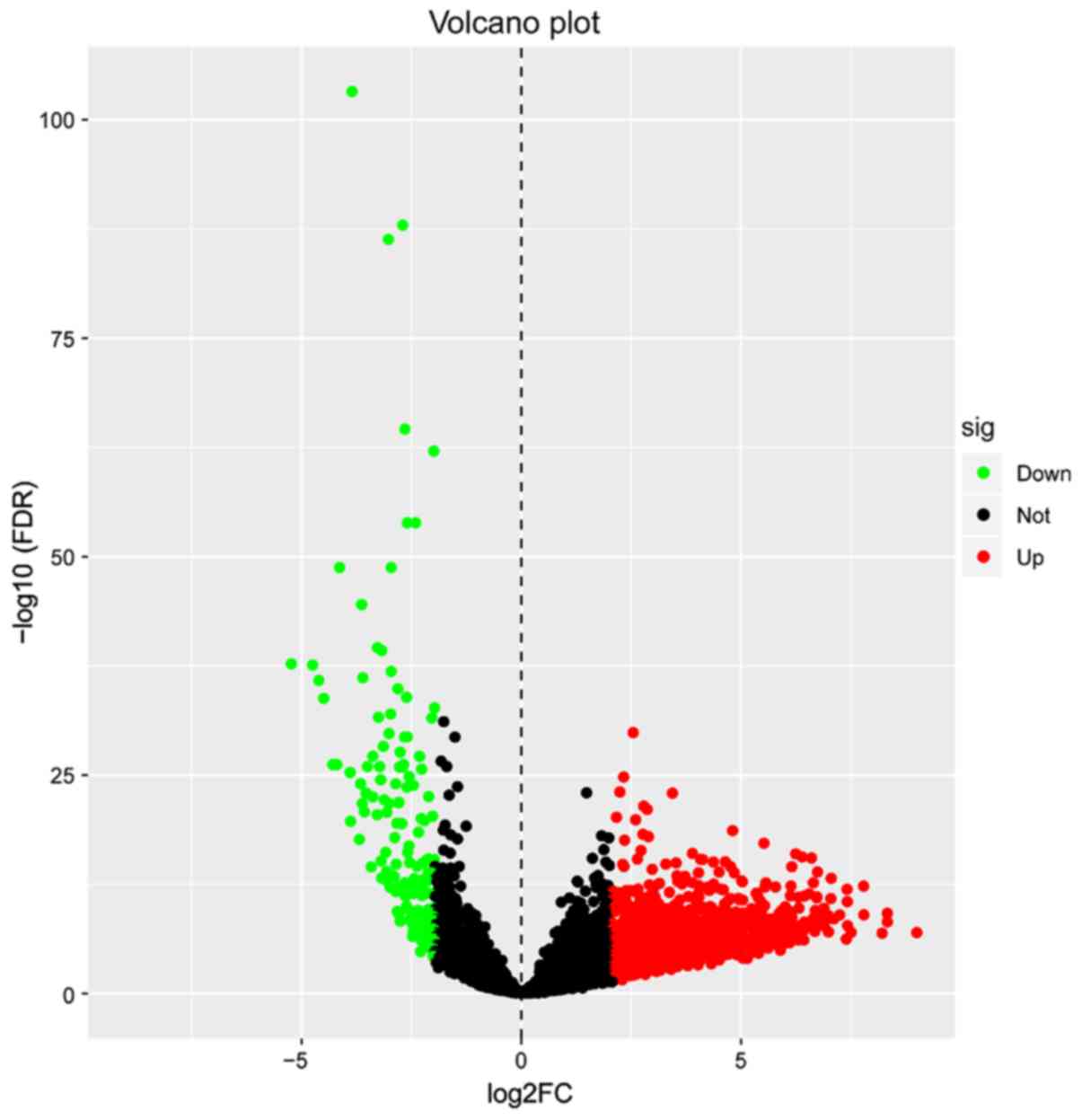
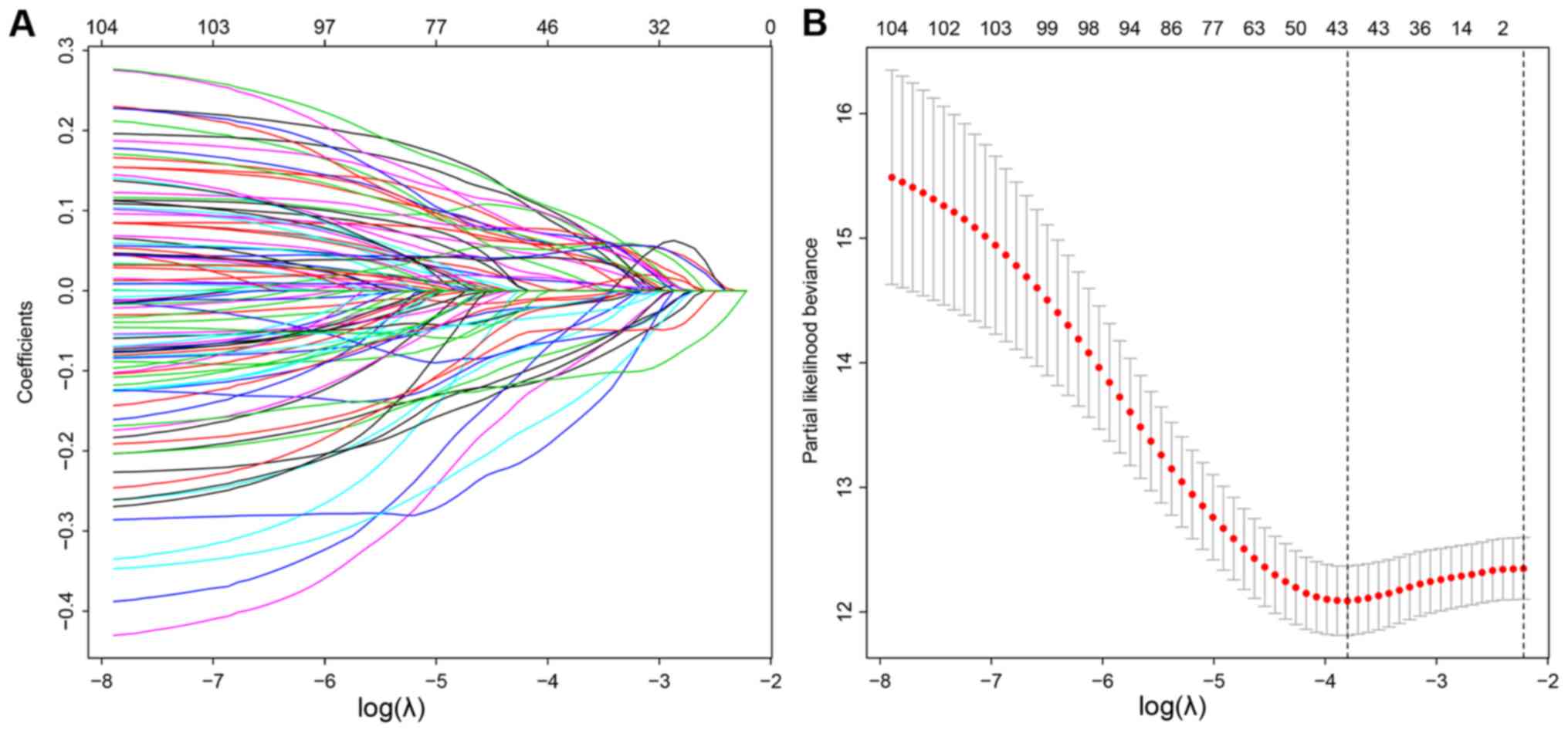
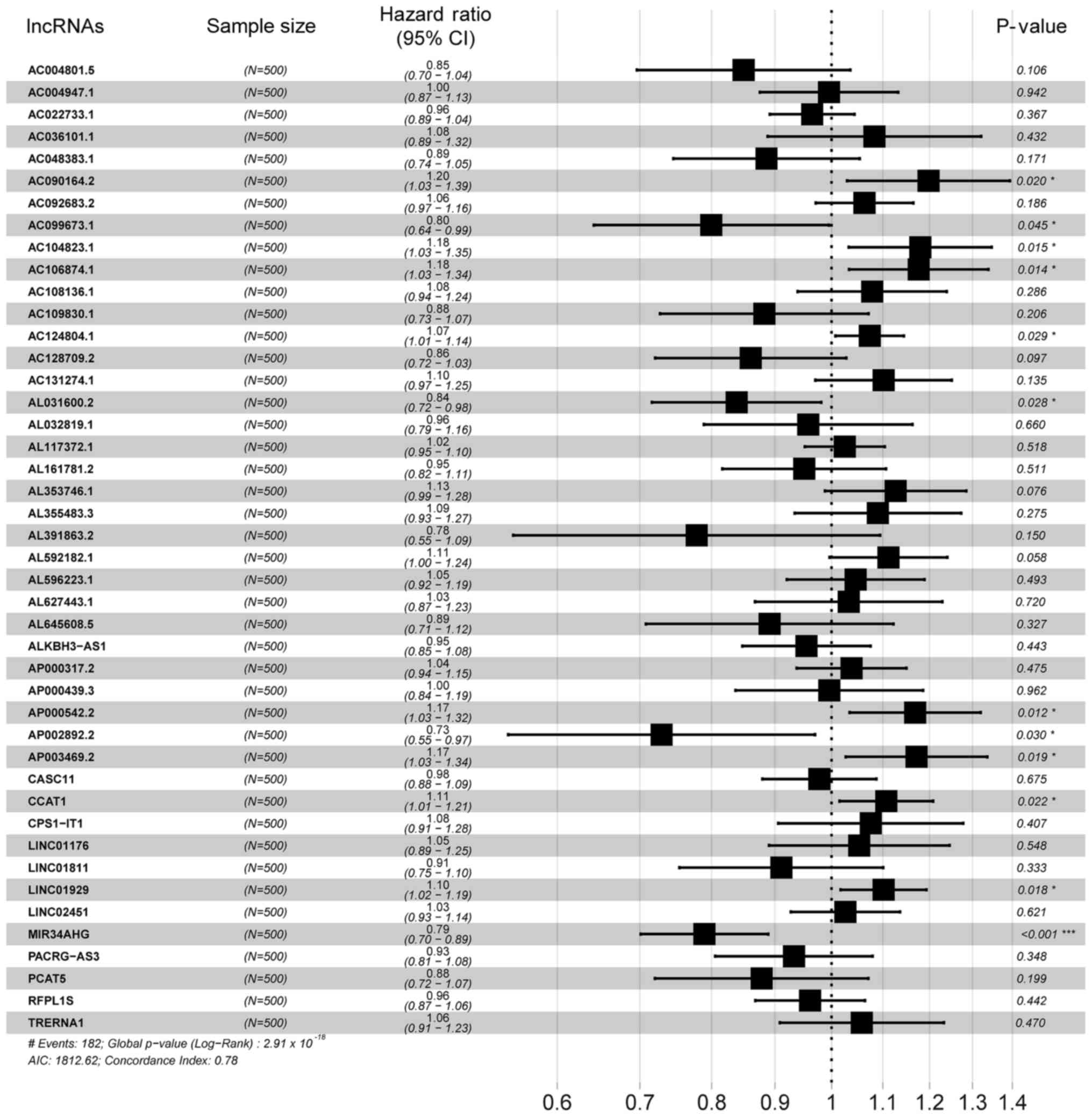
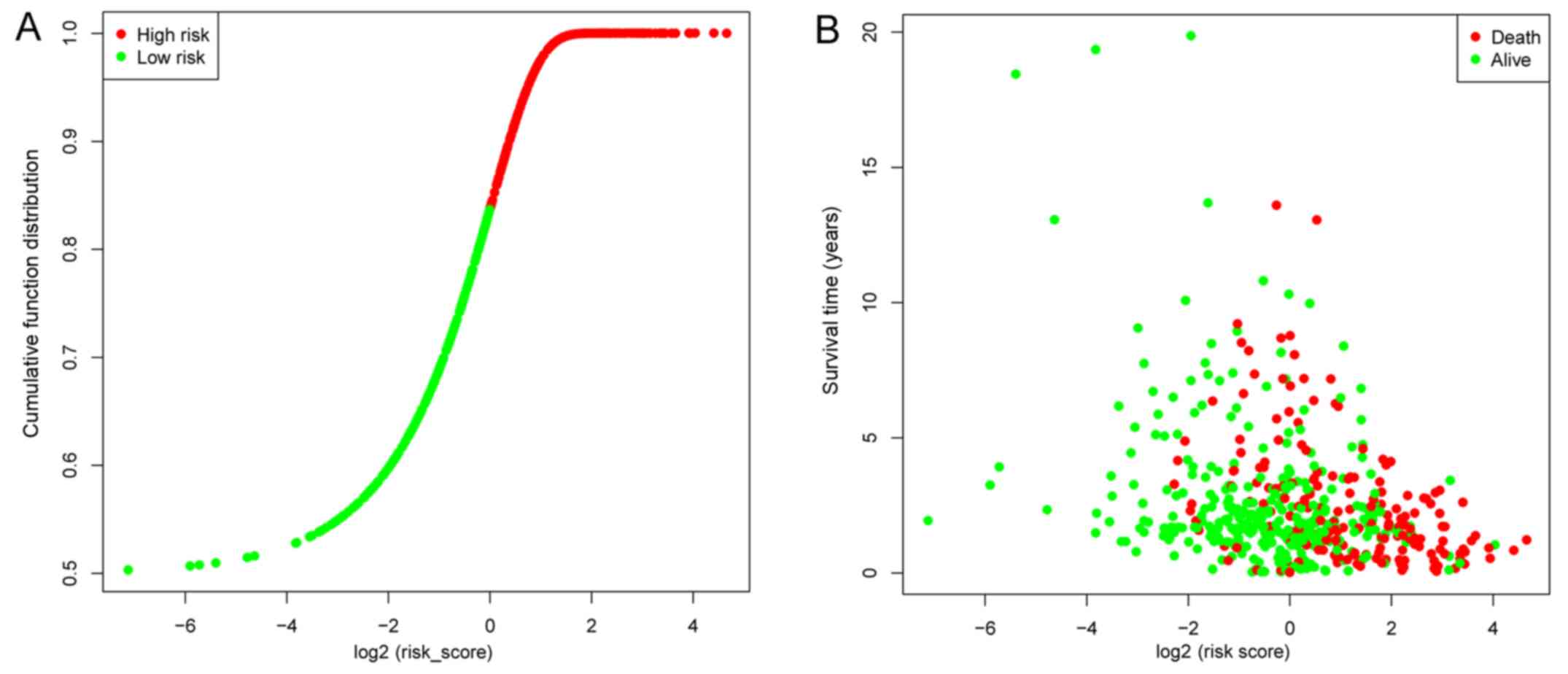
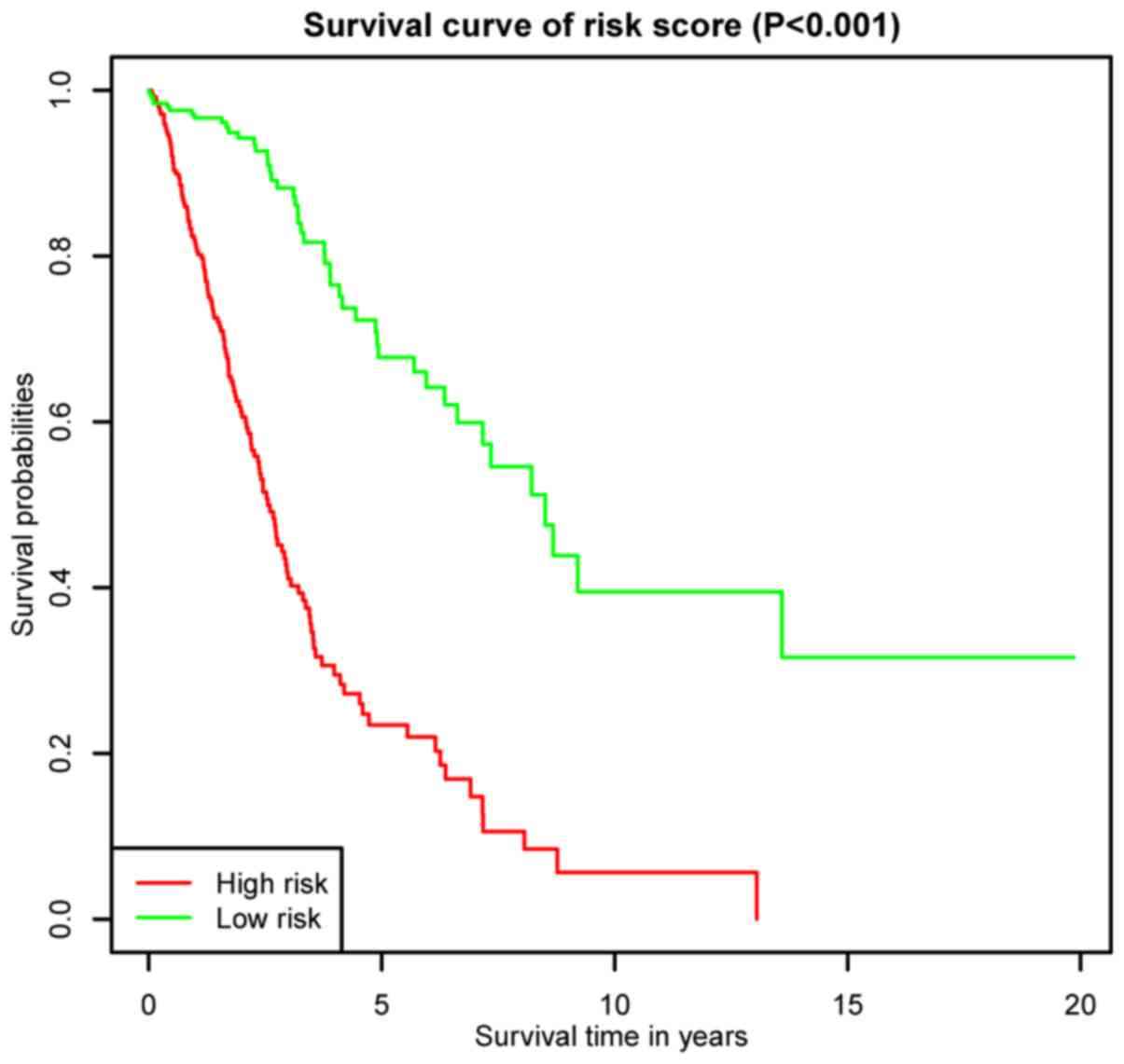
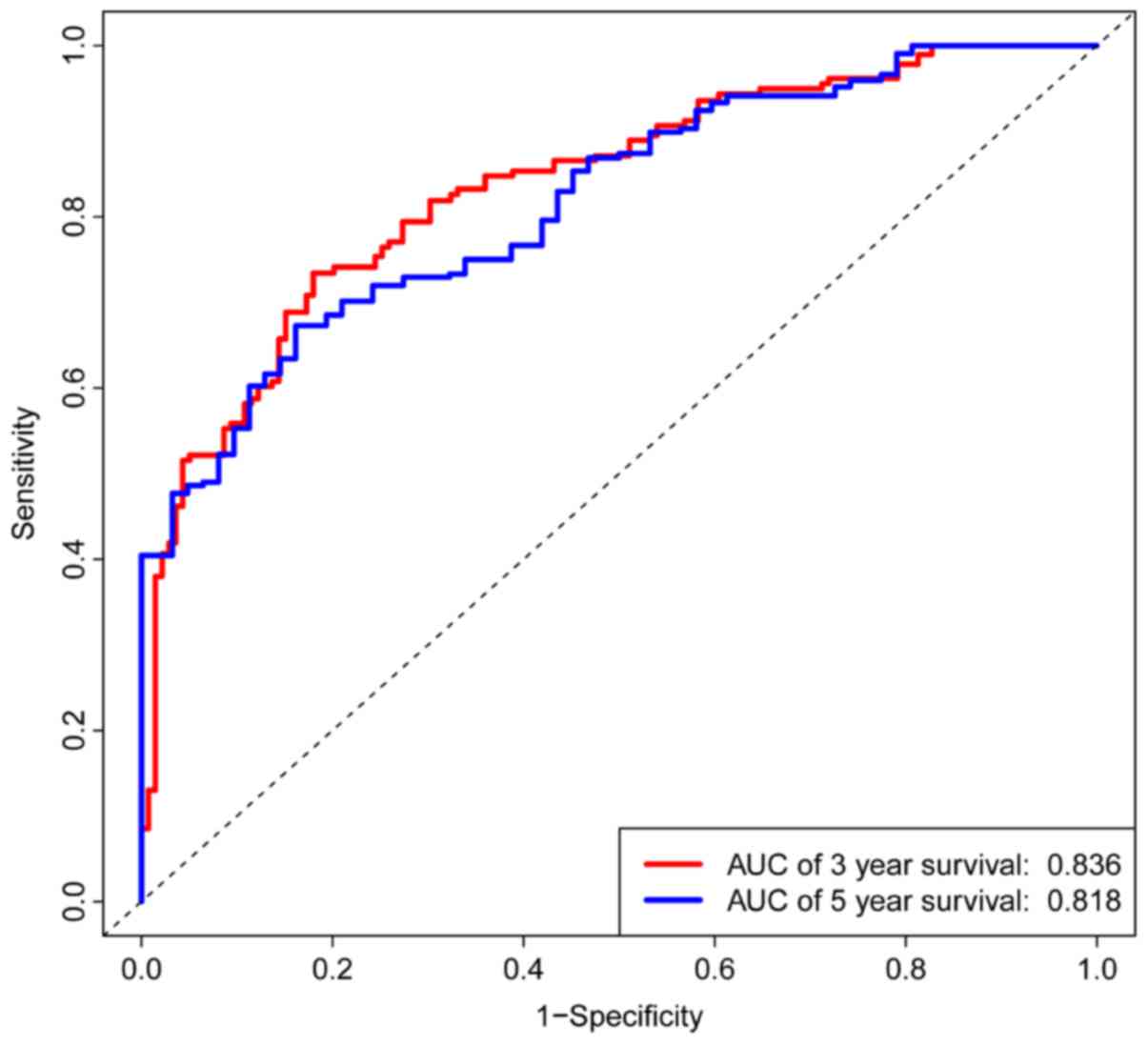
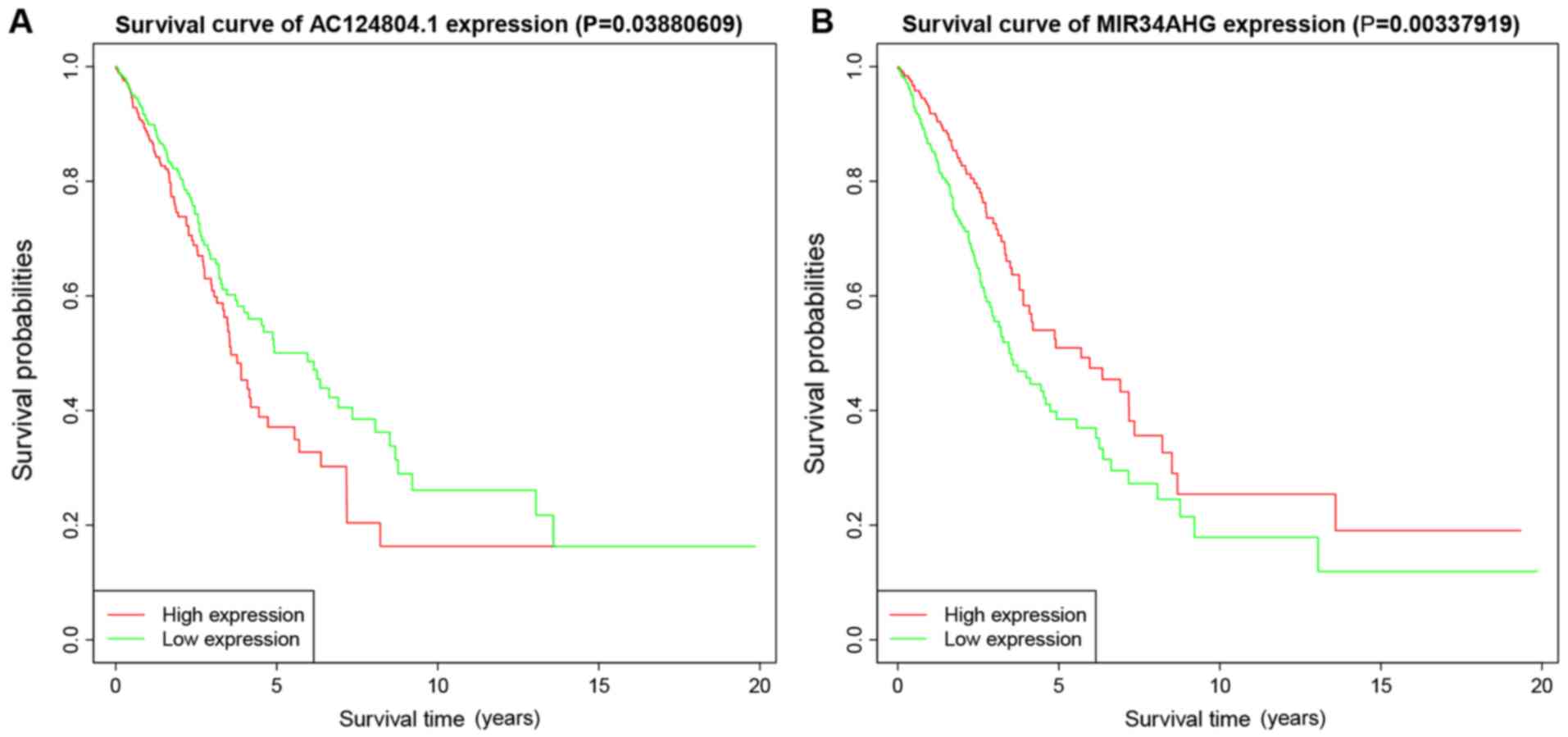
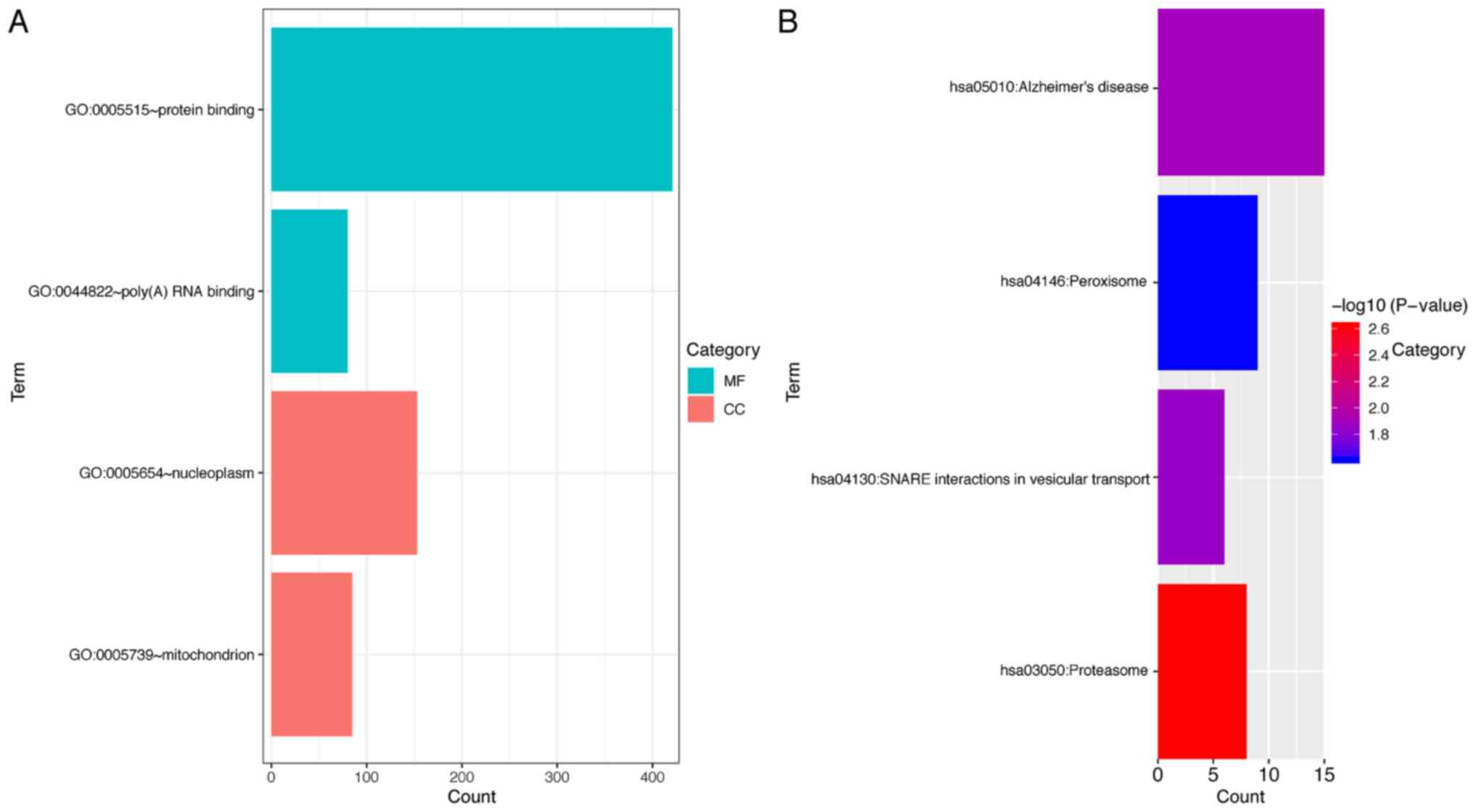
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Simó M, Vaquero L, Ripollés P,
Gurtubay-Antolin A, Jové J, Navarro A, Cardenal F, Bruna J and
Rodríguez-Fornells A: Longitudinal brain changes associated with
prophylactic cranial irradiation in lung cancer. J Thorac Oncol.
11:475–486. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shahabi S, Kumaran V, Castillo J, Cong Z,
Nandagopal G, Mullen DJ, Alvarado A, Correa MR, Saizan A, Goel R,
et al: LINC00261 is an epigenetically regulated tumor suppressor
essential for activation of the DNA damage response. Cancer Res.
79:3050–3062. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lu GS, Li M, Xu CX and Wang D: APE1
stimulates EGFR-TKI resistance by activating Akt signaling through
a redox-dependent mechanism in lung adenocarcinoma. Cell Death Dis.
9:11112018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Macheleidt IF, Dalvi PS, Lim SY, Meemboor
S, Meder L, Käsgen O, Müller M, Kleemann K, Wang L, Nürnberg P, et
al: Preclinical studies reveal that LSD1 inhibition results in
tumor growth arrest in lung adenocarcinoma independently of driver
mutations. Mol Oncol. 12:1965–1979. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hung MS, Lung JH, Lin YC, Fang YH, Huang
SY, Jiang YY, Hsieh MJ and Tsai YH: Comparative analysis of two
methods for the detection of EGFR mutations in plasma circulating
tumor DNA from lung adenocarcinoma patients. Cancers (Basel).
11:8032019. View Article : Google Scholar
|
|
7
|
Li S, Choi YL, Gong Z, Liu X, Lira M, Kan
Z, Oh E, Wang J, Ting JC, Ye X, et al: Comprehensive
characterization of oncogenic drivers in Asian lung adenocarcinoma.
J Thorac Oncol. 11:2129–2140. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang X, Min S, Liu H, Wu N, Liu X, Wang T,
Li W, Shen Y, Wang H, Qian Z, et al: Nf1 loss promotes Kras-driven
lung adenocarcinoma and results in Psat1-mediated glutamate
dependence. EMBO Mol Med. 11:e98562019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang L, Meng X, Zhu XW, Yang DC, Chen R,
Jiang Y and Xu T: Long non-coding RNAs in Oral squamous cell
carcinoma: Biologic function, mechanisms and clinical implications.
Mol Cancer. 18:1022019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yan Y, Xu Z, Li Z, Sun L and Gong Z: An
insight into the increasing role of lncRNAs in the pathogenesis of
gliomas. Front Mol Neurosci. 10:532017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu Z, Yan Y, Qian L and Gong Z: Long
non-coding RNAs act as regulators of cell autophagy in diseases
(Review). Oncol Rep. 37:1359–1366. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiang L, Wang R, Fang L, Ge X, Chen L,
Zhou M, Zhou Y, Xiong W, Hu Y, Tang X, et al: HCP5 is a
SMAD3-responsive long non-coding RNA that promotes lung
adenocarcinoma metastasis via miR-203/SNAI axis. Theranostics.
9:2460–2474. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ge X, Li GY, Jiang L, Jia L, Zhang Z, Li
X, Wang R, Zhou M, Zhou Y, Zeng Z, et al: Long noncoding RNA CAR10
promotes lung adenocarcinoma metastasis via miR-203/30/SNAI axis.
Oncogene. 38:3061–3076. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen J, Liu X, Xu Y, Zhang K, Huang J, Pan
B, Chen D, Cui S, Song H, Wang R, et al: TFAP2C-activated MALAT1
modulates the chemoresistance of docetaxel-resistant lung
adenocarcinoma cells. Mol Ther Nucleic Acids. 14:567–582. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Salavaty A, Rezvani Z and Najafi A:
Survival analysis and functional annotation of long non-coding RNAs
in lung adenocarcinoma. J Cell Mol Med. 23:5600–5617. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Serghiou S, Kyriakopoulou A and Ioannidis
JP: Long noncoding RNAs as novel predictors of survival in human
cancer: A systematic review and meta-analysis. Mol Cancer.
15:502016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shi X, Tan H, Le X, Xian H, Li X, Huang K,
Luo VY, Liu Y, Wu Z, Mo H, et al: An expression signature model to
predict lung adenocarcinoma-specific survival. Cancer Manag Res.
10:3717–3732. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sui J, Yang S, Liu T, Wu W, Xu S, Yin L,
Pu Y, Zhang X, Zhang Y, Shen B, et al: Molecular characterization
of lung adenocarcinoma: A potential four-long noncoding RNA
prognostic signature. J Cell Biochem. 120:705–714. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
R Core Team (2012), . R: A language and
environment for statistical computing. R Foundation for Statistical
Computing; Vienna: http://www.R-project.org/
|
|
20
|
RStudio Team (2015), . RStudio: Integrated
Development for R. RStudio, Inc.; Boston, MA: http://www.rstudio.com/
|
|
21
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Friedman J, Hastie T and Tibshirani R:
Regularization paths for generalized linear models via coordinate
descent. J Stat Softw. 33:1–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Therneau TM and Grambsch PM: Modeling
Survival Data: Extending the Cox Model. Statistics for Biology and
Health. Springer Science and Business Media; New York, NY: pp.
39–76. 2000, View Article : Google Scholar
|
|
24
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:32003.
View Article : Google Scholar
|
|
25
|
Yan Y, Xu Z, Qian L, Zeng S, Zhou Y, Chen
X, Wei J and Gong Z: Identification of CAV1 and DCN as potential
predictive biomarkers for lung adenocarcinoma. Am J Physiol Lung
Cell Mol Physiol. 316:L630–L643. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sivakumar S, San Lucas FA, Jakubek YA,
McDowell TL, Lang W, Kallsen N, Peyton S, Davies GE, Fukuoka J,
Yatabe Y, et al: Genomic landscape of allelic imbalance in
premalignant atypical adenomatous hyperplasias of the lung.
EBioMedicine. 42:296–303. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu C, Tian F, Liu J, Su M, Wu M, Zhu X and
Qian W: Circular RNA cMras inhibits lung adenocarcinoma progression
via modulating miR-567/PTPRG regulatory pathway. Cell Prolif.
52:e126102019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kerdidani D, Chouvardas P, Arjo AR,
Giopanou I, Ntaliarda G, Guo YA, Tsikitis M, Kazamias G, Potaris K,
Stathopoulos GT, et al: Wnt1 silences chemokine genes in dendritic
cells and induces adaptive immune resistance in lung
adenocarcinoma. Nat Commun. 10:14052019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Misono S, Seki N, Mizuno K, Yamada Y,
Uchida A, Sanada H, Moriya S, Kikkawa N, Kumamoto T, Suetsugu T, et
al: Molecular pathogenesis of gene regulation by the miR-150
duplex: miR-150-3p regulates TNS4 in lung adenocarcinoma. Cancers
(Basel). 11:6012019. View Article : Google Scholar
|
|
30
|
Abdel-Aziz A, Ahmed RA and Ibrahiem AT:
Expression of pRb, Ki67 and HER 2/neu in gastric carcinomas:
Relation to different histopathological grades and stages. Ann
Diagn Pathol. 30:1–7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ren Y, Zhao S, Jiang D, Feng X, Zhang Y,
Wei Z, Wang Z, Zhang W, Zhou QF, Li Y, et al: Proteomic biomarkers
for lung cancer progression. Biomarkers Med. 12:205–215. 2018.
View Article : Google Scholar
|
|
32
|
Wang P, Jin M, Sun CH, Yang L, Li YS, Wang
X, Sun YN, Tian LL and Liu M: A three-lncRNA expression signature
predicts survival in head and neck squamous cell carcinoma (HNSCC).
Biosci Rep. 38:BSR201815282018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Peng W, He D, Shan B, Wang J, Shi W, Zhao
W, Peng Z, Luo Q, Duan M, Li B, et al: LINC81507 act as a competing
endogenous RNA of miR-199b-5p to facilitate NSCLC proliferation and
metastasis via regulating the CAV1/STAT3 pathway. Cell Death Dis.
10:5332019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Machado AD, Gómez LM, Marchioni DM, Dos
Anjos FS, Molina MD, Lotufo PA, Benseñor IJ and Titan SM:
Association between dietary intake and coronary artery
calcification in non-dialysis chronic kidney disease: The PROGREDIR
Study. Nutrients. 10:3722018. View Article : Google Scholar
|
|
35
|
Kwon Y, Lee T, Lang A and Burnette D:
Assessment on latitudinal tree species richness using environmental
factors in the southeastern United States. PeerJ. 7:e67812019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Waldmann P, Ferenčaković M, Mészáros G,
Khayatzadeh N, Curik I and Sölkner J: AUTALASSO: An automatic
adaptive LASSO for genome-wide prediction. BMC Bioinformatics.
20:1672019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Follo C, Vidoni C, Morani F, Ferraresi A,
Seca C and Isidoro C: Amino acid response by Halofuginone in cancer
cells triggers autophagy through proteasome degradation of mTOR.
Cell Commun Signal. 17:392019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kaur S, Nag A, Gangenahalli G and Sharma
K: Peroxisome proliferator activated receptor gamma sensitizes
non-small cell lung carcinoma to gamma irradiation induced
apoptosis. Front Genet. 10:5542019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang YS, Tzeng HT, Tsai CH, Cheng HC, Lai
WW, Liu HS and Wang YC: VAMP8, a vesicle-SNARE required for
RAB37-mediated exocytosis, possesses a tumor metastasis suppressor
function. Cancer Lett. 437:79–88. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yun MR, Lim SM, Kim SK, Choi HM, Pyo KH,
Kim SK, Lee JM, Lee YW, Choi JW, Kim HR, et al: Enhancer remodeling
and MicroRNA alterations are associated with acquired resistance to
ALK inhibitors. Cancer Res. 78:3350–3362. 2018.PubMed/NCBI
|
|
41
|
Zhao K, Cheng J, Chen B, Liu Q, Xu D and
Zhang Y: Circulating microRNA-34 family low expression correlates
with poor prognosis in patients with non-small cell lung cancer. J
Thorac Dis. 9:3735–3746. 2017. View Article : Google Scholar : PubMed/NCBI
|