Introduction
Breast cancer (BC) is the one of the most common
malignancies in women worldwide, and the leading cause of
cancer-associated mortality (1). In
2018, ~40,000 people died of breast cancer (2). Long-term survival rates decrease from
90 to 5% following the development of distant metastasis or
recurrence, thus leading to poor prognosis in patients with BC
(3). Tamoxifen monotherapy poses as
an anti-estrogen agent in the mammary tissue and is considered
effective in decreasing the recurrence rate and improving the
overall survival (OS) of patients with BC (4). For patients with estrogen receptor
(ER)-positive BC, the response rate of tamoxifen monotherapy
decreased by 50–70 and 5–10% for patients with ER-negative BC.
Thus, the development of novel prognostic biomarkers is critical in
order to accurately assess the outcome and provide individualized
treatment strategies for patients with ER-negative BC.
Long non-coding RNAs (lncRNAs) are defined as RNA
transcripts >200 base pairs in length (5). Accumulating evidence demonstrates
lncRNA dysregulation have in multiple human diseases, particularly
different types of cancer (6–8), and
their expression is associated with cancer development and
metastasis (9). For example, lncRNA
Inhibiting Metastasis is upregulated in BC and is associated the
inhibition of cell proliferation and metastasis (10). Furthermore, lncRNA HOXA transcript
induced by TGFβ plays important roles in regulating the
epithelial-to-mesenchymal transition (EMT), invasion and metastasis
of BC (11). Several lncRNA
signatures have been developed for specific BC subtypes, based on
the aberrant expression of these lncRNAs in BC (12). The results of these studies have
demonstrated the potential role of lncRNAs as biomarkers for the
clinical outcome of ER-negative BC (13,14).
A number of prognosis-associated lncRNAs have been
identified as effective biomarkers via systematic, and subsequently
used as lncRNA signatures for the prognosis of BC (15). For example, Wang et al
(16) developed an 11-lncRNA
signature using Cox regression analysis. Furthermore, Sun et
al (17) developed an
eight-lncRNA signature based on a weighted co-expressed network and
competing endogenous RNA. Li et al (15) developed a five-lncRNA signature using
the microarray re-annotation method. The aforementioned studies
predominantly focused on all of the BC subtypes or patients with
triple negative BC; however, very few focused on the identification
of prognostic biomarkers for patients with ER-negative BC.
In order to develop an lncRNA signature for
prognosis, the present study obtained samples from patients with
ER-negative BC from multiple datasets archived in the Gene
Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). The nine-lncRNA
signature was developed using the Cox regression model, and
validated in independent cohorts using the lncRNA expression
profiles obtained using the microarray re-annotation method.
Functional analyses demonstrated that these signature lncRNAs play
important roles in tumor development and metastasis, which provides
further evidence for the predictive ability of the signature.
Materials and methods
Data acquisition and
pre-processing
A total of three datasets, GSE21653 (18,19),
GSE58812 (20) and GSE19615
(21) were retrieved from the GEO
database and measured using the Affymetrix Human Genome U133 Plus
2.0 Array platform (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL570).
The GSE21653 dataset contained 266 BC samples, among which 110 were
ER-negative BC samples and used as the discovery cohort. The
GSE58812 and GSE19615 datasets contained 107 and 45 ER-negative BC
samples, respectively, which were used as the validation cohorts.
In the present study, the gene expression profiles (CEL. files) for
each dataset, and the corresponding clinical information, were
downloaded. The robust multichip average algorithm was used for
background adjustment of the gene expression profiles. Each
probe-set ID was mapped to its Entrez gene ID using the
corresponding platform annotation files. In the case of multiple
probes being mapped to the same gene, the expression value for the
gene was summarized as the arithmetic mean value of the multiple
probes. In the instance that a probe was mapped to multiple or no
genes, this probe was deleted.
lncRNA probe re-annotation
Using the BLASTn tools (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&
PAGE_TYPE=BlastSearch&LINK_LOC=blasthome), the probe
annotation sequences supported by Affymetrix were aligned to the
human long non-coding transcript sequences and protein-coding
transcript sequences from the GENCODE database (Release 30;
http://www.gencodegenes.org/human/),
respectively. The results of the sequence alignment were filtered
as follows: Only the probes that matched to one long non-coding
transcript were reserved, and each transcript should identically
match with more than six probes. Gene expression profiles
containing 15,942 mRNAs and 1,631 lncRNAs were obtained using the
re-annotation method.
Survival analysis
The univariate Cox proportional hazards regression
model was used to identify lncRNAs significantly associated with
disease-free survival (DFS) of patients in the discovery cohort
(22), with P<0.05 as the
threshold. Subsequently, the multivariate Cox proportional hazards
regression model was used to obtain robust prognosis-associated
lncRNAs, which were used to develop the risk-score model according
to the following equation:
RiskScore=∑k=1nExpk*eHRk
Where n is the number of prognostic lncRNAs,
Expk is the expression value of the prognostic
lncRNAs and eHRk is the estimated
regression coefficient of the lncRNAs in the multivariate Cox
regression analysis.
The present study used a stepwise regression method
(23) in order to perform
multivariate regression analysis on 16 lncRNAs, from which nine
stable molecules were obtained. Stepwise regression is commonly
used to introduce variables, one by one, into the model. Following
each introduction of an explanatory variable, an F-test is
performed, and the explanatory variables that have been selected
are subjected to a t-test, respectively. When an explanatory
variable is no longer significant due to the introduction of a
subsequent explanatory variable, the former is deleted in order to
ensure that only the significant variables are included in the
regression equation prior to the introduction of each new variable.
This procedure is an iterative process until neither a significant
explanatory variable is selected into the regression equation, nor
an insignificant explanatory variable is removed from the
regression equation, in order to ensure that the resulting set of
explanatory variables is optimal. The present study used the R step
function to implement the stepwise regression process.
Consistency evaluation of DEGs
The reproducibility of the DEGs identified in the
different datasets (GSE21653, GSE58812 and GSE19615) was evaluated
using consistency analysis. The concordance score was calculated as
k/n × 100% for several DEG lists extracted separately randomly from
two of the three datasets (GSE21653, GSE58812 and GSE19615) which
shared n genes, of which k genes demonstrated the same deregulation
directions (up- or down-regulation). The score evaluates the
consistency of DEGs by randomly extracting two independent datasets
from two of the three datasets (GSE21653, GSE58812 and
GSE19615).
The probability of observing a concordance score of
k/n by chance was evaluated using the cumulative binomial
distribution model as follows:
p=1-∑i=0k-1(ni)P0i(1-P0)n-i
Where P0 (here, 0.5) is the
probability of one gene having the concordant association between
the two lists of genes by chance.
Kyoto encyclopedia of genes and
genomes (KEGG) enrichment analysis
KEGG pathway enrichment analysis was performed using
the R package clusterProfiler (24)
for DEGs and genes associated with the signature lncRNAs, and
visualized using the R package DOSE (25). For both analyses, P<0 .05 was
considered to indicate a statistically significant difference.
Statistical analysis
The median risk-score evaluated in each dataset by
the signature for ER-negative BC samples was used as the cut-off
(median score=26.1) in order to classify these samples into high-
and low-risk groups. Kaplan-Meier (KM) plots were generated in
order to assess DFS, followed by the log-rank test for the
statistical comparison of the two groups. The predictive
performance of the signature was further assessed using the pROC
package (version 1.15.3). All statistical analyses were performed
using the R software package (version 3.1.2;).
Results
Identification of prognosis-associated
lncRNAs
For the present study, the gene expression profiles
and clinical follow-up data of datasets GSE21653, GSE58812 and
GSE19615 were downloaded, resulting in a total of 262 ER-negative
BC samples. Subsequently, gene expression profiles containing
15,942 mRNAs and 1,631 lncRNAs were obtained via the probe
re-annotation method. The univariate Cox proportional hazards
regression model identified 16 prognosis-associated lncRNAs in the
GSE21653 dataset, among which five were protective factors and 11
were risk factors (P<0.05; Table
I). A number of lncRNAs have been reported to be associated
with the invasion and metastasis of various types of cancer. For
example, lncRNA Long Intergenic Non-Protein Coding RNA, P53 Induced
Transcript (LINC-PINT) has been reported to inhibit cancer cell
proliferation, invasion and migration in osteosarcoma by
downregulating micro (miR)-21 (26).
Furthermore, LINC-PINT is known to inhibit tumor cell invasion
through a highly conserved sequence element (27). The lncRNA LINC00324 has been
demonstrated to promote cell proliferation in gastric cancer by
binding with human antigen R and stabilizing FAM83B expression
(28). In addition, the lncRNA
Homeobox A11 antisense (HOXA11-AS) regulates the JAK-STAT signaling
pathway via the miR-15a-3p/STAT3 axis in order to promote liver
cancer growth and metastasis (29).
 | Table I.LncRNAs associated with risk of
post-surgery recurrence in ER-negative breast cancer. |
Table I.
LncRNAs associated with risk of
post-surgery recurrence in ER-negative breast cancer.
| lncRNA_ID | Symbol | Coefficient | HR | P-value |
|---|
| 101929340 | LOC101929340 | 1.1931 | 3.2973 |
1.61×10−04 |
| 100131067 | CKMT2-AS1 | 0.9910 | 2.6939 |
1.77×10−04 |
| 378805 | LINC-PINT | −0.7932 | 0.4524 |
1.85×10−04 |
| 284029 | LINC00324 | −1.4682 | 0.2303 |
3.23×10−04 |
| 101929504 | LINC02544 | 0.6051 | 1.8314 |
4.35×10−04 |
| 221883 | HOXA11-AS | 1.5265 | 4.6020 |
6.36×10−04 |
| 105221694 | BISPR | −0.6952 | 0.4990 |
7.27×10−04 |
| 29034 | CPS1-IT1 | 3.6059 | 36.8160 |
8.83×10−04 |
| 102724105 | ZNF528-AS1 | 1.0762 | 2.9335 |
1.25×10−03 |
| 100507463 | PSMB8-AS1 | −0.4446 | 0.6411 |
1.80×10−03 |
| 727915 | AGBL1-AS1 | 1.0646 | 2.8996 |
1.87×10−03 |
| 100131208 | SNAP25-AS1 | 1.9705 | 7.1743 |
2.33×10−03 |
| 84793 | FOXD2-AS1 | 0.9001 | 2.4599 |
4.14×10−03 |
| 100506211 | MIR210HG | 0.5215 | 1.6845 |
4.40×10−03 |
| 114614 | MIR155HG | −0.5074 | 0.6020 |
4.56×10−03 |
| 284930 | LOC284930 | 0.6582 | 1.9314 |
4.92×10−03 |
Development of the nine-lncRNA
signature
Using the multivariate Cox proportional hazards
regression model, a total of nine lncRNAs were identified from the
16 DFS-associated lncRNAs with optimal predictive performance in
the discovery cohort, which were defined as the nine-lncRNA
signature. The multivariate regression results of the nine
identified lncRNAs are presented in Table II. Subsequently, the risk-score
equations were formulated as follows: Risk-score=2.4651*exp
(CPS1-IT1) + 0.5640*exp (FOXD2-AS1) + 0.8631*exp
(HOXA11-AS)-1.4586*exp (LINC00324) + 0.8866*exp
(CKMT2-AS1)-0.6698*exp (BISPR) + 1.3965*exp (SNAP25-AS1) +
1.1144*exp (LOC101929340) + 0.4768*exp (LINC02544). The risk-score
for each ER-negative BC sample in the discovery cohort was
calculated for the nine-lncRNA signature. Receiver operating
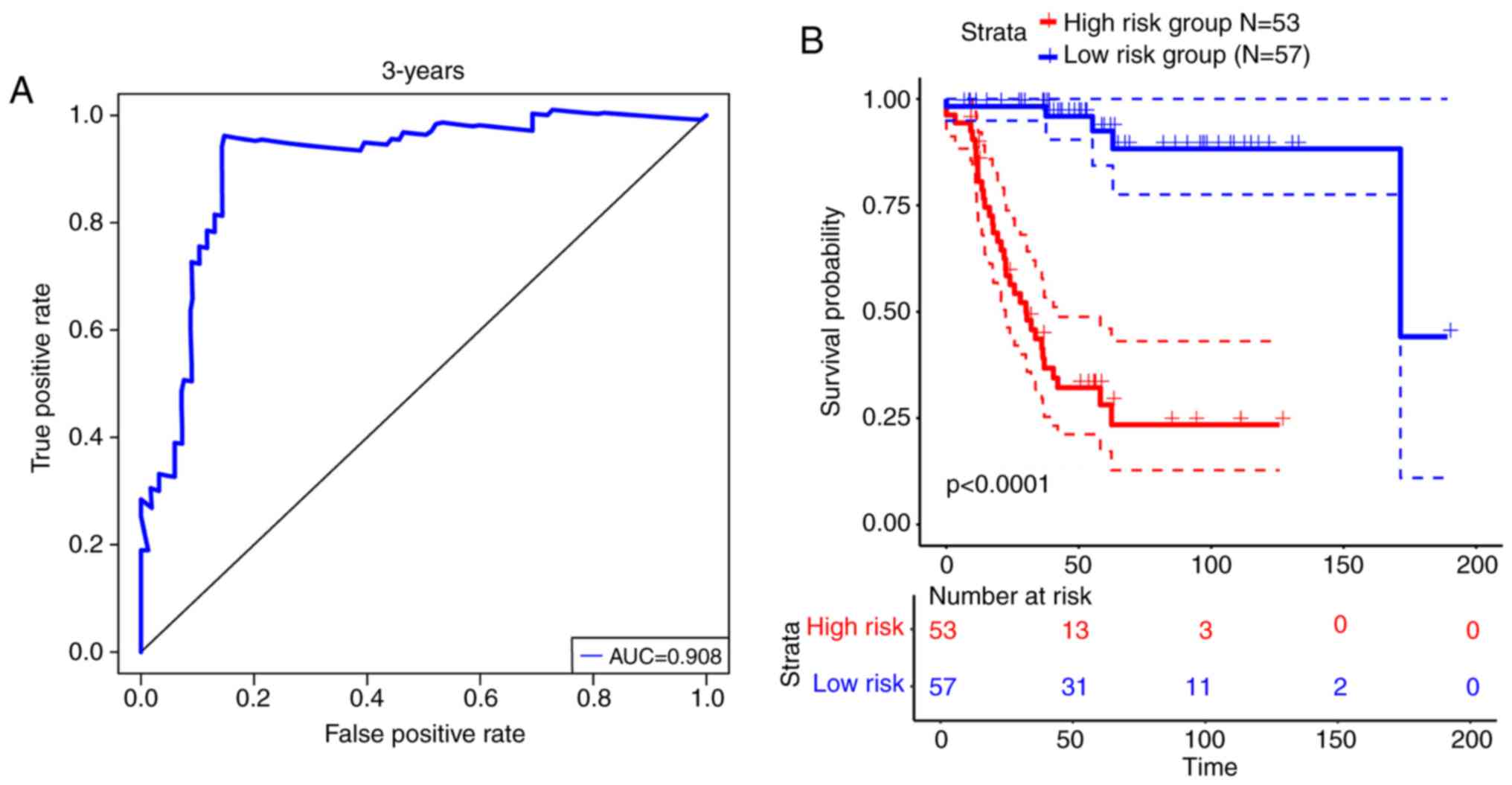
characteristic (ROC) curve analysis demonstrated that the 3-year
area under curve reached 0.908 (Fig.
1A). Samples were divided into high- and low-risk groups for
the nine-lncRNAs signature, with the median risk-scores as
threshold. The 53 patients in the high-risk group had significantly
worse DFS compared with the 57 patients in the low-risk group
[hazard ratio (HR)=2.718, 95% confidence interval (CI)=2.115–3.494,
P<0.0001; Fig. 1B].
 | Table II.Multivariate Cox regression analysis
of the nine lncRNAs. |
Table II.
Multivariate Cox regression analysis
of the nine lncRNAs.
| lncRNA | Coefficient | HR | z | P-value | Lower 0.95 | Upper 0.95 |
|---|
| CPS1-IT1 | 2.465 | 11.765 | 2.286 | 0.022 | 1.421 | 97.373 |
| FOXD2-AS1 | 0.564 | 1.758 | 1.682 | 0.093 | 0.911 | 3.391 |
| HOXA11-AS | 0.863 | 2.371 | 1.577 | 0.115 | 0.811 | 6.931 |
| LINC00324 | −1.459 | 0.233 | −2.879 | 0.004 | 0.086 | 0.628 |
| CKMT2-AS1 | 0.887 | 2.427 | 2.768 | 0.006 | 1.295 | 4.546 |
| SNAP25-AS1 | 1.397 | 4.041 | 2.080 | 0.038 | 1.084 | 15.064 |
| LOC101929340 | 1.114 | 3.048 | 2.681 | 0.007 | 1.350 | 6.882 |
| LINC02544 | 0.477 | 1.611 | 2.345 | 0.019 | 1.081 | 2.400 |
| BISPR | −0.670 | 0.512 | −2.480 | 0.013 | 0.301 | 0.869 |
Validation of the nine-lncRNA
signature
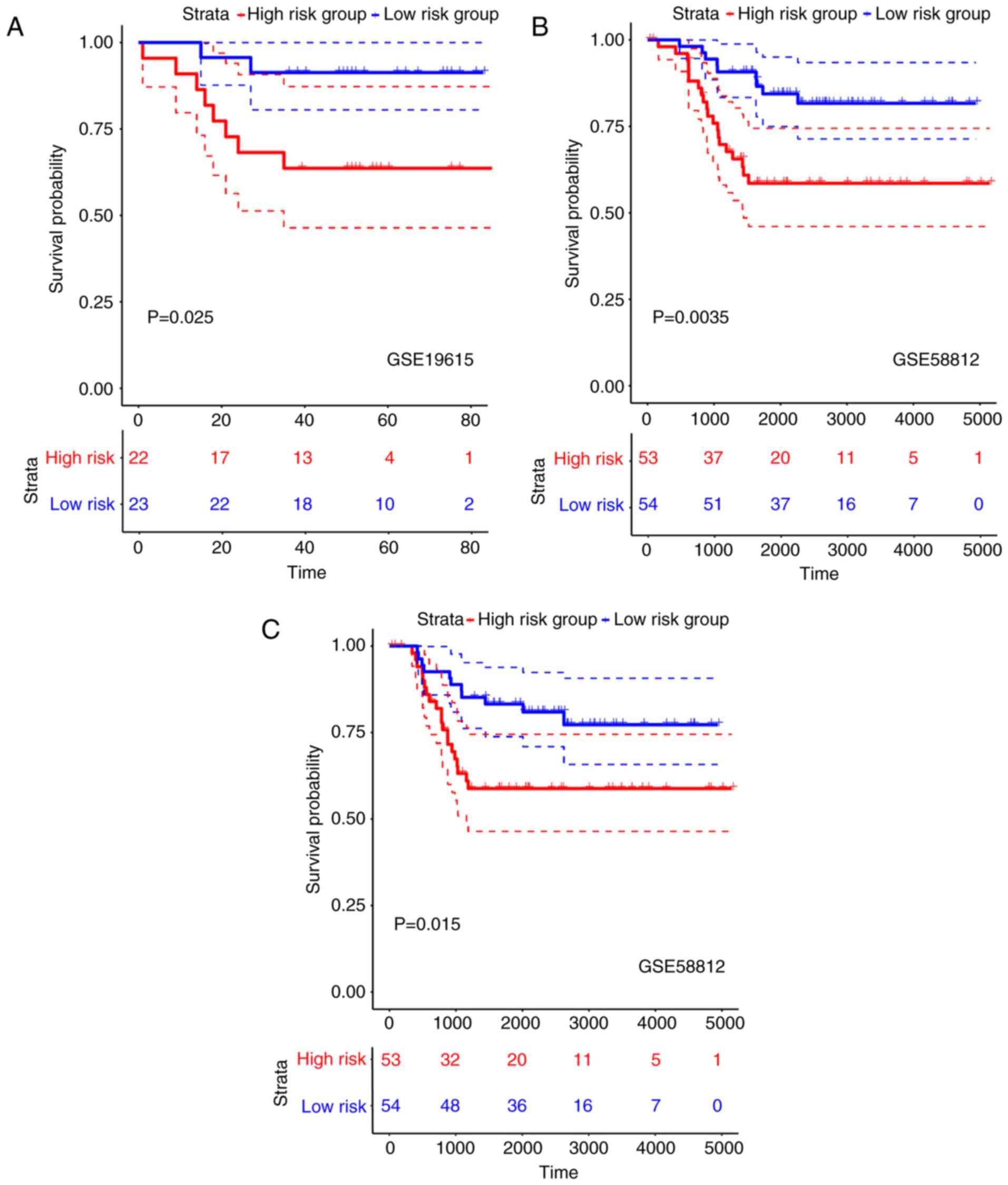
Risk-scores for samples in the validation cohort of
the GSE19615 dataset were calculated using the nine-lncRNA
signature. Subsequently, 23 and 22 samples were divided into low-
and high-risk groups, respectively, with the median risk-score as
the threshold. The DFS of patients in the high-risk group was
significantly worse than that of the patients in the low-risk group
(HR=1.499, 95% CI=0.950–2.365, P=0.025; Fig. 2A). Similarly, in the validation
cohort of the GSE58812 dataset, 54 and 53 samples were classified
into low- and high-risk groups, respectively. The OS rate of
patients in the high-risk group was significantly worse compared
with patients in the low-risk group (HR=1.316, 95% CI=1.094–1.582,
P=0.0035; Fig. 2B). Furthermore,
patients in the high-risk group demonstrated significantly worse
DFS than those in the low-risk group (HR=1.262, 95% CI=1.056–1.510,
P=0.015; Fig. 2C). Overall, the
nine-lncRNA characteristic risk-score κ had a good prognostic
stratification in both the training set (GSE21653) and the
independent validation sets.
Association between risk-score and the
expression levels of the signature lncRNAs
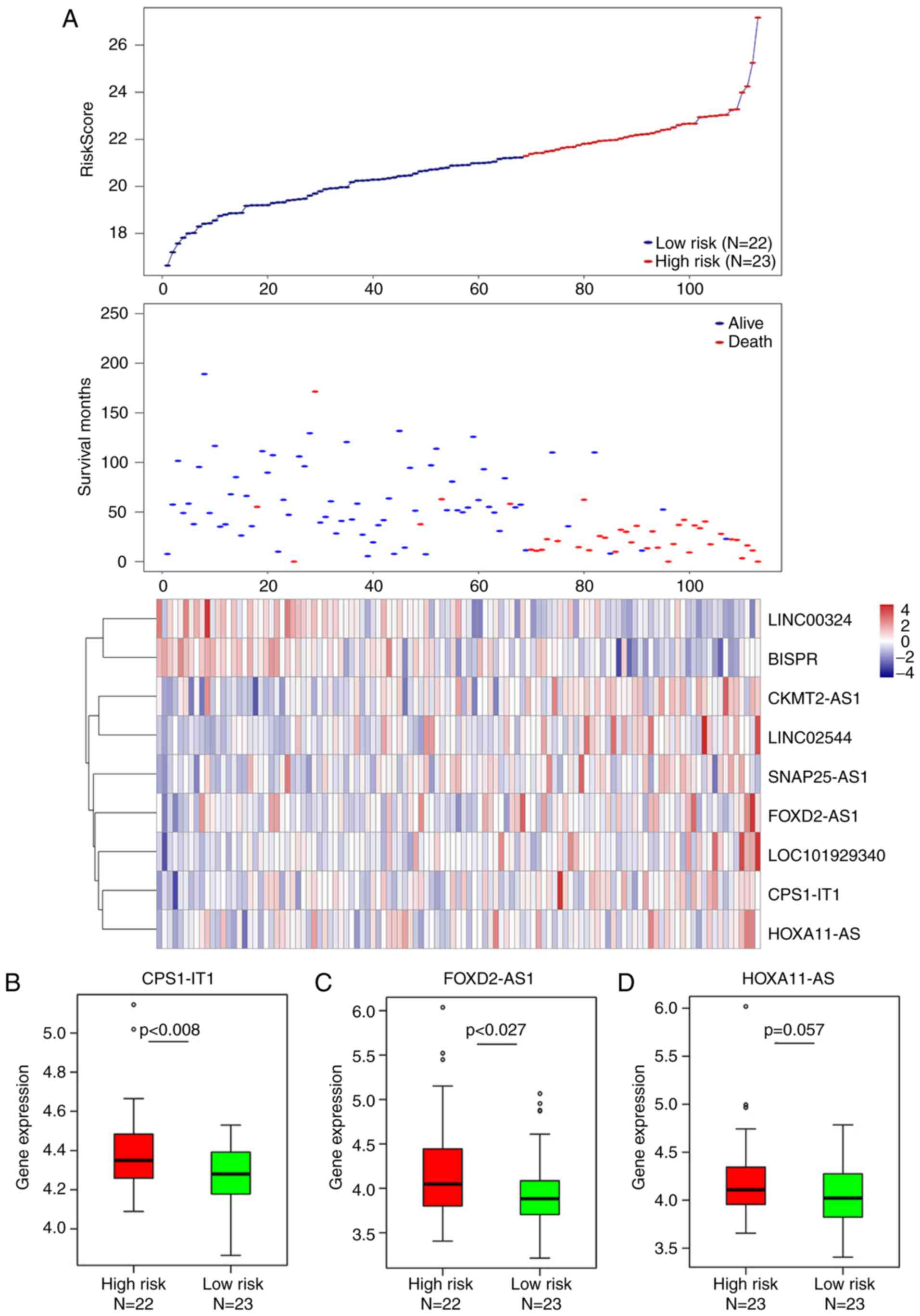
The distribution of risk-scores for each patient
calculated by the nine-lncRNA signature were analyzed. Together
with patients' survival status, this identified a total of seven
risk-associated lncRNAs (CKMT2-AS1, HOXA11-AS, CPS1-IT1,
LOC101929340, FOXD2-AS1, SNAP25-AS1 and LINC02544), which were
demonstrated to be overexpressed in patients with high risk-scores,
and two protective lncRNAs (BISPRL and INC00324) were expressed at
a lesser degree in patients with low risk-scores (Fig. 3A). Furthermore, eight of the nine
lncRNAs were demonstrated to be significantly differentially
expressed between the high- and low-risk groups classified by the
nine-lncRNAs signature (Fig. 3B, C and
E-J); only HOXA11-AS exhibited marginally differential
expression (P=0.057; Fig. 3D).
Association between the nine-lncRNA
signature and clinical features
It has been reported that lymph node status, age,
histodifferentiation, progesterone receptor (PR) status and human
epidermal growth factor receptor 2 (HER-2) status may influence the
prognosis of patients with BC (30,31).
Using the multivariate Cox regression model, the present study
demonstrated that the predictive performance of the nine-lncRNA
signature was independent of the aforementioned clinical features.
The results of the univariate and multivariate analyses for the
nine-lncRNA signature and other clinical features are presented in
Table III.
 | Table III.Univariate and multivariate Cox
regression analyses in the training set. |
Table III.
Univariate and multivariate Cox
regression analyses in the training set.
|
|
| Univariate
model | Multivariate
model |
|---|
|
|
|
|
|
|---|
| Factor | Sample, n | HR | P-value | HR | 95% CI | P-value |
|---|
| Nine-lncRNA risk
score, high risk vs. low risk | 55 vs. 55 | 2.7180 |
5.7×10−77 | 16.5578 | 4.908–55.854 | 4.05E-06 |
| Stage, N1 vs.
N0 | 50 vs. 58 | 2.9203 | 0.0011 | 2.6189 | 1.1417–6.007 | 0.023 |
| Grade, G3 vs G1 and
G2 | 78 vs. 33 | 1.2295 | 0.0444 | 1.0540 | 0.7268–2.805 | 0.874 |
| Age, ≥ 50 vs.
<50 years | 70 vs. 40 | 1.1959 | 0.5981 | 1.0051 | 0.9761–1.035 | 0.734 |
| PR, Pos vs.
Neg | 6 vs. 104 | 2.7364 | 0.0481 | 0.9480 | 0.2733–3.546 | 0.981 |
| HER-2, Pos vs.
Neg | 15 vs. 89 | 1.3656 | 0.4579 | 1.2110 | 0.3727–3.935 | 0.750 |
Nine-lncRNA signature-associated
biological processes
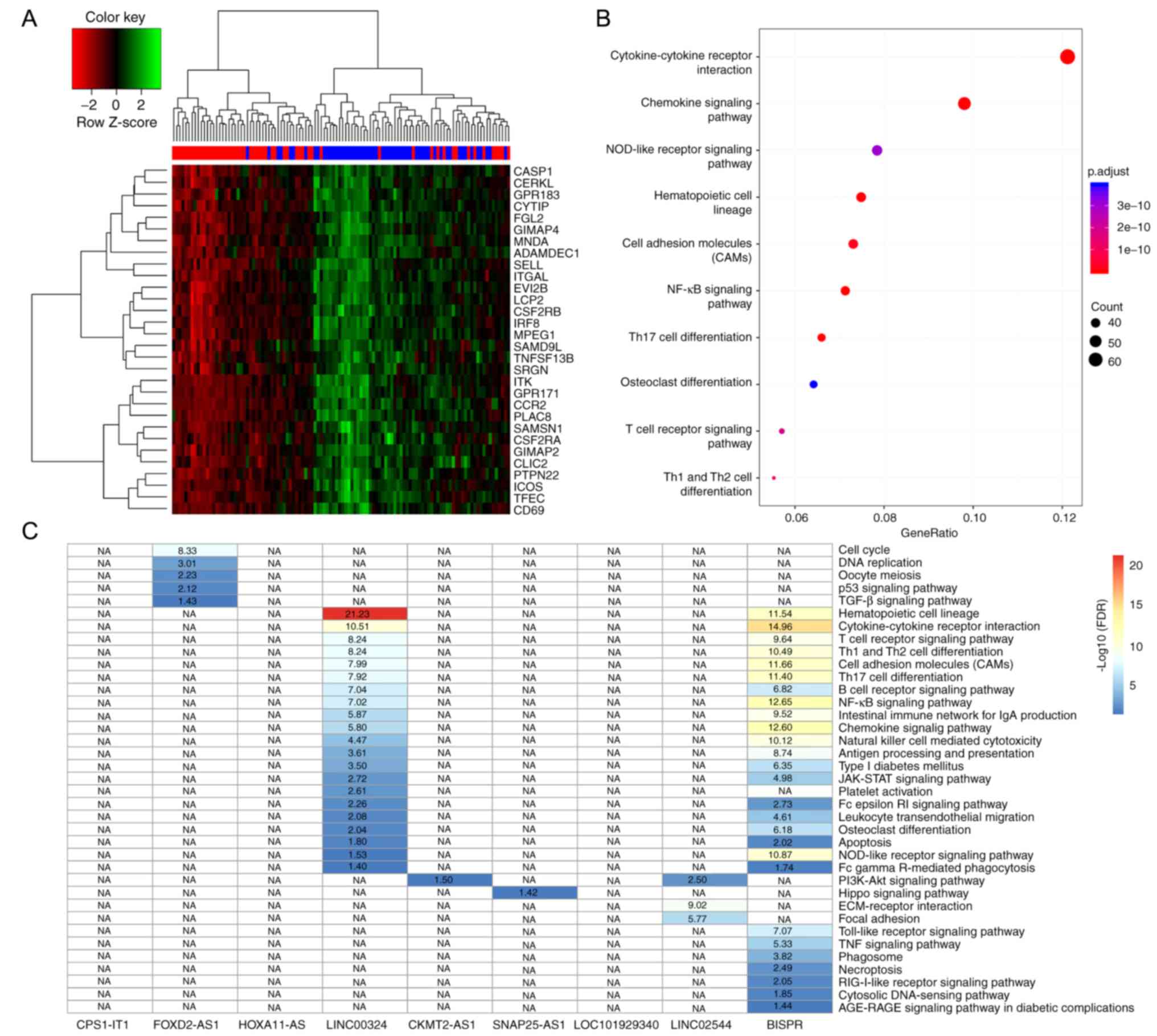
The present study identified 1,276 DEGs between the
57 low-risk samples and the 53 high-risk samples classified by the
nine-lncRNA signature in the discovery cohort (Table SI). As predicted, hierarchical
clustering analysis of the top 30 most significant DEGs (Fig. 4A) demonstrated that samples in the
high- and low-risk groups were divided into two clusters,
respectively. Furthermore, KEGG pathway function analysis
demonstrated that these DEGs were significantly enriched in 32
signaling pathways (Table SII),
including signaling pathways associated with cancer cell growth,
proliferation, migration and angiogenesis. The associated signaling
pathways in the present study included ‘cytokine-cytokine receptor
interaction’ and ‘cell adhesion molecules’ (CAMs), and signaling
pathways associated with the immune system, such as ‘T-cell
receptor signaling pathway’, ‘NF-κB signaling pathway’, ‘Th17 cell
differentiation’, and ‘Th1 and Th2 cell differentiation’. Immune
escape mechanisms have been reported to play an important role in
the development and metastasis of cancer (32), which indicated the dysregulation of
important carcinogenesis-associated biological processes between
the classified high- and low-risk groups in the present study. With
regards to the 110 samples assessed in the discovery cohort of the
present study, the expression levels of 4,195 mRNAs were
demonstrated to be significantly associated with the nine lncRNAs,
among which 3,159 mRNA-lncRNA pairs were positively associated and
1,036 mRNA-lncRNA pairs were negatively associated (Pearson's
correlation analysis, FDR<0.01) (Table SIII). These mRNAs were predicted to
be co-expressed with the corresponding lncRNAs. The present study
performed KEGG pathway function analysis using the co-expressed
mRNAs, for each lncRNAs. A total of 37 KEGG pathways (Table SIV) were obtained for six lncRNAs
(Fig. 4C), including
cancer-associated signaling pathways, such as ‘cell cycle’, ‘DNA
replication’ and ‘p53 signaling pathway’, and immune-associated
signaling pathways including, ‘Th1 and Th2 cell differentiation’,
‘B-cell receptor signaling pathway’ and ‘T-cell receptor signaling
pathway’. Overall, the nine lncRNAs were associated with the
carcinogenesis and development of patients with ER-negative BC.
Discussion
In recent years, a number of studies have
demonstrated that dysregulated lncRNAs play an important role in
the carcinogenesis, development and metastasis of different types
of tumor (33–35). A number of lncRNAs have been reported
to be associated with the recurrence, metastasis and resistance to
adjuvant therapy of patients with BC (36,37). For
example, lncRNA HOX Transcript Antisense Intergenic RNA has been
demonstrated to enhance ER signaling and confer tamoxifen
resistance in BC (38). Furthermore,
upregulation of the lncRNA SRY-box 2 has been demonstrated to
promote BC cell growth and invasion by activating the expression of
the lncRNA Plasmacytoma variant translocation 1 (39). The degradation of the histone-lysine
N-methyltransferase EZH2, mediated by lncRNA Antidifferentiation
noncoding RNA, has been indicated to decrease the invasion and
metastasis of BC (40). With the
development of genomic diagnostic tests such as Oncotype DX
(41) and Coloprint (42), high-throughput gene expression
profiles have become the principle method of identifying novel
prognostic biomarkers of cancer. Although several lncRNA signatures
have been developed for the prognosis of patients with BC (43,44),
there are currently none in use for clinical practice, particularly
for ER-negative BC. Thus, the present study used publicly available
gene expression profiles, together with the probe re-annotation
method, in order to develop a DFS-associated nine-lncRNA signature,
using the correlation analysis of the lncRNA expression data and
clinical information from patients with ER-negative BC. The
nine-lncRNA signature classified patients with ER-negative BC into
high- and low-risk groups with significantly different DFS, which
was validated in independent datasets. The results of the present
study provide evidence for the repeatability and clinical value of
the nine-lncRNA signature.
A number of signature lncRNAs have been reported to
be associated with different types of tumor (45,46). For
example, CPS1 Intronic Transcript 1 has been demonstrated to
suppress cell proliferation, invasion and metastasis in colorectal,
ovarian, lung and liver cancer (28–31,47–50).
Furthermore, upregulation of FOXD2 Adjacent Opposite Strand RNA 1
has been associated with poor prognosis in various types of tumor
(51–53). HOXA11-AS has been indicated to
promote BC invasion and metastasis by regulating EMT (54). Furthermore, LINC00324 acts as an
oncogene involved in the tumorigenesis and progression of gastric
and lung cancer (55). The lncRNA
BST2 Interferon Stimulated Positive Regulator has been demonstrated
to promote the progression of thyroid papillary carcinoma by
regulating miR-21-5p (56). Despite
numerous studies, the biological functions of several lncRNAs
remain unclear, thus further research is required.
In the present study, DEGs between the high- and
low-risk groups were identified in the discovery and validation
cohorts, and high consistency scores were obtained between these
cohorts (Table SV). KEGG pathway
function analysis demonstrated that these DEGs were significantly
enriched in signaling pathways associated with cancer cell growth,
proliferation, migration and angiogenesis, such as
‘cytokine-cytokine receptor interaction’ and ‘CAMs’, and signaling
pathways associated with the immune system, such as ‘T-cell
receptor signaling pathway’, ‘NF-κB signaling pathway’, ‘Th17 cell
differentiation’ and ‘Th1 and Th2 cell differentiation’.
Furthermore, genes co-expressed with these signature lncRNAs were
significantly enriched in signaling pathways associated with cancer
cell proliferation, migration and angiogenesis, such as ‘cell
cycle’, ‘DNA replication’ and ‘p53 signaling pathway’, and
signaling pathways associated with the immune system, such as ‘Th1
and Th2 cell differentiation’, ‘B-cell receptor signaling pathway’
and ‘T-cell receptor signaling pathway’. Altogether, the nine
lncRNAs assessed in the present study were associated with the
carcinogenesis and development of tumors, which provides evidence
for the prognostic ability of the nine-lncRNA signature.
Although candidate lncRNA biomarkers involved in
tumorigenesis were identified by bioinformatics analysis, some
limitations exist in the present study. Firstly, since clinical
follow-up information was missing, certain factors, including the
presence of patient's other health status, including ER, PR and
HER2 expression, were not considered to determine prognostic
biomarkers. Secondly, the signature obtained by bioinformatics
analysis was insufficient for clinical practice and requires
external experimental verification, such as in vitro/in
vivo validation. Therefore, further work with complete clinical
information and a larger sample size is required for future genetic
and experimental studies.
Overall, the present study used the available
microarray data and clinical information to develop a nine-lncRNA
signature, in order to promote personalized treatment for patients
with ER-negative BC. Functional analysis demonstrated that this
signature was involved in several signaling pathways associated
with cancer recurrence and metastasis, which provides evidence for
the prediction of post-surgery relapse risk for patients with
ER-negative BC. Large-scale prospective studies are required in the
future, in order to further evaluate and validate the robustness of
the signature prior to its clinical application. Furthermore,
future studies are required in order to understand the molecular
mechanisms underlying the nine-lncRNA signature.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by the International
Peace Maternity and Child Health Hospital (grant no. GFY 9307).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
ZW designed the present study. ZW, JW, LL and QH
were responsible for acquisition, analysis and interpretation of
data. ZW conducted the bioinformatics analyses. ZW and QH were
responsible for drafting the manuscript and revising it critically.
MW contributed to the conception of the present study and was
responsible for approving the version to be published. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
lncRNA
|
long non-coding RNAs
|
|
BC
|
breast cancer
|
|
ER
|
estrogen receptor
|
|
ROC
|
receiver operating characteristic
|
|
DEGs
|
differentially expressed genes
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
DFS
|
disease-free survival
|
|
HR
|
hazard ratio
|
|
CI
|
confidence interval
|
|
GEO
|
Gene Expression Omnibus
|
|
FDR
|
false discovery rate
|
References
|
1
|
DeSantis C, Ma J, Bryan L and Jemal A:
Breast cancer statistics, 2013. CA Cancer J Clin. 64:52–62. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ma M, Huang W and Kong D: IL-17 inhibits
the accumulation of myeloid-derived suppressor cells in breast
cancer via activating STAT3. Int Immunopharmacol. 59:148–156. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shandley LM, Spencer JB, Fothergill A,
Mertens AC, Manatunga A, Paplomata E and Howards PP: Impact of
tamoxifen therapy on fertility in breast cancer survivors. Fertil
Steril. 107:243–252.e245. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sanchez Calle A, Kawamura Y, Yamamoto Y,
Takeshita F and Ochiya T: Emerging roles of long non-coding RNA in
cancer. Cancer Sci. 109:2093–2100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mitra SA, Mitra AP and Triche TJ: A
central role for long non-coding RNA in cancer. Front Genet.
3:172012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gibb EA, Vucic EA, Enfield KS, Stewart GL,
Lonergan KM, Kennett JY, Becker-Santos DD, MacAulay CE, Lam S,
Brown CJ and Lam WL: Human cancer long non-coding RNA
transcriptomes. PLoS One. 6:e259152011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sas-Chen A, Aure MR, Leibovich L, Carvalho
S, Enuka Y, Körner C, Polycarpou-Schwarz M, Lavi S, Nevo N,
Kuznetsov Y, et al: LIMT is a novel metastasis inhibiting lncRNA
suppressed by EGF and downregulated in aggressive breast cancer.
EMBO Mol Med. 8:1052–1064. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Richards EJ, Zhang G, Li ZP, Permuth-Wey
J, Challa S, Li Y, Kong W, Dan S, Bui MM, Coppola D, et al: Long
non-coding RNAs (LncRNA) regulated by transforming growth factor
(TGF) β: LncRNA-hit-mediated TGFβ-induced epithelial to mesenchymal
transition in mammary epithelia. J Biol Chem. 290:6857–6867. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao W, Luo J and Jiao S: Comprehensive
characterization of cancer subtype associated long non-coding RNAs
and their clinical implications. Sci Rep. 4:65912014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu SJ, Xie J, Li Y, Yu B, Ma Q and Liu BQ:
Identification of lncRNAs-gene interactions in transcription
regulation based on co-expression analysis of RNA-seq data. Math
Biosci Eng. 16:7112–7125. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bao S, Zhao H, Yuan J, Fan D, Zhang Z, Su
J and Zhou M: Computational identification of mutator-derived
lncRNA signatures of genome instability for improving the clinical
outcome of cancers: A case study in breast cancer. Brief Bioinform.
Oct 28–2019.(Epub ahead of print). View Article : Google Scholar
|
|
15
|
Li J, Wang W, Xia P, Wan L, Zhang L, Yu L,
Wang L, Chen X, Xiao Y and Xu C: Identification of a five-lncRNA
signature for predicting the risk of tumor recurrence in patients
with breast cancer. Int J Cancer. 143:2150–2160. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang K, Li J, Xiong YF, Zeng Z, Zhang X
and Li HY: A potential prognostic long noncoding RNA signature to
predict recurrence among ER-positive breast cancer patients treated
with tamoxifen. Sci Rep. 8:31792018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sun M, Wu D, Zhou K, Li H, Gong X, Wei Q,
Du M, Lei P, Zha J, Zhu H, et al: An eight-lncRNA signature
predicts survival of breast cancer patients: A comprehensive study
based on weighted gene co-expression network analysis and competing
endogenous RNA network. Breast Cancer Res Treat. 175:59–75. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sabatier R, Finetti P, Adelaide J, Guille
A, Borg JP, Chaffanet M, Lane L, Birnbaum D and Bertucci F:
Down-regulation of ECRG4, a candidate tumor suppressor gene, in
human breast cancer. PLoS One. 6:e276562011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sabatier R, Finetti P, Cervera N,
Lambaudie E, Esterni B, Mamessier E, Tallet A, Chabannon C, Extra
JM, Jacquemier J, et al: A gene expression signature identifies two
prognostic subgroups of basal breast cancer. Breast Cancer Res
Treat. 126:407–420. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jézéquel P, Loussouarn D,
Guérin-Charbonnel C, Campion L, Vanier A, Gouraud W, Lasla H,
Guette C, Valo I, Verrièle V and Campone M: Gene-expression
molecular subtyping of triple-negative breast cancer tumours:
Importance of immune response. Breast Cancer Res. 17:432015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Y, Zou L, Li Q, Haibe-Kains B, Tian R,
Li Y, Desmedt C, Sotiriou C, Szallasi Z, Iglehart JD, et al:
Amplification of LAPTM4B and YWHAZ contributes to chemotherapy
resistance and recurrence of breast cancer. Nat Med. 16:214–218.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guo JC, Wu Y, Chen Y, Pan F, Wu ZY, Zhang
JS, Wu JY, Xu XE, Zhao JM, Li EM, et al: Protein-coding genes
combined with long noncoding RNA as a novel transcriptome molecular
staging model to predict the survival of patients with esophageal
squamous cell carcinoma. Cancer Commun (Lond). 38:42018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zapata I, Moraes LE, Fiala EM,
Zaldivar-Lopez S, Couto CG, Rowell JL and Alvarez CE: Risk-modeling
of dog osteosarcoma genome scans shows individuals with
Mendelian-level polygenic risk are common. BMC Genomics.
20:2262019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu G, Wang LG, Yan GR and He QY: DOSE: An
R/Bioconductor package for disease ontology semantic and enrichment
analysis. Bioinformatics. 31:608–609. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu W: LncRNA LINC-PINT inhibits cancer
cell proliferation, invasion, and migration in osteosarcoma by
downregulating miRNA-21. Cancer Biother Radiopharm. 34:258–263.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Marín-Béjar O, Mas AM, González J,
Martinez D, Athie A, Morales X, Galduroz M, Raimondi I, Grossi E,
Guo S, et al: The human lncRNA LINC-PINT inhibits tumor cell
invasion through a highly conserved sequence element. Genome Biol.
18:2022017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zou Z, Ma T, He X, Zhou J, Ma H, Xie M,
Liu Y, Lu D, Di S and Zhang Z: Long intergenic non-coding RNA 00324
promotes gastric cancer cell proliferation via binding with HuR and
stabilizing FAM83B expression. Cell Death Dis. 9:7172018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang S, Zhang S, He Y, Huang X, Hui Y and
Tang Y: HOXA11-AS regulates JAK-STAT pathway by miR-15a-3p/STAT3
axis to promote the growth and metastasis in liver cancer. J Cell
Biochem. 120:15941–15951. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen X, Li X, Fan Z, Li J, Xie Y, Wang T
and Ouyang T: Ultrasound as a replacement for physical examination
in clinical staging of axillary lymph nodes in breast cancer
patients. Thorac Cancer. 11:48–54. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shui R, Liang X, Li X, Liu Y, Li H, Xu E,
Zhang Z, Lian Y, Guo S, Yao M, et al: Hormone receptor and human
epidermal growth factor receptor 2 detection in invasive breast
carcinoma: A retrospective study of 12,467 patients from 19 chinese
representative clinical centers. Clin Breast Cancer. Aug
23–2019.(Epub ahead of print). PubMed/NCBI
|
|
32
|
Montes P, Bernal M, Campo LN,
González-Ramírez AR, Jiménez P, Garrido P, Jurado M, Garrido F,
Ruiz-Cabello F and Hernández F: Tumor genetic alterations and
features of the immune microenvironment drive myelodysplastic
syndrome escape and progression. Cancer Immunol Immunother.
68:2015–2027. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jiang L, Zhao XH, Mao YL, Wang JF, Zheng
HJ and You QS: Long non-coding RNA RP11-468E2.5 curtails colorectal
cancer cell proliferation and stimulates apoptosis via the JAK/STAT
signaling pathway by targeting STAT5 and STAT6. J Exp Clin Cancer
Res. 38:4652019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cao W, Gao W, Zheng P, Sun X and Wang L:
Medroxyprogesterone acetate causes the alterations of endoplasmic
reticulum related mRNAs and lncRNAs in endometrial cancer cells.
BMC Med Genomics. 12:1632019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Verhoeven RJA, Tong S, Mok BW, Liu J, He
S, Zong J, Chen Y, Tsao SW, Lung ML and Chen H: Epstein-barr virus
BART long non-coding RNAs function as epigenetic modulators in
nasopharyngeal carcinoma. Front Oncol. 9:11202019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bao G, Huang J, Pan W, Li X and Zhou T:
Long noncoding RNA CERS6-AS1 functions as a malignancy promoter in
breast cancer by binding to IGF2BP3 to enhance the stability of
CERS6 mRNA. Cancer Med. 9:278–289. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tang T, Guo C, Xia T, Zhang R, Zen K, Pan
Y and Jin L: LncCCAT1 promotes breast cancer stem cell function
through activating WNT/β-catenin Signaling. Theranostics.
9:7384–7402. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xue X, Yang YA, Zhang A, Fong KW, Kim J,
Song B, Li S, Zhao JC and Yu J: LncRNA HOTAIR enhances ER signaling
and confers tamoxifen resistance in breast cancer. Oncogene.
35:2746–2755. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang Y, Zhou J, Wang Z, Wang P and Li S:
Upregulation of SOX2 activated LncRNA PVT1 expression promotes
breast cancer cell growth and invasion. Biochem Biophys Res Commun.
493:429–436. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li Z, Hou P, Fan D, Dong M, Ma M, Li H,
Yao R, Li Y, Wang G, Geng P, et al: The degradation of EZH2
mediated by lncRNA ANCR attenuated the invasion and metastasis of
breast cancer. Cell Death Differ. 24:59–71. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Siow ZR, De Boer RH, Lindeman GJ and Mann
GB: Spotlight on the utility of the Oncotype DX® breast
cancer assay. Int J Womens Health. 10:89–100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tan IB and Tan P: Genetics: Genetics: An
18-gene signature (ColoPrint®) for colon cancer
prognosis. Nat Rev Clin Oncol. 8:131–133. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
He Y, Li X, Meng Y, Fu S, Cui Y, Shi Y and
Du H: A prognostic 11 long noncoding RNA expression signature for
breast invasive carcinoma. J Cell Biochem. 120:16692–16702.
2019.PubMed/NCBI
|
|
44
|
Guo W, Wang Q, Zhan Y, Chen X, Yu Q, Zhang
J, Wang Y, Xu XJ and Zhu L: Transcriptome sequencing uncovers a
three-long noncoding RNA signature in predicting breast cancer
survival. Sci Rep. 6:279312016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhao K, Wang M, Kang H and Wu A: A
prognostic five long-noncoding RNA signature for patients with
rectal cancer. J Cell Biochem. Nov 10–2019.(Epub ahead of print).
View Article : Google Scholar
|
|
46
|
Lv J, Guo Y, Yan L, Lu Y, Liu D and Niu J:
Development and validation of a five-lncRNA signature with
prognostic value in colon cancer. J Cell Biochem. Nov 3–2019.(Epub
ahead of print). View Article : Google Scholar
|
|
47
|
Zhang W, Yuan W, Song J, Wang S and Gu X:
LncRNA CPS1-IT1 suppresses EMT and metastasis of colorectal cancer
by inhibiting hypoxia-induced autophagy through inactivation of
HIF-1α. Biochimie. 144:21–27. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang W, Yuan W, Song J, Wang S and Gu X:
LncRna CPS1-IT1 suppresses cell proliferation, invasion and
metastasis in colorectal cancer. Cell Physiol Biochem. 44:567–580.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang YS, Ma LN, Sun JX, Liu N and Wang H:
Long non-coding RNA CPS1-IT1 is a positive prognostic factor and
inhibits epithelial ovarian cancer tumorigenesis. Eur Rev Med
Pharmacol Sci. 21:3169–3175. 2017.PubMed/NCBI
|
|
50
|
Wang TH, Yu CC, Lin YS, Chen TC, Yeh CT,
Liang KH, Shieh TM, Chen CY and Hsueh C: Long noncoding RNA
CPS1-IT1 suppresses the metastasis of hepatocellular carcinoma by
regulating HIF-1α activity and inhibiting epithelial-mesenchymal
transition. Oncotarget. 7:43588–43603. 2016.PubMed/NCBI
|
|
51
|
Rong L, Zhao R and Lu J: Highly expressed
long non-coding RNA FOXD2-AS1 promotes non-small cell lung cancer
progression via Wnt/β-catenin signaling. Biochem Biophys Res
Commun. 484:586–591. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen G, Sun W, Hua X, Zeng W and Yang L:
Long non-coding RNA FOXD2-AS1 aggravates nasopharyngeal carcinoma
carcinogenesis by modulating miR-363-5p/S100A1 pathway. Gene.
645:76–84. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Bao J, Zhou C, Zhang J, Mo J, Ye Q, He J
and Diao J: Upregulation of the long noncoding RNA FOXD2-AS1
predicts poor prognosis in esophageal squamous cell carcinoma.
Cancer Biomark. 21:527–533. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li W, Jia G, Qu Y, Du Q and Liu B and Liu
B: Long non-coding RNA (LncRNA) HOXA11-AS promotes breast cancer
invasion and metastasis by regulating epithelial-mesenchymal
transition. Med Sci Monit. 23:3393–3403. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Pan ZH, Guo XQ, Shan J and Luo SX:
LINC00324 exerts tumor-promoting functions in lung adenocarcinoma
via targeting miR-615-5p/AKT1 axis. Eur Rev Med Pharmacol Sci.
22:8333–8342. 2018.PubMed/NCBI
|
|
56
|
Zhang H, Cai Y, Zheng L, Zhang Z, Lin X
and Jiang N: LncRNA BISPR promotes the progression of thyroid
papillary carcinoma by regulating miR-21-5p. Int J Immunopathol
Pharmacol. 32:20587384187726522018. View Article : Google Scholar : PubMed/NCBI
|