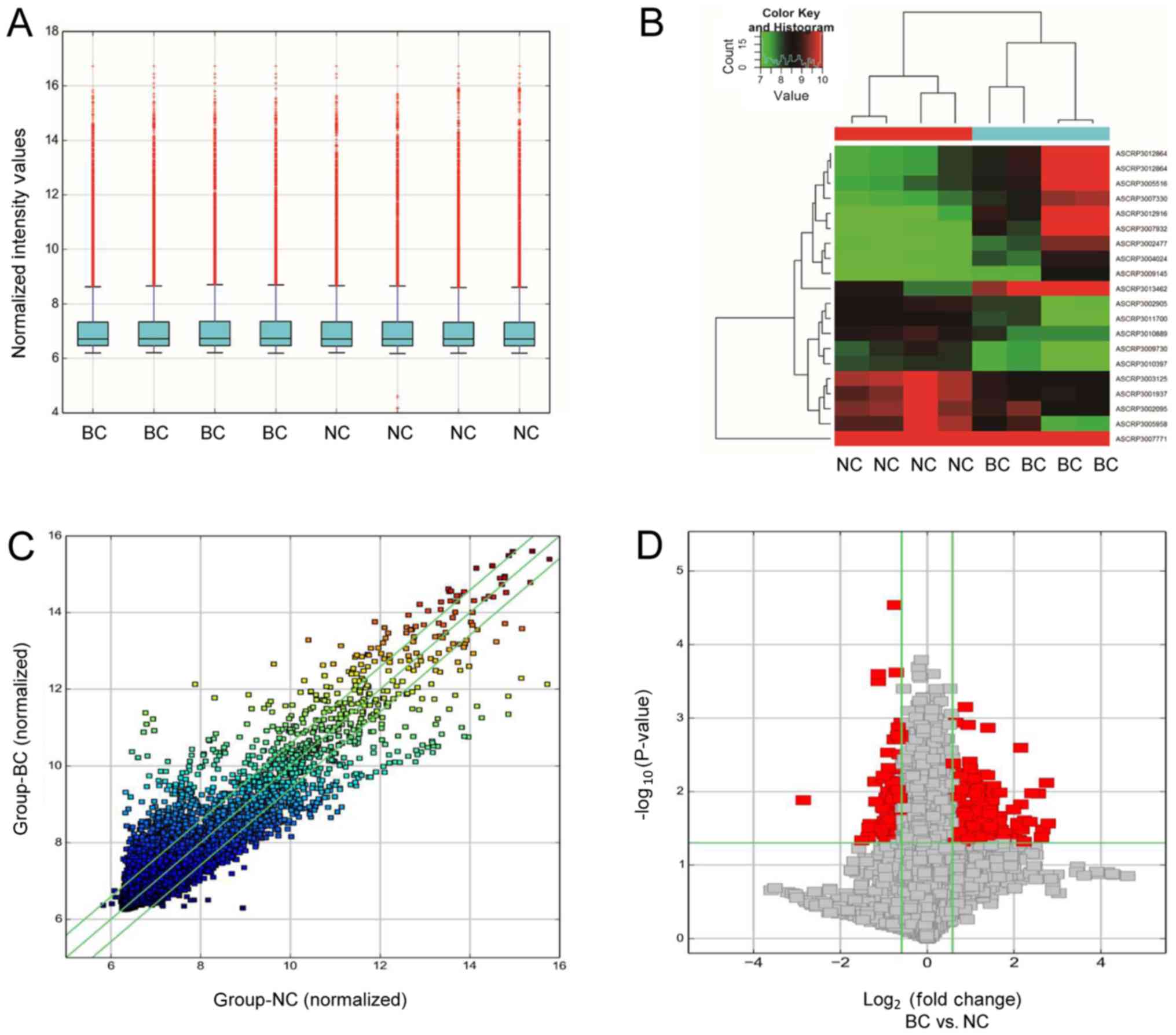
box plot was drawn for visualizing the distribution the intensities
of all the datasets following normalization, and observed that the
distribution of log2 ratios was similar in all the
tested samples (Fig. 1A).
Hierarchical clustering was used for generating an overview of
circRNA expression profiles between the two groups. The circRNA
expression patterns were distinguishable between experiment and
control groups (Fig. 1B). Scatter
plots were performed to evaluate differential circRNA expression
between two different conditions. Differentially changed circRNAs
were identified through fold change filtering (Fig. 1C). Volcano plots were used to
visualize the difference in the expression of circRNAs in bladder
cancer tissues and paired adjacent normal tissues. The volcano plot
filtering displayed differentially expressed circRNAs between the
two groups (Fig. 1D).
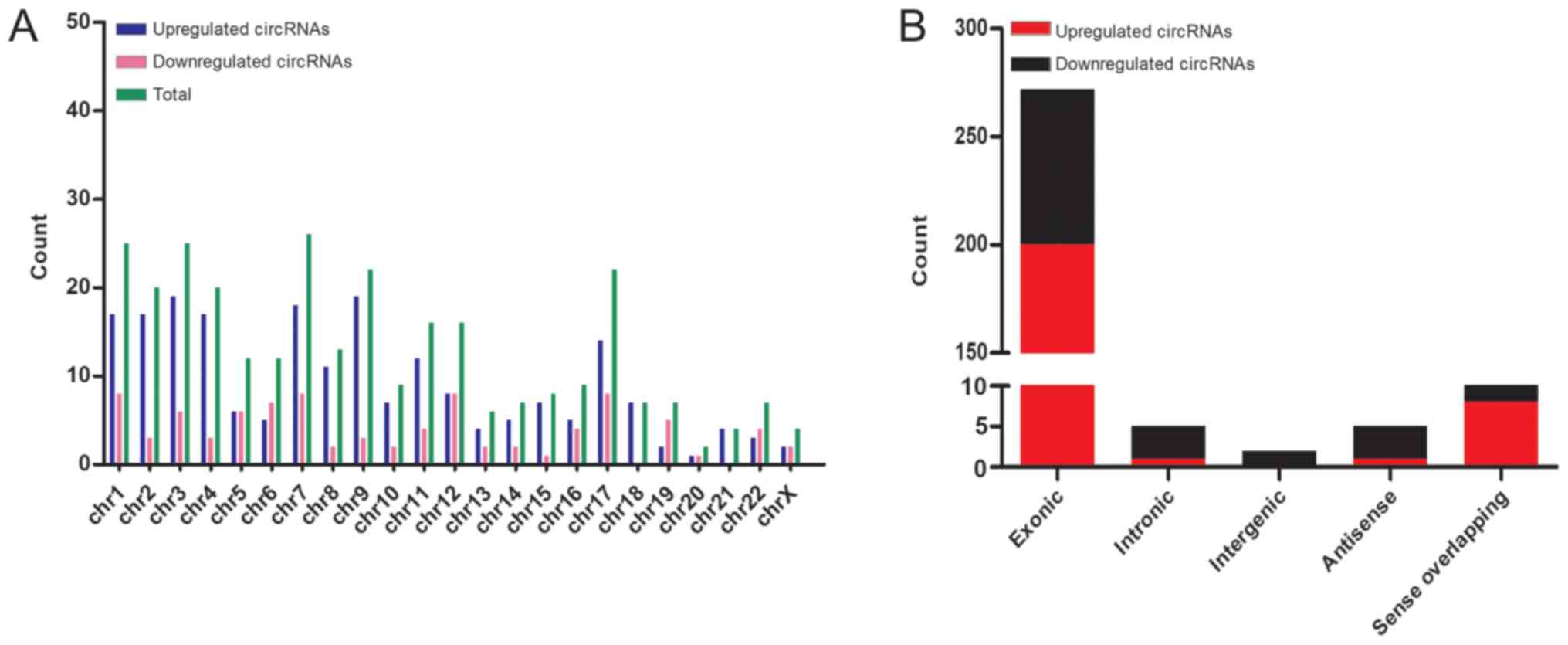
Following circRNA microarray analysis, significant
differential expression was observed in 299 circRNAs, of which 89
circRNAs were downregulated and 210 circRNAs were upregulated
(Table SII). The results from
microarray demonstrated the distribution of dysregulated circRNAs
on human chromosomes. The upregulated and downregulated circRNAs
were further summarized and the differentially expressed circRNAs
on human chromosomes were delineated (Fig. 2A). Most of the dysregulated circRNAs
were transcribed by exons (Fig.
2B).
Validation of the differential
expression levels of circRNAs
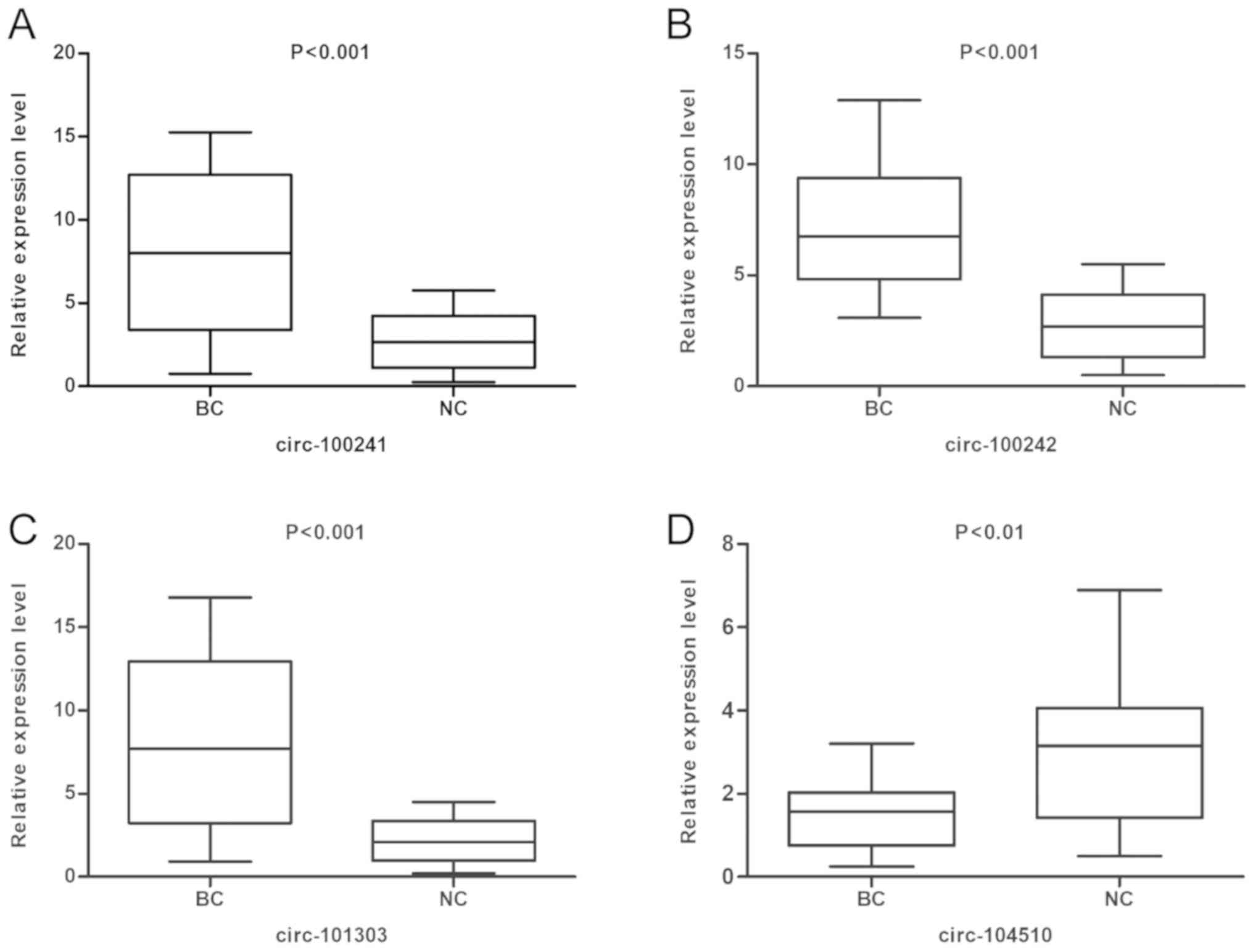
A total of four dysregulated circRNAs were randomly
selected to confirm the microarray findings by using RT-qPCR. The
results demonstrated that hsa_circ_100241, hsa_circ_100242 and
hsa_circ_101303 were validated as significantly upregulated in
bladder cancer samples, whereas hsa_circ_104510 was significantly
downregulated in bladder cancer samples compared with adjacent
normal tissues (Fig. 3). These
results were consistent with findings from the microarray
analysis.
Construction of the circRNA/miRNA
interaction network
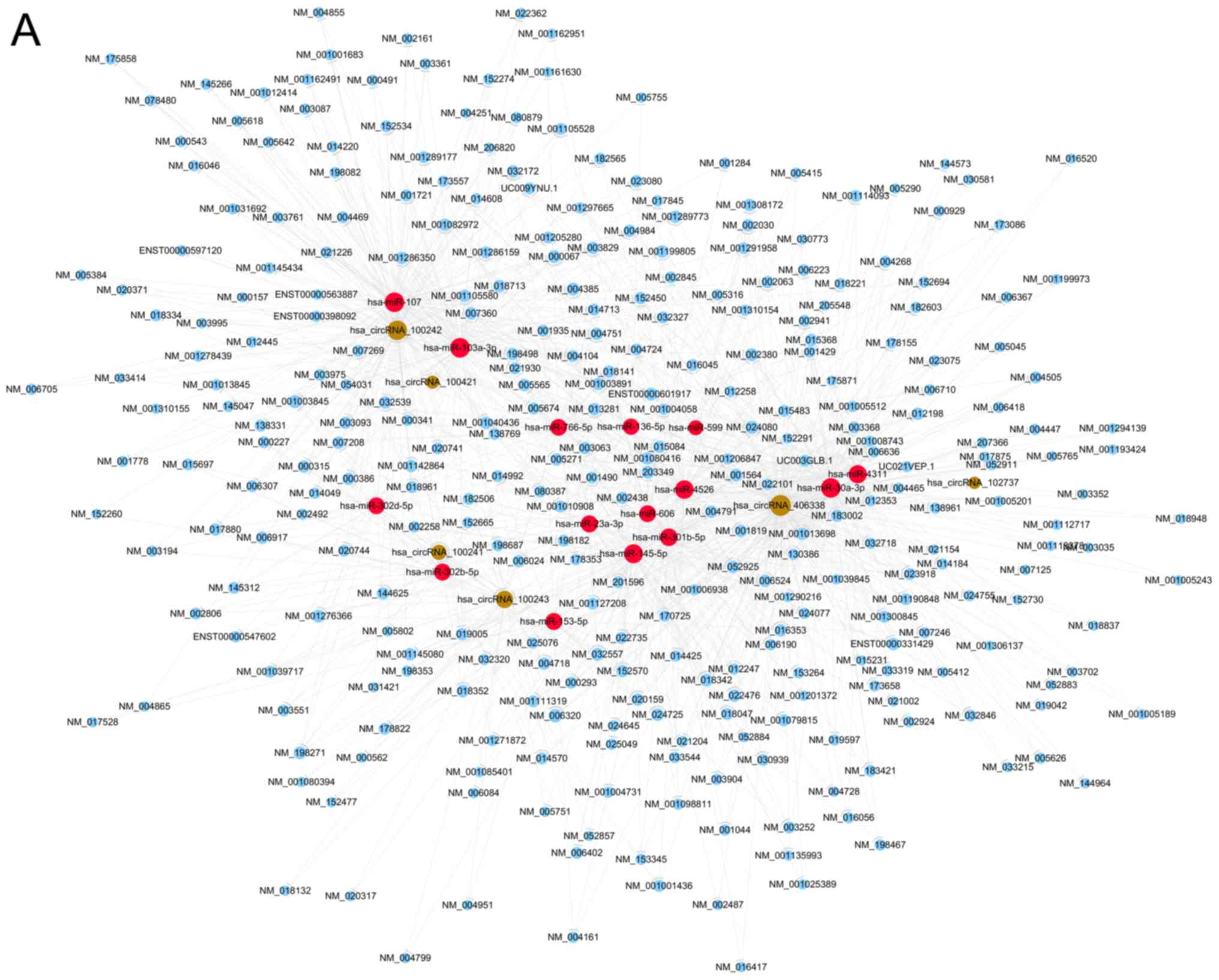
The circRNA/miRNA interaction was theoretically
predicted through Arraystar's home-made miRNA target prediction
software based on TargetScan and miRanda. Cytoscape was used to
depict an entire network of circRNA/miRNA interaction (Fig. 4A). The part of the graph was then
enlarged to clearly display several circRNAs and their target
miRNAs in bladder cancer (Fig. 4B).
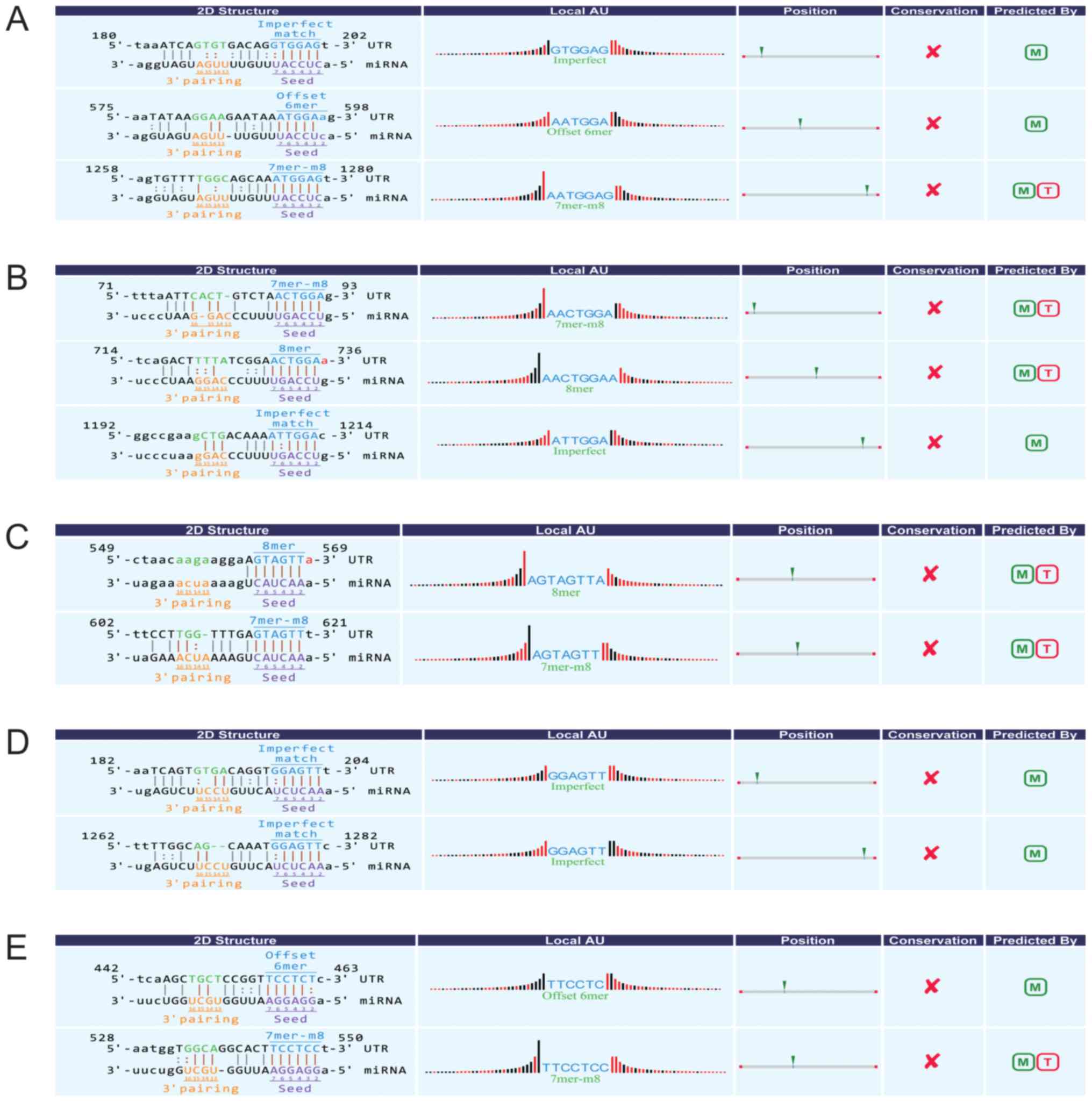
One confirmed circRNA (hsa_circ_100242) was annotated in detail
with circRNA/miRNA interaction information, indicating that
hsa_circ_100242 had microRNA response element of miR-145-5p
(Fig. 5).
Bioinformatics analyses
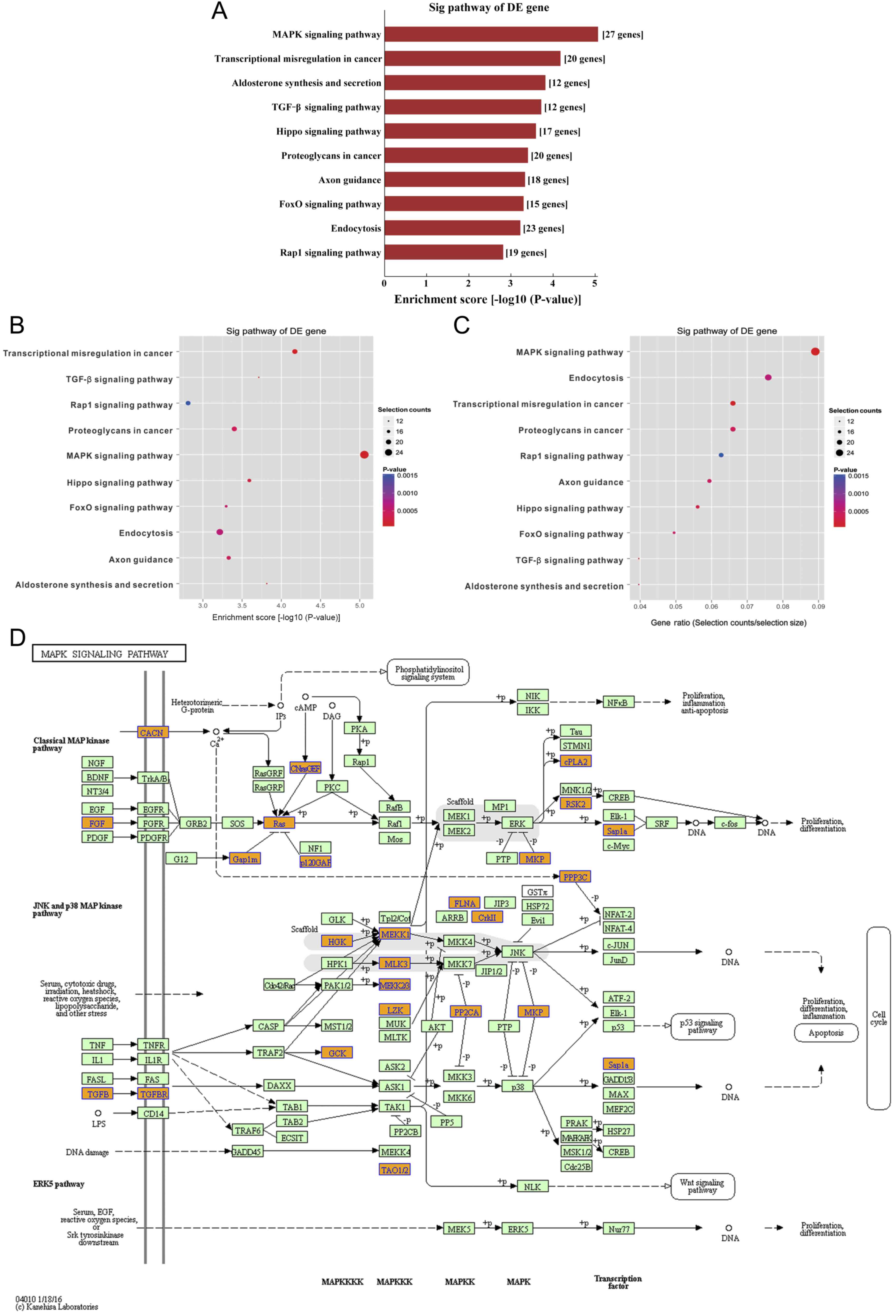
The results from KEGG pathway analysis demonstrated
that 10 signaling pathways were associated with the dysregulated
circRNAs, including ‘MAPK pathway’, ‘transcriptional misregulation
in cancer’, ‘aldosterone synthesis and secretion’, ‘TGF-β pathway’,
‘Hippo pathway’, ‘proteoglycans in cancer’, ‘axon guidance’, ‘FoxO
signaling in cancer’, ‘endocytosis’ and ‘Rap1 pathway’. In
particular, ‘MAPK signaling pathway’ was the most important pathway
of the dysregulated circRNAs in bladder cancer samples (Fig. 6).
Discussion
The present study determined the expression profiles
of circRNAs in advanced bladder cancer samples by using circRNA
microarray. The analysis revealed 210 upregulated circRNAs and 89
downregulated circRNAs in bladder cancer tissues. Furthermore, the
‘MAPK signaling pathway’ was the most significant pathway for the
differentially expressed circRNAs in bladder cancer. According to
the results from circRNA microarray and our previous study
(20), hsa_circ_100242 may be
involved in bladder cancer initiation and progression by sponging
miR-145.
Previous studies have reported that circRNAs could
be considered as ceRNA molecules or miRNA sponges and serve a
crucial role in certain diseases (such as pathological hypertrophy
and glioma) by influencing gene expression (7,10,21–24).
For instance, Wang et al (21) demonstrated that heart-related
circRNAs (HRCRs) could affect miR-223 activity in cardiac
hypertrophy by acting as an endogenous miR-223 sponge. HRCRs may
therefore represent novel therapeutic targets for treating cardiac
hypertrophy. In the present study, circRNA/miRNA interactions were
also predicted on the basis of conserved seed sequence matches
using miRanda and TargetScan tools. In addition, the circRNA/miRNA
interaction network that was constructed in this study may provide
novel evidence for further research on ceRNAs involving
dysregulated circRNAs.
The present study demonstrated that hsa_circ_100242
was significantly upregulated in bladder cancer samples compared
with matched normal samples. Our previous study demonstrated that
miR-145 could directly target the 3′-untranslated region of the
type 1 insulin-like growth factor receptor (IGF-IR) in human
bladder cancer sample (20). In
addition, results from small interfering RNA- and miR-145-mediated
IGF-IR knockdown experiments revealed that miR-145 could promote
bladder cancer cell apoptosis and inhibit its cell proliferation
and migration via suppression of IGF-IR expression (20). Subsequently, hsa_circ_100242 may be
implicated in bladder cancer initiation and progression by
targeting miR-145-5p. The role of hsa_circ_100242 in bladder cancer
requires therefore further investigation.
The present study presented some limitations.
Firstly, bladder cancer and matched normal tissues were all
collected from one hospital. Secondly, the small sample size of
bladder cancer tissues used for the microarray analysis was not
large enough to draw any definitive conclusions. Thirdly, only the
expression profiles of circRNAs in bladder cancer and matched
normal samples were compared. Of equal importance is the
differential circRNA expression profiles in bladder cancer patients
of different genders, stages or grades. In future investigation, a
higher number of tissues from various hospitals should therefore be
collected to carry out a deeper analysis. Fourthly, circRNAs
expression in blood and urine might be also crucial for the
identification of potent biomarkers for earlier diagnosis of
bladder cancer. Blood and urine samples from patients with bladder
cancer are therefore currently being collected for future
investigation. Experiments in bladder cancer cell lines will also
be performed. Furthermore, following RNA interference and
transfection of adenovirus vector plasmid in bladder cancer cell
lines, the role of circRNA in bladder cancer and the determination
of the regulatory mechanism of circRNA on biological activity of
bladder cancer cells will be further investigated.
The present study demonstrated that circRNAs were
differentially expressed in bladder cancer tissues compared with
matched normal samples. Subsequently, pathway analysis was
performed, and binding miRNAs were predicted for the dysregulated
circRNAs of bladder cancer tissues. The results revealed that ‘MAPK
signaling pathway’ was the most important pathway of the
differentially expressed circRNAs in bladder cancer samples. In
addition, hsa_circ_100242 may be involved in bladder cancer
initiation and progression by sponging miR-145. These findings may
lay the foundation for future studies on circRNAs in bladder
cancer.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural
Science Foundation of China (grant no. 81702503).
Availability of data and materials
Not applicable.
Authors' contributions
ZZ and XZ: Conception and design of the study, and
final approval of the manuscript. FC, JL and JW: Acquisition,
analysis and interpretation of data.
Ethics approval and consent to
participate
The study was granted an exemption from requiring
ethic approval from the Ethics Committee of our institution.
Patients provided informed consent prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pasquinelli AE: MicroRNAs and their
targets: Recognition, regulation and an emerging reciprocal
relationship. Nat Rev Genet. 13:271–282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li Y and Wang X: Role of long noncoding
RNAs in malignant disease (Review). Mol Med Rep. 13:1463–1469.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guo S, Xu X, Ouyang Y, Wang Y, Yang J, Yin
L, Ge J and Wang H: Microarray expression profile analysis of
circular RNAs in pancreatic cancer. Mol Med Rep. 17:7661–7671.
2018.PubMed/NCBI
|
|
10
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu BH, Zhang BB, Liu XQ, Zheng S, Dong KR
and Dong R: Expression profiling identifies circular RNA signature
in hepatoblastoma. Cell Physiol Biochem. 45:706–719. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qian Y, Lu Y, Rui C, Qian Y, Cai M and Jia
R: Potential significance of circular RNA in human placental tissue
for patients with preeclampsia. Cell Physiol Biochem. 39:1380–1390.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang X, Yuan W, Tao J, Li P, Yang C, Deng
X, Zhang X, Tang J, Han J, Wang J, et al: Identification of
circular RNA signature in bladder cancer. J Cancer. 8:3456–3463.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li M, Liu Y, Zhang X, Liu J and Wang P:
Transcriptomic analysis of high-throughput sequencing about
circRNA, lncRNA and mRNA in bladder cancer. Gene. 677:189–197.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li P, Yang X, Yuan W, Yang C, Zhang X, Han
J, Wang J, Deng X, Yang H, Li P, et al: CircRNA-cdr1as exerts
anti-oncogenic functions in bladder cancer by sponging
MicroRNA-135a. Cell Physiol Biochem. 46:1606–1616. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhong Z, Huang M, Lv M, He Y, Duan C,
Zhang L and Chen J: Circular RNA MYLK as a competing endogenous RNA
promotes bladder cancer progression through modulating VEGFA/VEGFR2
signaling pathway. Cancer Lett. 403:305–317. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xie F, Li Y, Wang M, Huang C, Tao D, Zheng
F, Zhang H, Zeng F, Xiao X and Jiang G: Circular RNA BCRC-3
suppresses bladder cancer proliferation through miR-182-5p/p27
axis. Mol Cancer. 17:1442018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gou Q, Wu K, Zhou JK, Xie Y, Liu L and
Peng Y: Profiling and bioinformatic analysis of circular RNA
expression regulated by c-Myc. Oncotarget. 8:71587–71596. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhong Z, Lv M and Chen J: Screening
differential circular RNA expression profiles reveals the
regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in
bladder carcinoma. Sci Rep. 6:309192016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu Z, Xu T, Wang L, Wang X, Zhong S, Xu C
and Shen Z: MicroRNA-145 directly targets the insulin-like growth
factor receptor I in human bladder cancer cells. FEBS Lett.
588:3180–3185. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang K, Long B, Liu F, Wang JX, Liu CY,
Zhao B, Zhou LY, Sun T, Wang M, Yu T, et al: A circular RNA
protects the heart from pathological hypertrophy and heart failure
by targeting miR-223. Eur Heart J. 37:2602–2611. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jin P, Huang Y, Zhu P, Zou Y, Shao T and
Wang O: CircRNA circHIPK3 serves as a prognostic marker to promote
glioma progression by regulating miR-654/IGF2BP3 signaling. Biochem
Biophys Res Commun. 503:1570–1574. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li Y, Wan B, Liu L, Zhou L and Zeng Q:
Circular RNA circMTO1 suppresses bladder cancer metastasis by
sponging miR-221 and inhibiting epithelial-to-mesenchymal
transition. Biochem Biophys Res Commun. 508:991–996. 2019.
View Article : Google Scholar : PubMed/NCBI
|