Introduction
Lung cancer is a leading cause of cancer-associated
mortality worldwide, with 85–90% of cases classified as non-small
cell lung cancer (NSCLC) (1). As one
type of NSCLC, the lung squamous cell carcinoma (LSCC) subtype
accounts for 25–30% of all lung cancer cases (2). Despite advances in the targeted
treatment of patients with NSCLC, patients with LSCC do not often
benefit from them. For example, the epidermal growth factor
receptor (EGFR) has been reported as a treatment target for NSCLC
(3); however, LSCC rarely responds
to EGFR kinase inhibitors (4). In
pure LSCC, EGFR mutations do not occur; however, they do appear in
mixed adenosquamous carcinoma (5).
Therefore, further studies of the genetic etiology of LSCC are
required.
Mutations in phosphatidylethanolamine-binding
protein 4 (PEBP4) have frequently been reported in numerous types
of cancer (6), and PEBP4 has been
suggested as an important treatment target for ovarian (7), prostate (6) and rectal tumors (8). Our previous study observed a possible
role for PEBP4 in NSCLC progression through the PI3K/Akt/mTOR
signaling pathway (9). However, to
the best of our knowledge, no studies have reported a direct
association between PEBP4 and LSCC. To address this issue, the
present study conducted a systematic review and meta-analysis to
examine the gene expression changes of PEBP4 in LSCC. The results
were subsequently integrated with a literature-based pathway
analysis to examine possible functional pathways through which
PEBP4 may exert effects on LSCC. The aim of this study was to gain
comprehensive knowledge of the variations in the gene expression
levels of PEBP4 in LSCC, and to understand the influence of its
expression variance on LSCC using functional pathway analysis.
Materials and methods
Data selection
A systematic search was conducted on expression
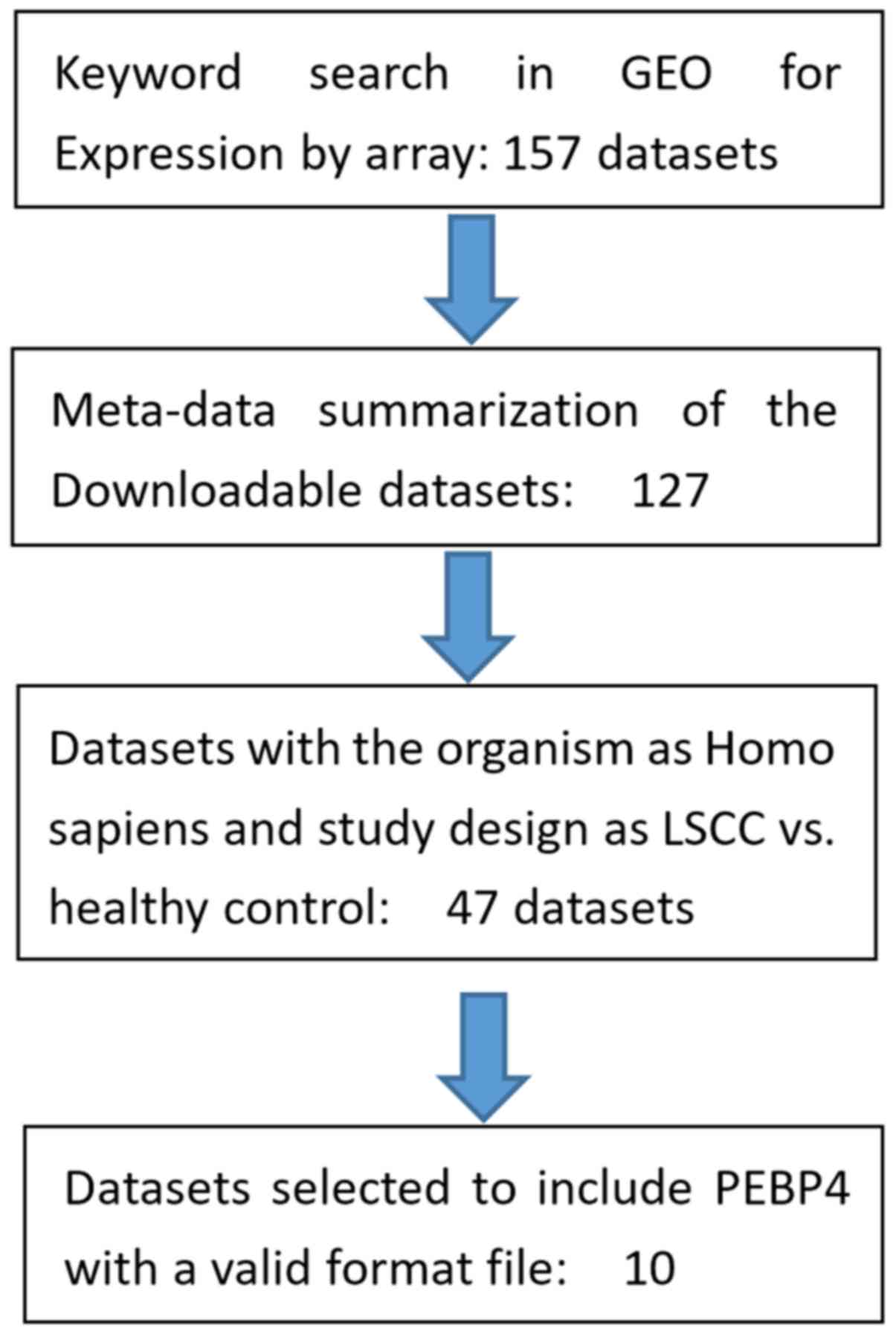
datasets from the Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo). Fig. 1 shows the workflow for expression
data selection for the meta-analysis. In total, 157 studies were
identified based on a keyword search using ‘lung squamous cell
carcinoma’. A total of 10 out of these 157 studies satisfied the
selection criteria of this study and were included in the
meta-analysis, as presented in Table
I (10–19). The selection criteria were as
follows: i) The data organism was Homo sapiens; ii) the data
type was RNA expression detected by array; iii) the study design
was limited to LSCC vs. healthy cases; and iv) the data included
gene expression of PEBP4. For the 10 studies included, there were
574 samples in total, comprising 255 LSCC cases and 319 controls.
Despite no date limitation in the systematic review, all data
collected were between 1 and 10 years old (2008–2017), as
determined using the following formula: Current year-collection
date + 1.
 | Table I.The 10 studies used in the present
meta-analysis. |
Table I.
The 10 studies used in the present
meta-analysis.
| Study name | Dataset GEO ID | Control (n) | Case (n) | Country | Data type | (Refs.) |
|---|
| Nazarov et
al | GSE84784 | 9 | 9 | Luxembourg | Expression by
array | (10) |
| Tong et
al | GSE67061 | 8 | 69 | China | Expression by
array | – |
| Mascaux et
al | GSE33479 | 27 | 14 | USA | Expression by
array | – |
| Rousseaux et
al | GSE30219 | 14 | 61 | France | Expression by
array | (11) |
| Girard et
al | GSE32036 | 59 | 12 | USA | Expression by
array | (12,13) |
| Kuner et
al | GSE27489 | 10 | 10 | Germany | Expression by
array | (14) |
| Philipsen et
al | GSE19188 | 65 | 27 | Netherlands | Expression by
array | (15) |
| Ishikawa et
al | GSE2088 | 30 | 48 | Japan | Expression by
array | (16) |
| Takahashi et
al | GSE11969 | 5 | 35 | Japan | Expression by
array | (17,18) |
| Boelens et
al | GSE12428 | 28 | 34 | Netherlands | Expression by
array | (19) |
The selection of the data covered all LSCC
expression array datasets from the GEO, which is owned by the
National Institute of Health. The datasets are publicly available,
and no permission or confirmation is required for their use. In
addition, the dataset extraction had no selection bias in terms of
publication journals, owner affiliations and authors. All authors
agreed on the data selection criteria to avoid any subjective bias.
In addition, the original data were used, rather than the processed
results of each dataset, to perform the analysis, in order to avoid
any potential noise caused by individual data processing.
Meta-analysis models
The fixed-effect and random-effects models (20) were employed to study the effect size
of PEBP4 in LSCC. For each expression dataset, the log fold-change
(LFC) was calculated for the LSCC samples and used as the index of
effect size in the meta-analysis. The expression data were
normalized and log2-transformed, if not ready done so in the
original dataset. Results from both models were reported and
compared. The heterogeneity of the meta-analysis was analyzed to
study the variance within and between the different studies, where
between-study variance [Tau-squared (τ2)] was
calculated. All analysis was conducted by an individually-developed
MATLAB (version R2017a; http://www.mathworks.com/products/matlab.html)
meta-analysis package. The additional detailed results of the
meta-analysis are available online (http://gousinfo.com/database/Data_Genetic/LSCC_PEBP4.xlsx).
Multiple linear regression (MLR)
analysis
MLR analysis was employed to study the possible
influence of three factors on the gene expression alterations in
LSCC: Sample size, population region and study date. P-values and
95% confidence interval (CI) were reported for each of the factors.
The analysis was done in MATLAB (R 2017a) using the ‘regress’
statistical analysis package.
Pathway analysis
Literature-based functional pathway analysis was
conducted using Pathway Studio (version 12.1.0.9; www.pathwaystudio.com) to study the potential
functional pathways associated with PEBP4 in LSCC. The results were
presented as a network graph with the corresponding supporting
association list of references.
Results
Meta-analysis results
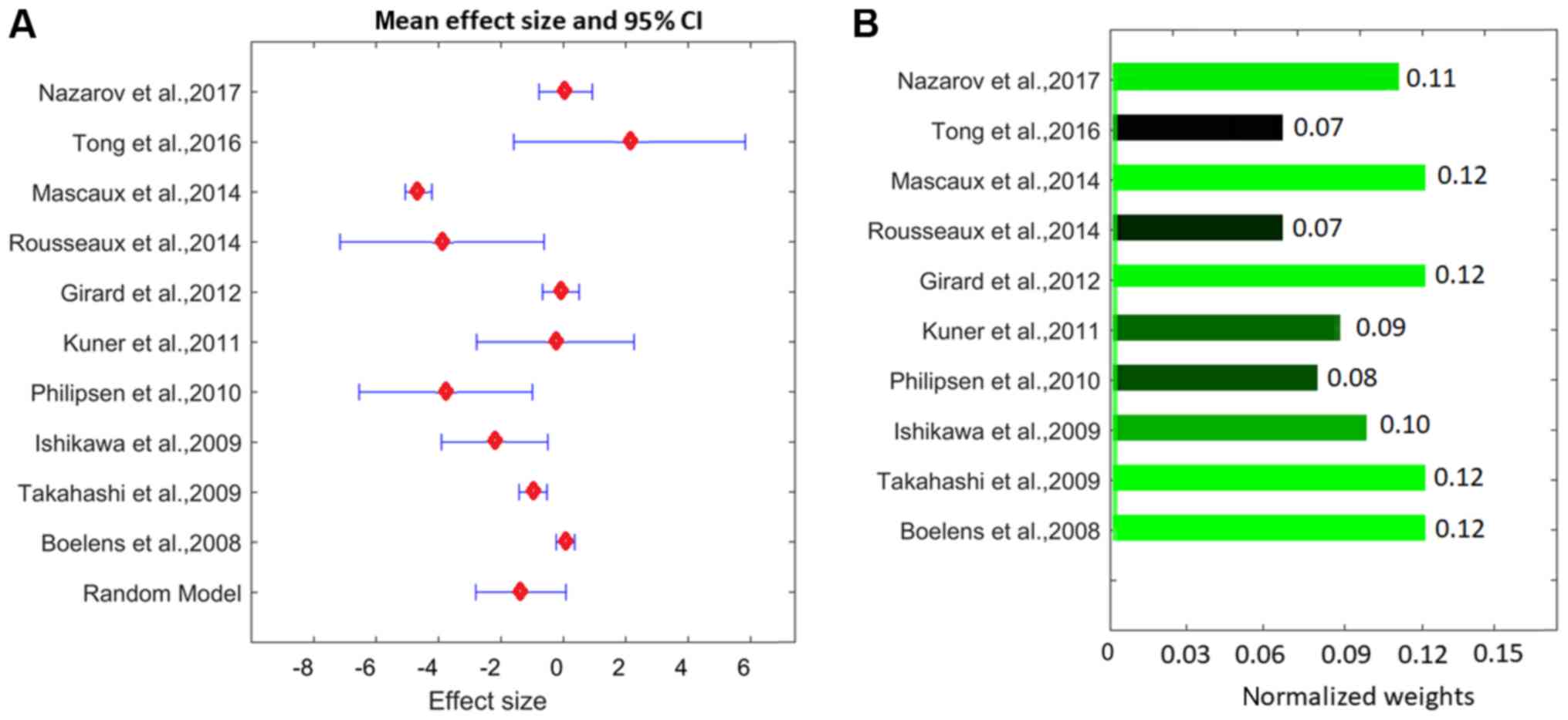
The effect sizes and associated statistics from the
ten studies, and the meta-analysis results for the PEBP4 gene are
presented in Fig. 2 and Table II. The results indicated that the
weights from the random-effects model and the fixed-effect model
were different for the PEBP4 gene (Table II), which suggested that
between-study variance existed and the random-model should be used
for the analysis.
 | Table II.The effects of the two models for
phosphatidylethanolamine binding protein 4. |
Table II.
The effects of the two models for
phosphatidylethanolamine binding protein 4.
| Study name | Effect size | Lower limit of 95%
CI | Upper limit of 95%
CI | Z-value | P-value | Weight of
fixed-effect model | Weight of
random-effects model | (Refs.) |
|---|
| Nazarov et
al, 2017 | 0.08 | −0.77 | 0.93 | 0.18 | 0.43 | 5.26 | 0.21 | (10) |
| Tong et al,
2016 | 2.13 | −1.58 | 5.84 | 1.12 | 0.13 | 0.28 | 0.12 | – |
| Mascaux et
al, 2014 | −4.63 | −5.06 | −4.20 | −21.00 | <0.001 | 21.10 | 0.22 | – |
| Rousseaux et
al, 2014 | −3.88 | −7.15 | −0.60 | −2.30 | 0.01 | 0.36 | 0.14 | (11) |
| Girard et
al, 2012 | −0.07 | −0.66 | 0.51 | −0.30 | 0.40 | 11.20 | 0.22 | (12,13) |
| Kuner et al,
2011 | −0.25 | −2.78 | 2.27 | −0.20 | 0.42 | 0.60 | 0.16 | (14) |
| Philipsen et
al, 2010 | −3.76 | −6.54 | −1.00 | −2.70 | <0.001 | 0.50 | 0.15 | (15) |
| Ishikawa et
al, 2009 | −2.20 | −3.90 | −0.50 | −2.50 | 0.01 | 1.32 | 0.19 | (16) |
| Takahashi et
al, 2009 | −0.97 | −1.41 | −0.50 | −4.20 | <0.001 | 19.20 | 0.22 | (17,18) |
| Boelens et
al, 2008 | 0.072 | −0.22 | 0.37 | 0.48 | 0.32 | 43.90 | 0.22 | (19) |
| Fixed model | −1.15 | −1.34 | −1.00 | −12.00 | <0.001 | – | – | – |
| Random-effects
model | −1.36 | −2.80 | 0.09 | −1.80 | 0.03 | – | – | – |
Heterogeneity analysis indicated that the
between-study variance (τ2) was calculated as 4.56,
indicating a significant between-study variance. The total variance
(Q) was 354.43, with an expected variance ‘df’ (under the
assumption that all studies have the same effect size) of 9. This
resulted in an ISq of 97.46, indicating that >97.46% of
variances were due to between-study variance; and
P<1×10−320 for the hypothesis that Q was from
within-study variances only. These results suggested a significant
between-study variance of the effect size (LFC). Therefore, a
random-effects model was indicated to be more appropriate for this
study, which estimates the mean of effect sizes from different
studies. The following discussion focused on the results from the
random-effects model only.
The LFC from the meta-analysis was −1.80 [95% CI:
(−2.80, 0.09); P=0.03; Fig 2]. These
results suggested that, on average, PEBP4 presented significantly
decreased expression levels in cases of LSCC in the ten studies
involved. However, there were significant between-study variances
(P<1×10−324; see LSCC_PEBP4→ Ref for pathway
analysis), with three studies exhibiting increased expression
levels, including LFC=2.13, 0.080 and 0.072, calculated from
datasets GSE67061 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE67061),
GSE84784 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE84784)
and GSE12428 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE12428),
respectively; this variability in results requires an analysis to
study the influence of potential factors affecting the results. For
more results, please refer to LSCC_PEBP4→ sumResults).
MLR analysis
Results from the MLR models indicated that
population region was a significant influencing factor for the
expression fold-change of PEBP4 (P=0.0064), as presented in
Table III. Conversely, the sample
size and study date indicated no significant influence
(P>0.35).
 | Table III.Multiple linear regression analysis
results. |
Table III.
Multiple linear regression analysis
results.
| Parameter | Sample size | Population
region | Study date |
|---|
| Beta | 0.0020 | 0.88 | 0.066 |
| LowLimit | −0.060 | 0.020 | −0.44 |
| UpLimit | 0.063 | 1.74 | 0.57 |
| P-value | 0.46 | 0.01 | 0.35 |
Pathway analysis results
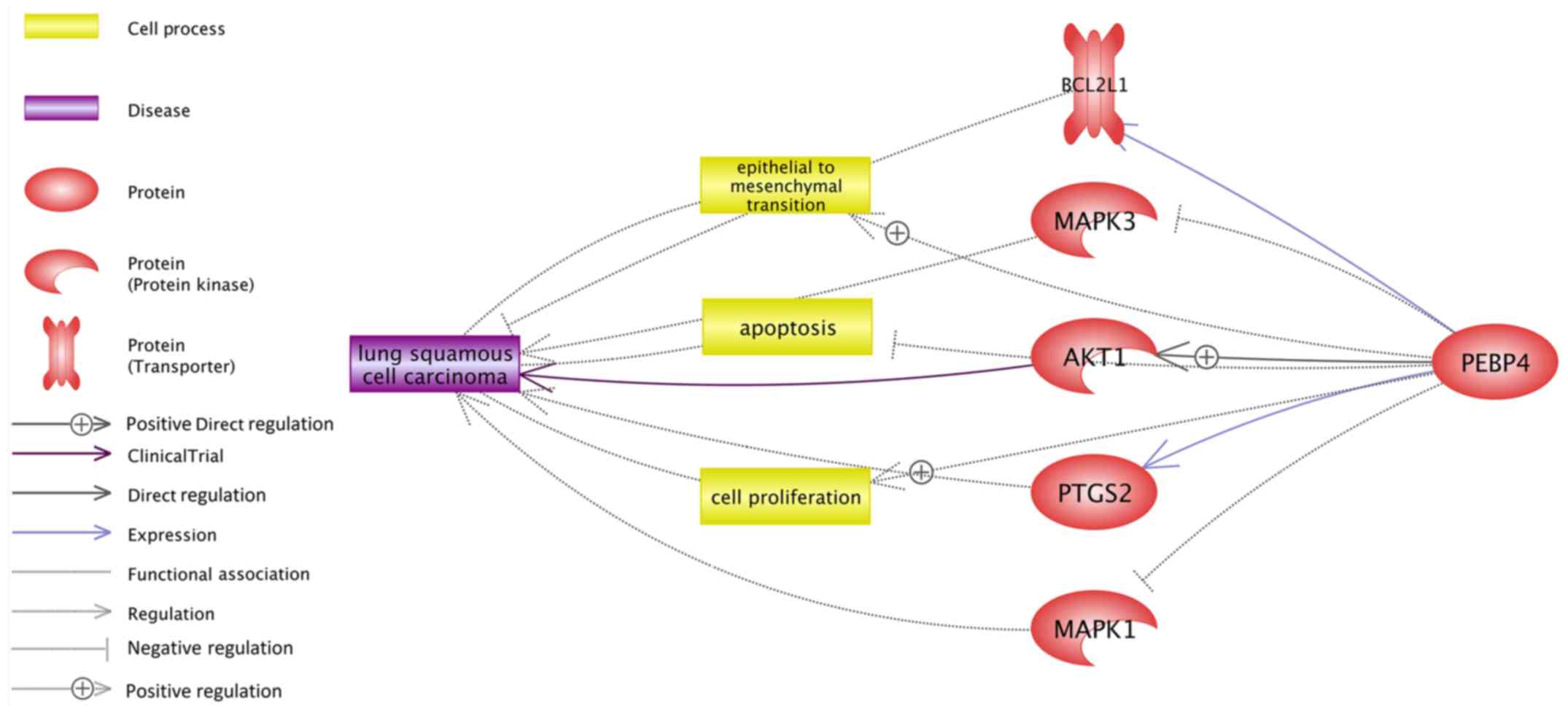
Pathway analysis using Pathway Studio (www.pathwaystudio.com) was conducted to identify
possible pathways through which PEBP4 may exert an effect on LSCC.
As shown in Fig. 3, there were
various pathways that linked PEBP4 with LSCC through different
entities, including proteins/genes, cell processes and functional
class. Five potential genetic pathways were identified, as
indicated in Fig. 3. Among these
five pathways, three suggested an LSCC-promoting effect of PEBP4,
including PEBP4→BCL2L1→LSCC, PEBP4→AKT1→LSCC and
PEBP4→prostaglandin-endoperoxide synthase 2 (PTGS2)→LSCC. In
addition, two genetic pathways indicated a treatment-resistant
effect of PEBP4 for LSCC, including PEBP4-->MAPK3→LSCC and
PEBP4→MAPK1→LSCC. Three cellular processing pathways were also
identified, which were associated with cell proliferation,
apoptosis and epithelial-mesenchymal transition (Fig. 3). Each relationship (edge) in
Fig. 3 had ≥1 supporting reference,
assisting in understanding the potential mechanism underlying the
effects of PEBP4 on the pathogenesis of LSCC. For example, it has
been reported that increased expression levels of PEBP4 can inhibit
the activity of MAPK3 (7), which is
already recognized as a therapeutic target for LSCC (6); notably, a PEBP4→MAPK3→LSCC pathway was
present in Fig. 3. These findings
suggested an anti-drug-effect pathway of PEBP4 in the treatment of
LSCC. In total, there were eight pathways composed of 16
relationships (edges), and these relationships were supported by
163 references. The full list of these relations and the
corresponding supporting references are presented in LSCC_PEBP4→
Ref for pathway analysis, which is available online (http://gousinfo.com/database/Data_Genetic/LSCC_PEBP4.xlsx).
Discussion
During the past few years, PEBP4 has been identified
as a contributor to the development of numerous types of cancer
(6–9,20,21). Our
previous study indicated that overexpression of PEBP4 enhances
NSCLC cell proliferation and the invasive ability of cancer cells,
while inhibiting apoptosis (9).
Another study indicated that PEBP4 promotes the
epithelial-to-mesenchymal transition by activating the sonic
hedgehog signaling pathway in NSCLC (22).
To the best of our knowledge, no study has reported
a direct association between LSCC and PEBP4. In the present study,
a meta-analysis and literature-based functional pathway analysis
was conducted, to examine the possible influence of PEBP4 on LSCC
and the potential underlying mechanisms.
The present meta-analysis used gene expression
datasets available in the GEO (www.ncbi.nlm.nih.gov/geo), which passed data selection
criteria. A total of 10 human gene expression datasets were
included with a design of LSCC vs. control. These datasets had a
publication-age gap of ≤10 years (2008–2017), and were from
different regions, including France, USA, Netherlands, Japan,
Germany, Luxembourg and China.
Meta-analysis indicated that there was a decreased
activity of PEBP4 in terms of LFC, with a significant between-study
variance (LSCC_PEBP4→ Reference Table). One possible reason may lie
in the data generation of different studies, as they used different
platforms, including GPL570 for GSE30219 and GPL6884 for GSE32036,
and sample sources, including lung samples in GSE67061 and
bronchial biopsy samples in GSE33479 (LSCC_PEBP4→ Data). Other
explanations could be the existence of multiple influential factors
that may lead to increased or decreased expression of PEBP4 under
different circumstances. MLR analysis confirmed this and indicated
that the sample population region (country) was a significant
influential factor. MLR results also indicated that the sample size
and study ages had no significant influence on the expression
variance of PEBP4 in the patients in the 10 LSCC groups included in
this study; however, the sample sizes varied from 9 to 69, and the
publication years were between 2008 and 2017. This suggested that,
under similar circumstances (e.g., same population region), the
expression of PEBP4 may not be significantly changed in LSCC cases
from the past 10 years.
Despite between-study differences in terms of PEBP4
expression levels, the present meta-analysis integrated information
from independent but related studies by using a random-effects
model, thus assisting in improving the reliability of the results.
The meta-analysis results indicated that increased and decreased
activity of PEBP4 may occur in LSCC. Therefore, it is necessary to
examine the possible consequences in the case of varied PEBP4
activity. To address this issue, a pathway analysis was conducted
using the shortest path functionality of Pathway Studio (www.pathwaystudio.com). This function identifies
directed pathways through which PEBP4 may exert functional
influence on LSCC. Each relationship (edge) had one or more
supporting references (LSCC_PEBP4→ Ref for pathway analysis).
Multiple potential pathways were identified, through
which PEBP4 may promote the pathological development of LSCC.
Notably, overexpression of PEBP4 promotes the activity of PTGS2,
AKT1 and BCL2L1 (23,24), and these three genes serve an
important role in the development of LSCC (25,26).
These pathways indicated that the overexpression of PEBP4 may
promote the development of LSCC. In addition, PEBP4 inhibits MAPK3
and MAPK1 (23), whereas the
activation of these two genes has been suggested as therapeutic
targets for the treatment of LSCC (20).
At a cellular processing level, three potential
LSCC-promoting pathways were identified. Firstly, the prognosis of
LSCC is associated with LSCC cancer cell proliferation, apoptosis
and epithelial-to-mesenchymal transition (27). Secondly, it has been reported that
PEBP4 may enhance NSCLC cancer cell proliferation (28), inhibit apoptosis of numerous cancer
cells (8), and promote the
epithelial-to-mesenchymal transition in multiple cancer cells
(6,28). These pathways may assist in
understanding the potential associations between PEBP4 and
LSCC.
There are numerous limitations of this study that
require further investigation. Firstly, due to limited meta-data,
only three potential factors were studied. Considering the
significant between-study variance of PEBP4 expression levels in
LSCC, more influential factors are expected and should be examined,
including age, sex and the presence of co-morbidities. Secondly,
the functional pathways analysis was literature-based. Although
supported by previous studies, specific experiments should be
conducted to test these pathways. Finally, in the meta-analysis,
only expression by array datasets from the GEO were used. In the
GEO database, besides expression by array, there are multiple other
types of expression data, including expression profiling by single
nucleotide polymorphism array and by high throughput sequencing.
However, to avoid noise brought by the use of different data types,
this study only used expression profiling by array data.
Meta-analysis using other types of data could be conducted in the
future to confirm the results of this study.
In the present study, PEBP4 demonstrated overall
decreased expression levels in the meta-analysis, which was in
accordance with most of the studies (7 out of 10) employed.
Integrating results from the pathway analysis, this study indicated
that in the majority of LSCC cases, the influence of PEBP4 on LSCC
can be neglected. However, the expression levels of PEBP4
demonstrated strong heterogeneity, due to numerous influential
factors, including population and region, with possible increased
PEBP4 activity occurring in patients with LSCC. In such cases, the
potential influence of PEBP4 on LSCC should be considered as it may
promote the development of LSCC and lead to drug resistance.
Acknowledgements
Not applicable.
Funding
This study was partly supported by the 2017 Fifth
Provincial ‘333 Project’ Research Project; by non-small cell lung
cancer research funding, Jiangsu Provincial Planning Commission
(grant no. H201657); by clinical diagnosis and treatment of small
lung lesions and normative research, Wuxi City Health Planning
Commission (grant no. MS201625); and by Thirteen Five Science and
Education in Jiangsu Province.
Availability of data and materials
All data analyzed during this study is included in
this published article. The results of the meta-analysis are
available online at http://gousinfo.com/database/Data_Genetic/LSCC_PEBP4.xlsx.
The GEO datasets are stated in LSCC_PEBP4→GEO datasets, including
the study name, GEO ID and URL for the datasets.
Authors' contributions
GY and NZ contributed to the data collection,
analysis and manuscript writing. BH and YM contributed to the data
collection and analysis. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
World Health Organization International
Agency for Research on Cancer. GLOBOCAN 2012, . Estimated Cancer
Incidence, Mortality and Prevalence Worldwide in 2012. http://globocan.iarc.fr/Default.aspxJan
27–2015
|
|
2
|
Heist RS, Mino-Kenudson M, Sequist LV,
Tammireddy S, Morrissey L, Christiani DC, Engelman JA and Iafrate
AJ: FGFR1 amplification in squamous cell carcinoma of the lung. J
Thorac Oncol. 7:1775–1780. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pirker R: What is the best strategy for
targeting EGF receptors in non-small-cell lung cancer? Future
Oncol. 11:153–167. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
D'Angelo SP, Pietanza MC, Johnson ML,
Riely GJ, Miller VA, Sima CS, Zakowski MF, Rusch VW, Ladanyi M and
Kris MG: Incidence of EGFR exon 19 deletions and L858R in tumor
specimens from men and cigarette smokers with lung adenocarcinomas.
J Clin Oncol. 29:2066–2070. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rekhtman N, Paik PK, Arcila ME, Tafe LJ,
Oxnard GR, Moreira AL, Travis WD, Zakowski MF, Kris MG and Ladanyi
M: Clarifying the spectrum of driver oncogene mutations in
biomarker-verified squamous carcinoma of lung: Lack of EGFR/KRAS
and presence of PIK3CA/AKT1 mutations. Clin Cancer Res.
18:1167–1176. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li W, Dong Y, Zhang B, Kang Y, Yang X and
Wang H: PEBP4 silencing inhibits hypoxia-induced
epithelial-to-mesenchymal transition in prostate cancer cells.
Biomed Pharmacother. 81:1–6. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li P, Wang X, Li N, Kong H, Guo Z, Liu S
and Cao X: Anti-apoptotic hPEBP4 silencing promotes TRAIL-induced
apoptosis of human ovarian cancer cells by activating ERK and JNK
pathways. Int J Mol Med. 18:505–510. 2006.PubMed/NCBI
|
|
8
|
Qiu J, Yang G, Lin A, Shen Z, Wang D and
Ding L: Human phosphatidylethanolamine-binding protein 4 promoted
the radioresistance of human rectal cancer by activating Akt in an
ROS-dependent way. PLoS One. 9:e900622014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yu G, Huang B, Chen G and Mi Y:
Phosphatidylethanolamine-binding protein 4 promotes lung cancer
cells proliferation and invasion via PI3K/Akt/mTOR axis. J Thorac
Dis. 7:1806–1816. 2015.PubMed/NCBI
|
|
10
|
Nazarov PV, Muller A, Kaoma T, Nicot N,
Maximo C, Birembaut P, Tran NL, Dittmar G and Vallar L: RNA
sequencing and transcriptome arrays analyses show opposing results
for alternative splicing in patient derived samples. BMC Genomics.
18:4432017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rousseaux S, Debernardi A, Jacquiau B,
Vitte AL, Vesin A, Nagy-Mignotte H, Moro-Sibilot D, Brichon PY,
Lantuejoul S, Hainaut P, et al: Ectopic activation of germline and
placental genes identifies aggressive metastasis-prone lung
cancers. Sci Transl Med. 5:186ra662013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Byers LA, Diao L, Wang J, Saintigny P,
Girard L, Peyton M, Shen L, Fan Y, Giri U, Tumula PK, et al: An
epithelial-mesenchymal transition gene signature predicts
resistance to EGFR and PI3K inhibitors and identifies Axl as a
therapeutic target for overcoming EGFR inhibitor resistance. Clin
Cancer Res. 19:279–290. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schuster K, Venkateswaran N, Rabellino A,
Girard L, Peña-Llopis S and Scaglioni PP: Nullifying the CDKN2AB
locus promotes mutant K-ras lung tumorigenesis. Mol Cancer Res.
12:912–923. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kahn N, Meister M, Eberhardt R, Muley T,
Schnabel PA, Bender C, Johannes M, Keitel D, Sültmann H, Herth FJ
and Kuner R: Early detection of lung cancer by molecular markers in
endobronchial epithelial-lining fluid. J Thorac Oncol. 7:1001–1008.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hou J, Aerts J, den Hamer B, van Ijcken W,
den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens
JA, Hoogsteden HC, et al: Gene expression-based classification of
non-small cell lung carcinomas and survival prediction. PLoS One.
5:e103122010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fujiwara T, Hiramatsu M, Isagawa T,
Ninomiya H, Inamura K, Ishikawa S, Ushijima M, Matsuura M, Jones
MH, Shimane M, et al: ASCL1-coexpression profiling but not single
gene expression profiling defines lung adenocarcinomas of
neuroendocrine nature with poor prognosis. Lung Cancer. 75:119–125.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Takeuchi T, Tomida S, Yatabe Y, Kosaka T,
Osada H, Yanagisawa K, Mitsudomi T and Takahashi T: Expression
profile-defined classification of lung adenocarcinoma shows close
relationship with underlying major genetic changes and
clinicopathologic behaviors. J Clin Oncol. 24:1679–988. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Matsuyama Y, Suzuki M, Arima C, Huang QM,
Tomida S, Takeuchi T, Sugiyama R, Itoh Y, Yatabe Y, Goto H and
Takahashi T: Proteasomal non-catalytic subunit PSMD2 as a potential
therapeutic target in association with various clinicopathologic
features in lung adenocarcinomas. Mol Carcinog. 50:301–309. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Boelens MC, van den Berg A, Fehrmann RS,
Geerlings M, de Jong WK, te Meerman GJ, Sietsma H, Timens W, Postma
DS and Groen HJ: Current smoking-specific gene expression signature
in normal bronchial epithelium is enhanced in squamous cell lung
cancer. J Pathol. 218:182–191. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Borenstein M, Hedges LV, Higgins JP and
Rothstein HR: A basic introduction to fixed-effect and
random-effects models for meta-analysis. Res Synth Methods.
1:97–111. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li F, Fang Z, Zhang J, Li C, Liu H, Xia J,
Zhu H, Guo C, Qin Z, Li F, et al: Identification of TRA2B-DNAH5
fusion as a novel oncogenic driver in human lung squamous cell
carcinoma. Cell Res. 26:1149–1164. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jian W, Bai Y, Li X, Kang J, Lei Y and Xue
Y: Phosphatidylethanolamine-binding protein 4 promotes the
epithelial-to-mesenchymal transition in non-small cell lung cancer
cells by activating the sonic hedgehog signaling pathway. J Cell
Biochem. 120:5386–5395. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
An LP, Maeda T, Sakaue T, Takeuchi K,
Yamane T, Du PG, Ohkubo I and Ogita H: Purification, molecular
cloning and functional characterization of swine
phosphatidylethanolamine-binding protein 4 from seminal plasma.
Biochem Biophys Res Commun. 423:690–696. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li H, Wang X, Li N, Qiu J, Zhang Y and Cao
X: hPEBP4 resists TRAIL-induced apoptosis of human prostate cancer
cells by activating Akt and deactivating ERK1/2 pathways. J Biol
Chem. 282:4943–4950. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li H, Chen F, Yu L, Liu M, Chen H, Zhang Y
and Liu X: Expression of inflammation related enzymes during
experimental rat lung carcinogenesis. Zhonghua Zhong Liu Za Zhi.
24:316–319. 2002.PubMed/NCBI
|
|
26
|
Weeden CE, Ah-Cann C, Holik AZ, Pasquet J,
Garnier JM, Merino D, Lessene G and Asselin-Labat ML: Dual
inhibition of BCL-XL and MCL-1 is required to induce tumour
regression in lung squamous cell carcinomas sensitive to FGFR
inhibition. Oncogene. 37:4475–4488. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Aruga N, Kijima H, Masuda R, Onozawa H,
Yoshizawa T, Tanaka M, Inokuchi S and Iwazaki M:
Epithelial-mesenchymal transition (EMT) is correlated with
patient's prognosis of lung squamous cell carcinoma. Tokai J Exp
Clin Med. 43:5–13. 2018.PubMed/NCBI
|
|
28
|
Yu G, Shen Z, Chen G, Teng X, Hu Y and
Huang B: PEBP4 enhanced HCC827 cell proliferation and invasion
ability and inhibited apoptosis. Tumour Biol. 34:91–98. 2013.
View Article : Google Scholar : PubMed/NCBI
|