Introduction
Nasopharyngeal carcinoma (NPC) is an epithelial
malignancy of the head and neck that is prevalent in China, with a
high incidence rate (1). A number of
factors are associated with the development and progression of NPC,
including Epstein-Barr virus infection, genetic components and
environmental factors (2). The
prognosis of NPC is poor due to recurrence and distant metastasis;
however, the use of chemoradiotherapy and intensity-modulated
radiotherapy has increased the 5-year survival rate to ~70% between
January 2003 and December 2006 in Guangzhou, China (3,4).
Therefore, it is critical to understand the molecular mechanisms
that underlie the progression of NPC, as this may promote the
development of novel therapeutic strategies.
MicroRNAs (miRNAs/miRs) are a class of non-coding
RNAs that are 19–25 nucleotides long (5,6).
Previous studies have demonstrated that miRNAs regulate a number of
biological roles, including proliferation, apoptosis, migration and
differentiation (7–9). Recent studies have demonstrated that
miRNAs are abnormally expressed in numerous human diseases,
particularly cancer. miRNAs can serve either as oncogenes or tumor
suppressors during the progression of tumors (7,9). A
number of mature miRNAs have been demonstrated to be abnormally
expressed in NPC, including miR-135a (10), miR-23a (11) and miR-203a-3p (12). This suggests that abnormally
expressed miRNAs can contribute to the development and progression
of NPC. miR-212 is abnormally expressed in numerous types of tumor,
including non-small cell lung cancer (13), gastric cancer (14), NPC (15) and pancreatic cancer (16). Previous studies have indicated that
miR-212 may serve a key role in cancer progression (13–16).
E74-like factor 3 (ELF3) is a transcription factor
of the epithelial-specific subfamily, which serves an important
role in a variety of pathophysiological processes in cancer
(17). Previous studies have
indicated that ELF3 is involved in cell proliferation,
differentiation and migration in numerous types of human cancer. In
colorectal cancer, ELF3 is overexpressed and promotes proliferation
and invasion by enhancing β-catenin signaling (18). In hepatocellular carcinoma, ELF3 has
been demonstrated to repress E-cadherin and promote the
epithelial-mesenchymal transition by suppressing miR-141-3p,
thereby activating zinc finger E-box binding homeobox 1 (19). However, to the best of our knowledge,
the role of ELF3 in NPC remains unknown.
The present study demonstrated that miR-212 was
downregulated in NPC cells and tissues. In addition, reduced
expression levels of miR-212 were identified to be associated with
clinical features of patients with NPC. Furthermore, overexpression
of miR-212 suppressed cell proliferation in vitro. ELF3 was
revealed to be downregulated by a direct interaction of its
3′-untranslated region (3′-UTR) with miR-212. The identified
miR-212/ELF3 axis may provide novel evidence regarding the
molecular mechanisms of NPC, and may reveal a novel therapeutic
target for NPC.
Materials and methods
Tissues and cell lines
A total of 30 normal nasopharyngeal epithelial
tissue specimens and paired freshly frozen NPC biopsy specimens
were collected at the Rui Jin Hospital Affiliated to Shanghai
Jiaotong University (Shanghai, China) between August 2016 and
October 2017. The specimens were confirmed by histopathological
examination. No patients with NPC had received radiotherapy or
chemotherapy prior to biopsy. Written informed consent was obtained
from all patients. The clinicopathological features and
demographics of all patients are presented in Table I. The present study was approved by
the Ethics Review Committee of Rui Jin Hospital Affiliated to
Shanghai Jiaotong University (Shanghai, China).
 | Table I.Association between
clinicopathological characteristics and expression levels of
miR-212 and ELF3 in nasopharyngeal carcinoma. |
Table I.
Association between
clinicopathological characteristics and expression levels of
miR-212 and ELF3 in nasopharyngeal carcinoma.
| A, miR-212 |
|---|
|
|---|
|
|
| Expression level |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristics | Number, n | High, n (%) | Low, n (%) | χ2 | P-value |
|---|
| Sex |
|
|
| 1.099 | 0.295 |
| Male | 22 | 13 (59.1) | 9 (40.9) |
|
|
|
Female | 8 | 3 (37.5) | 5 (62.5) |
|
|
| Age (years) |
|
|
| 0.136 | 0.713 |
| ≤45 | 13 | 7 (53.8) | 6 (46.2) |
|
|
|
>45 | 17 | 8 (47.1) | 9 (52.9) |
|
|
| Clinical stage |
|
|
| 4.800 | 0.029 |
| I–II | 10 | 6 (60.0) | 4 (40.0) |
|
|
|
III–IV | 20 | 4 (20.0) | 16 (80.0) |
|
|
| Local or distant
metastasis |
|
|
| 8.623 | 0.003 |
| No | 11 | 9 (81.8) | 2 (18.2) |
|
|
|
Yes | 19 | 5 (26.3) | 14 (73.7) |
|
|
|
| B, ELF3 |
|
|
|
| Expression
level |
|
|
|
|
|
|
|
|
|
Characteristics | Number,
n | High, n
(%) | Low, n
(%) |
χ2 | P-value |
|
| Sex |
|
|
| 2.058 | 0.151 |
|
Male | 22 | 12 (54.5) | 10 (45.5) |
|
|
|
Female | 8 | 2 (25.0) | 6 (75.0) |
|
|
| Age (years) |
|
|
| 1.033 | 0.310 |
|
≤45 | 13 | 6 (46.2) | 7 (53.8) |
|
|
|
>45 | 17 | 11 (64.7) | 6 (35.3) |
|
|
| Clinical stage |
|
|
| 6.429 | 0.011 |
|
I–II | 10 | 4 (40.0) | 6 (60.0) |
|
|
|
III–IV | 20 | 17 (85.0) | 3 (15.0) |
|
|
| Local or distant
metastasis |
|
|
| 9.726 | 0.002 |
| No | 11 | 3 (27.3) | 8 (72.7) |
|
|
|
Yes | 19 | 16 (84.2) | 3 (15.8) |
|
|
The human immortalized nasopharyngeal epithelial
cell line NP69 and the NPC cell line SUNE-1 were obtained from the
Shanghai Cell Bank of Chinese Academy of Sciences. Each cell line
was cultured in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (HyClone; GE Healthcare Life Sciences) as
well as 100 U/ml penicillin and 100 µg/ml streptomycin. All cells
were maintained in a humidified incubator at 37°C with 5%
CO2.
Cell transfection
The miR-212 mimic, inhibitor, ELF3 small interfering
RNA (siRNA) and negative controls (mimic-NC, inhibitor-NC or
siRNA-NC) were obtained from Guangzhou RiboBio Co, Ltd. and
transfected at a concentration of 100 nM. ELF3 plasmid and the
empty control vector were purchased from Shanghai GenePharma Co.,
Ltd. NPC cells were grown in six-well plates and transfected with
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. At
48 h post-transfection, cells were harvested for reverse
transcription-quantitative PCR (RT-qPCR) analysis. The following
sequences were used: miR-212 mimic (sense,
5′-ACCUUGGCUCUAGACUGCUUACU-3′; and antisense,
5′-UAAGCAGUCUAGAGCCAAGGUUU-3′), mimic negative control miRNA
(sense, 5′-UUCUCCGAACGUGUCACGUTT-3′; and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′), miR-212 inhibitor
(5′-GGCCGUGACUGGAGACUGUUA-3′), inhibitor-NC
(5′-CAGUACUUUUGUGUAGUACAA-3′), and ELF3 siRNA
(5′-GCUACCAAGUGGAGAAGAATT-3′) NC forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse
5′-ACGUGACACGUUCGGAGAATT-3′).
Additionally, ELF3 plasmid or empty vector was
transfected into cells in the presence or absence of the miR-212
mimic using Lipofectamine 2000. The cells were then cultured for 24
h in six-well plates. The transfected cells were incubated in
complete medium at 37°C. RT-qPCR and western blot analysis were
used to evaluate the transfection efficacy.
RT-qPCR
Total RNA was extracted from the cultured cells and
tissue samples using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
cDNA was synthesized using the SYBR PrimeScript RT-PCR kit (Takara
Bio, Inc.) by incubating the mixture at 37°C for 15 min, 85°C for 5
sec at 4°C. When the temperature reaches 4°C this process ends. The
ABI 7500 system (Applied Biosystems; Thermo Fisher Scientific,
Inc.) was used for PCR and the thermocycling conditions were as
follows: 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec
and 60°C for 34 sec. The relative expression was calculated via the
comparative cycle threshold method and the levels of miR-212 and
ELF3 were normalized using the U6 and GAPDH internal reference
genes, respectively. The following primer sequences were used:
miR-212 (forward, 5′-GGTAACAGTCTCCAGTCA-3′ and reverse,
5′-GCAATTGCACTGGATACG-3′), U6 (forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-AACGCTTCACGAATTTGCGT-3′), GAPDH (forward,
5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse, 5′-TGGTGAAGACGCCAGTGGA-3′),
ELF3 forward, (5′-GGCCCAGACCAAGCCTTAAT-3′ and reverse,
5′-CACTGAAAGCCAGGGCAAAC-3′). Analysis of relative gene expression
data was conducted using RT-qPCR and the 2−ΔΔCq method
(20). All PCR experiments were
performed in triplicate.
Western blot analysis
NPC cells were lysed and proteins were extracted
with RIPA buffer (Beyotime Institute of Biotechnology).
Subsequently, the concentrations of proteins were detected by using
a BCA Protein Assay Kit (Beyotime Institute of Biotechnology). A
total of 15 µg of each protein sample were separated on 10%
SDS-PAGE and transferred onto PVDF membranes. The membranes were
blocked with 5% non-fat milk in TBST buffer for 2 h at room
temperature, and incubated with primary antibodies against ELF3
(1:1,000; cat. no. AF6780; Beyotime Institute of Biotechnology) and
GAPDH (1:2,000; cat. no. MB9231; Biogot Technology Co, Ltd.)
overnight at 4°C. Subsequently, the membranes were incubated with a
secondary antibody conjugated to horseradish-peroxidase for 2 h at
room temperature. The secondary antibody used for ELF3 was an
anti-rabbit one (1:1,000; cat. no. A0208; Beyotime Institute of
Biotechnology) and for GAPDH was an anti-mouse one (1:1,000; cat.
no. A0216; Beyotime Institute of Biotechnology). The targeted
proteins in the membrane were detected using the Ncm-ECL Ultra kit
(New Cell & Molecular Biotech Co., Ltd). Western blotting data
were analyzed using ImageJ (version 1.8.0; National Institutes of
Health).
Luciferase assay
Site-directed mutagenesis was achieved in the
miR-212 binding site of ELF3 mRNA using the QuikChange Lightning
Site-Directed Mutagenesis kit (Agilent Technologies, Inc.) and
TargetScan version 7.2 (http://www.targetscan.org/vert_72/) and miRanda
(http://www.microrna.org/microrna/home.do) databases.
The wild type (WT) or mutant (MUT) 3′-UTR fragment of ELF3 mRNA was
then sub-cloned into a pGL3 luciferase vector (Promega Corporation)
by PCR, which was co-transfected with either miR-212 mimic or
mimic-NC into NPC cells for 36 h in 96-well plates using
Lipofectamine 2000. Subsequently, a dual luciferase assay (Promega
Corporation) was performed. Renilla (Promega Corporation)
activity was used as the internal control.
Cell proliferation assay
Cell proliferation was determined using a cell
counting kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) assay.
Briefly, CCK-8 solution was added to the transfected cells in
96-well plates (1.0×103 cells/well) at days 0–5 and
incubated for 4 h at 37°C. The absorbance of cells was detected at
450 nm using an ELISA plate reader (BioTek Instruments, Inc.). All
experiments were performed in triplicate.
Statistical analysis
All data are presented as the mean ± SEM. The
experiments were repeated 3 times. Differences were assessed by a
paired or unpaired Student's t-test for two group comparisons, as
appropriate, one-way ANOVA followed by Tukey's post hoc test for
multiple comparisons and Chi-square test. Pearson correlation
analysis was used to determine the association between miR-212 and
ELF3 expression. The statistical analyses were conducted using SPSS
18.0 software (SPSS, Inc.). P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-212 is downregulated in NPC cells
and tissues
First, the present study investigated the expression
levels of miR-212 in tissue samples from patients with NPC to
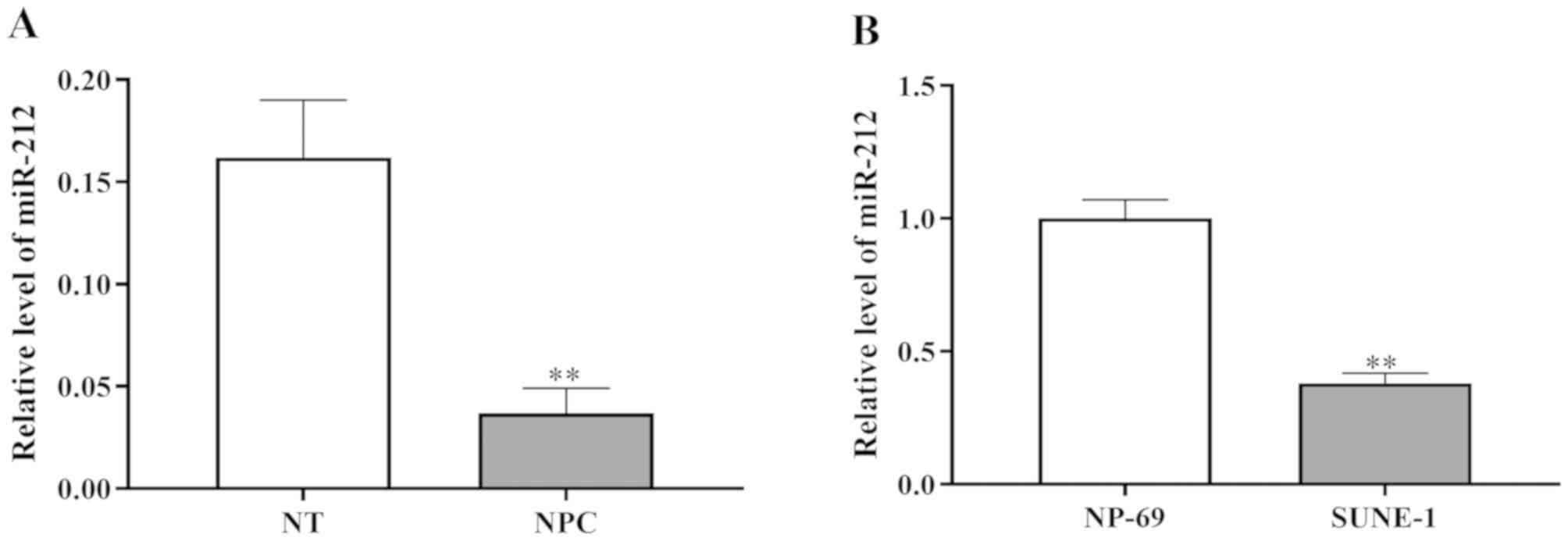
determine the role of miR-212 in NPC. As shown in Fig. 1A, the expression levels of miR-212
were identified to be significantly lower in NPC tissue samples
compared with normal tissue samples.
In addition, the expression level of miR-212 in the
human NPC cell line SUNE-1 was investigated. RT-qPCR revealed that
miR-212 expression was significantly downregulated in SUNE-1 cells
compared with the normal nasopharyngeal epithelial cell line NP69
(Fig. 1B). In summary, miR-212 was
revealed to be downregulated in NPC cells and tissues.
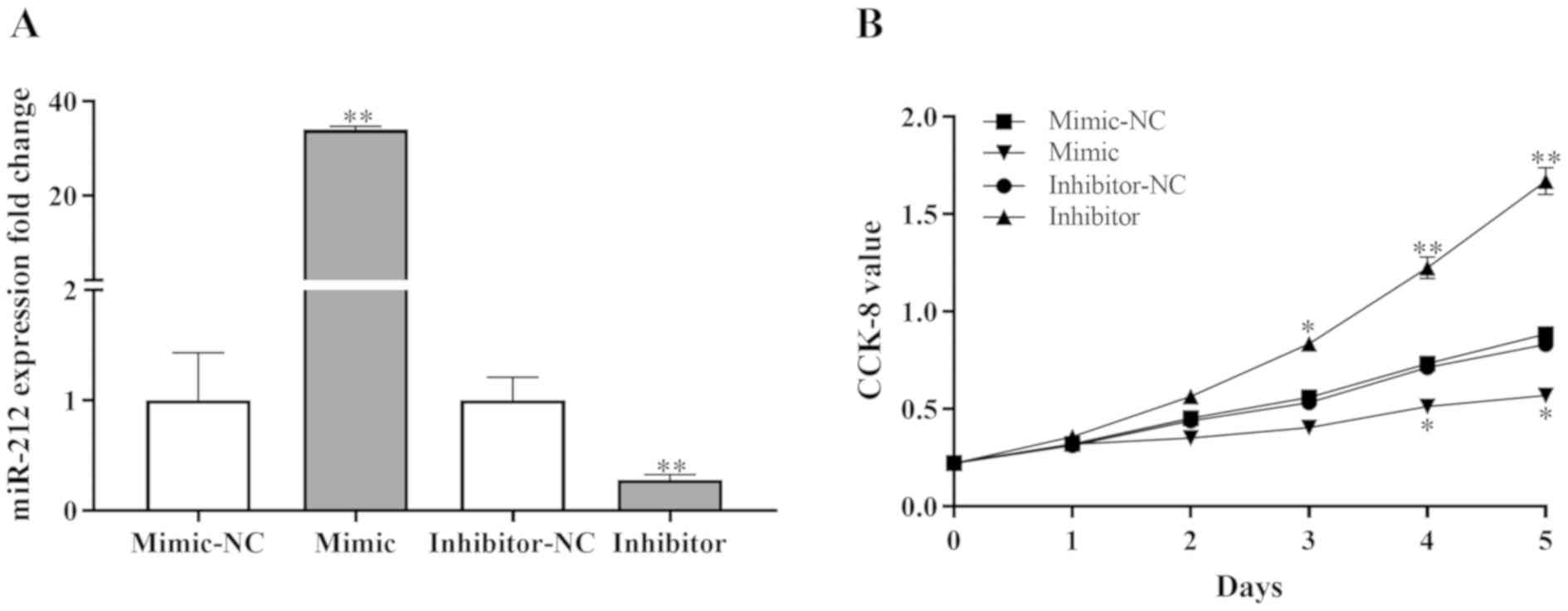
miR-212 inhibits NPC cell
proliferation in vitro
miR-212 mimic, inhibitor and negative controls were
used to evaluate the potential effects of miR-212 on NPC cell
proliferation in vitro. The SUNE-1 cell line was selected as
these cells demonstrated significantly different miR-212 expression
levels compared with normal cells (Fig.
1B). Using RT-qPCR, the efficiency of miR-212 mimic and
inhibitor transfection was assessed. Compared with negative
controls, the miR-212 mimic significantly enhanced the expression
levels of miR-212, and the miR-212 inhibitor significantly reduced
the expression levels of miR-212 in SUNE-1 cells (Fig. 2A).
A CCK-8 assay was performed to examine the effect of
miR-212 on the proliferation of SUNE-1 cells. The results revealed
that miR-212 mimic significantly inhibited proliferation and the
miR-212 inhibitor significantly increased proliferation compared
with negative controls (Fig. 2B).
These results indicated that miR-212 may suppress cell
proliferation in NPC.
ELF3 is a direct target of miR-212 in
NPC
To further elucidate the underlying mechanism of
miR-212 in NPC cells, possible targets of miR-212 were predicted
using the TargetScan and miRanda (http://www.microrna.org/microrna/home.do) databases.
To further confirm the predictions, a luciferase reporter assay was
performed in SUNE-1 cells. 3′-UTR regions of ELF3, which contained
the predicted WT binding site of miR-212 or the MT site, were
cloned into a luciferase vector (Fig.
3A). The results revealed that overexpression of miR-212
significantly decreased the luciferase activity of the WT ELF3
3′-UTR (Fig. 3B), whereas no effect
was observed for the MT ELF3 3-UTR. In addition, transfection with
miR-212 inhibitor increased the luciferase activity of WT ELF3
3′-UTR (Fig. 3C) and had no effect
on the luciferase activity of the MT ELF3 3′-UTR.
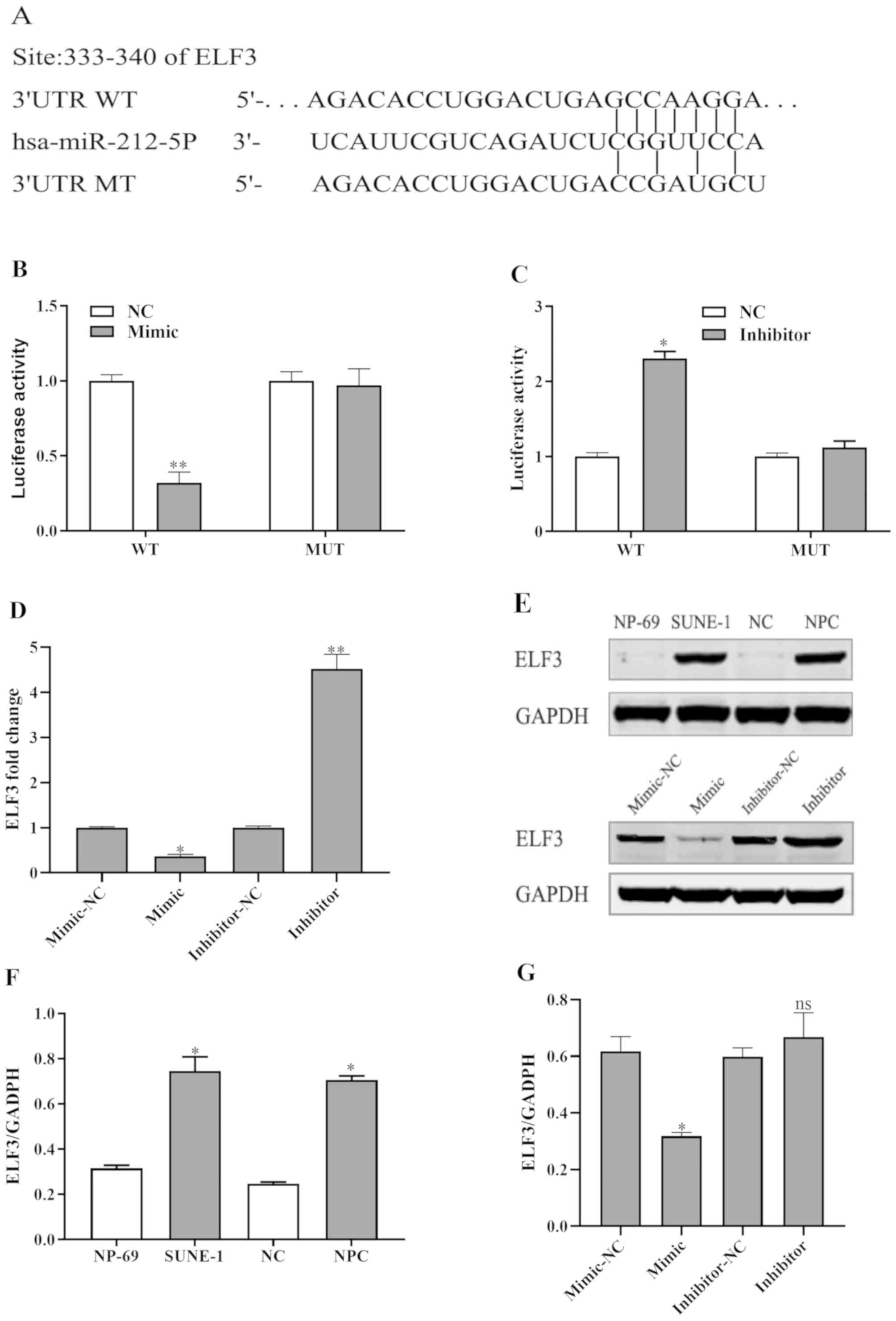 | Figure 3.ELF3 is a direct target of miR-212.
(A) Putative binding site of miR-212 at 333–340 bp of the 3′-UTR of
ELF3. (B) Luciferase activity assay. miR-212 mimic significantly
inhibited the luciferase activity of WT 3′-UTR of ELF3 but had no
marked influence on MT 3′-UTR of ELF3. SUNE-1 cells were
co-transfected with the WT or MT ELF3 3′-UTR construct, along with
the miR-212 inhibitor or NC. At 48 h after transfection, the cells
were lysed for the dual-luciferase reporter assay. (C) miR-212
inhibitor significantly increased the luciferase activity of WT
3′-UTR of ELF3 but had no marked influence on MT 3′-UTR of ELF3.
(D) ELF3 expression fold change was detected by reverse
transcription-quantitative PCR. (E) ELF3 protein expression levels
detected by western blotting. (F) Western blot analysis using
ImageJ v1.8.0 software for NPC tissues and SUNE-1 cells. (G)
miR-212 mimic decreased ELF3 expression and miR-212 inhibitor
increased ELF3 expression. *P<0.05, **P<0.01 vs. the
respective control group. 3′-UTR, 3′-untranslated region; ELF3,
E74-like factor 3; miR-212, microRNA-212; MT, mutant; NC, negative
control; ns, not significant; WT, wild type. |
Furthermore, the miR-212 mimic decreased the protein
and mRNA expression levels of ELF3 and the miR-212 inhibitor
increased the protein and mRNA expression levels of ELF3 compared
with negative controls in SUNE-1 cells (Fig. 3D and E). Additionally, ImageJ
software was used to assess ELF3 expression in NPC tissues and
SUNE-1 cells (Fig. 3F and G). These
results demonstrated that miR-212 could downregulate ELF3 by
directly targeting its 3′-UTR.
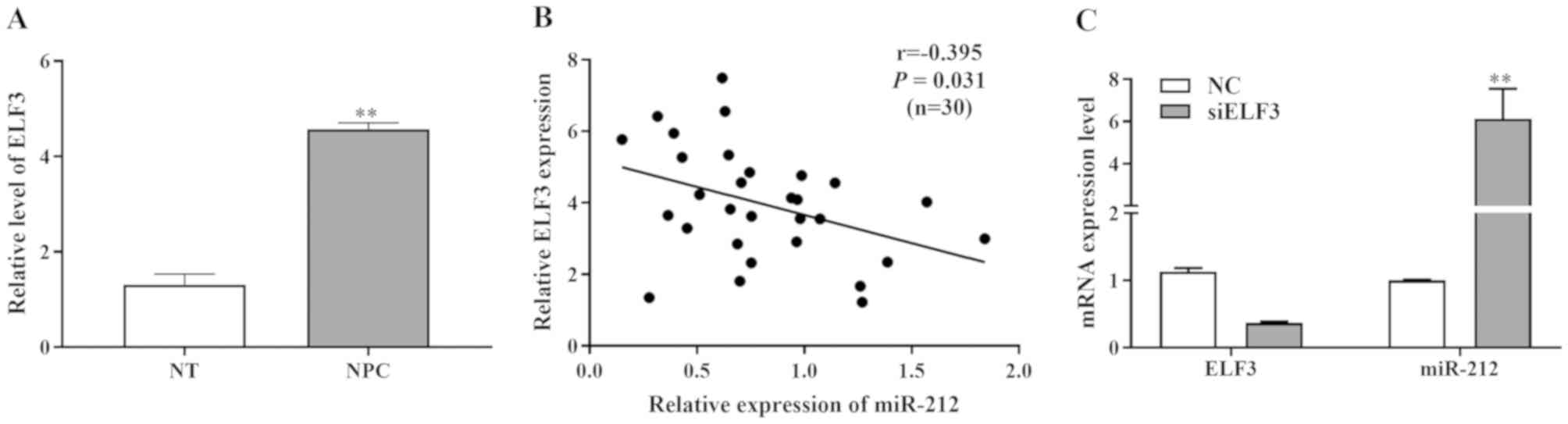
miR-212 expression is inversely
correlated with the expression level of ELF3
To investigate the expression levels of ELF3 in NPC,
30 freshly frozen NPC tissues and normal tissue specimens were
analyzed. Using RT-qPCR, it was identified that the ELF3 expression
was markedly increased in NPC tissues compared with normal adjacent
tissues (Fig. 4A).
In addition, a negative correlation was revealed
between the miR-212 and ELF3 expression levels in NPC tissues
(r=−0.395; P=0.031; Fig. 4B). The
downregulation of miR-212 was found to inversely correlate with the
clinical stage and local or distant metastasis, whereas the
upregulation of ELF3 was directly associated with the clinical
stage and local or distant metastasis in patients with HCC
(Table I). miR-212 and ELF3
expression levels were detected by RT-qPCR following transfection
of ELF3 overexpression vectors, ELF3 siRNA and their respective
negative controls into SUNE-1 cells (Fig. 4C). Overexpression of ELF3
significantly reduced miR-212 mRNA expression, however transfection
of siELF3 significantly increased miR-212 mRNA expression. These
results indicated an inverse association between miR-212 and ELF3
expression in NPC.
Discussion
Dysregulation of mature miRNAs has been reported in
numerous types of cancer. A number of studies have reported that
miRNAs can serve roles as tumor suppressors and oncogenes. Aberrant
expression levels of miRNAs and genes have been detected in NPC in
a number of studies; several miRNAs are upregulated in NPC,
including miR-27a-3p (21), miR-93
(22) and miR-125b (23), and certain miRNAs are downregulated
in NPC, including miR-135a (10),
miR-203a-3p (12) and miR-185
(24). The present study
demonstrated that the expression levels of miR-212 were
significantly lower in NPC tissues and cell lines compared with
normal controls. This indicated that miR-212 was downregulated in
NPC, which is consistent with other studies regarding lung cancer
(13), gastric cancer (14), hepatocellular carcinoma (25), prostate cancer (26) and NPC (15). The results of the present study
suggested that miR-212 served as a tumor suppressor gene in NPC. In
addition, aberrant expression of miR-212 has been demonstrated to
regulate tumor migration and invasion (15). These results indicate that miR-212
may exert pivotal pathological and biological roles, which may
contribute to the progression and growth of NPC.
miRNAs exert their roles by regulating downstream
target genes via binding to the 3′-UTR. Several studies have
revealed a number of target genes of miR-212, including SOX4
(13), paxillin (14), Kruppel like factor 4 (27) and SOX5 (28). The present study identified ELF3 as a
direct target of miR-212 using a luciferase assay and
bioinformatics analysis. miR-212 mimic markedly decreased the ELF3
expression level and miR-212 inhibitor increased the ELF3
expression level. In addition, transfection of SUNE-1 cells with
siELF3 significantly reduced ELF3 mRNA expression, but increased
miR-212 expression. Furthermore, it was demonstrated that the
expression level of miR-212 was low in NPC cells and tissues, and
miR-212 inhibited cell proliferation, possibly by targeting ELF3 in
NPC. miR-212 expression was inversely correlated with ELF3 protein
expression in NPC tissues. Additionally, the relative expression
levels of ELF3 and miR-212 were associated with metastasis and TNM
stage in NPC.
In conclusion, the present study provided an
improved understanding regarding the pathogenesis of NPC. Increased
miR-212 expression was identified to serve a positive role in NPC
by inhibiting the proliferation of NPC cells via ELF3. This may
provide a novel therapeutic target for NPC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YK performed the molecular biology experiments and
drafted the manuscript. YC collected tissue samples and performed
the statistical analysis. MT conceived the design of the study and
drafted the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The human study was approved by the Ethics Committee
of Rui Jin Hospital Affiliated to Shanghai Jiaotong University.
(Shanghai, China). The patients provided written informed consent
prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Han P, Wang X, Liang F, Liu Y, Qiu X, Xu
Y, Chen R, Yu S and Huang X: Osteoradionecrosis of the skull base
in nasopharyngeal carcinoma: Incidence and risk factors. Int J
Radiat Oncol Biol Phys. 102:552–555. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lin K, Qiu F, Chen S, He X, Peng S and
Chen H: Lack of association between the distribution of ABO blood
groups and nasopharyngeal carcinoma in a population of Southern
China. J Cancer Res Ther. 14:785–788. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nakanishi Y, Wakisaka N, Kondo S, Endo K,
Sugimoto H, Hatano M, Ueno T, Ishikawa K and Yoshizaki T:
Progression of understanding for the role of Epstein-Barr virus and
management of nasopharyngeal carcinoma. Cancer Metastasis Rev.
36:435–447. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lin Z, Yang Z, He B, Wang D, Gao X, Tam SY
and Wu VWC: Pattern of radiation-induced thyroid gland changes in
nasopharyngeal carcinoma patients in 48 months after radiotherapy.
PLoS One. 13:e02003102018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hezaveh K, Kloetgen A, Bernhart SH,
Mahapatra KD, Lenze D, Richter J, Haake A, Bergmann AK, Brors B,
Burkhardt B, et al: Alterations of microRNA and microRNA-regulated
messenger RNA expression in germinal center B-cell lymphomas
determined by integrative sequencing analysis. Haematologica.
101:1380–1389. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dallaire A and Simard MJ: The implication
of microRNAs and endo-siRNAs in animal germline and early
development. Dev Biol. 416:18–25. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chai C, Wu H, Wang B, Eisenstat DD and
Leng RP: MicroRNA-498 promotes proliferation and migration by
targeting the tumor suppressor PTEN in breast cancer cells.
Carcinogenesis. 39:1185–1196. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gorbea C, Mosbruger T and Cazalla D: A
viral Sm-class RNA base-pairs with mRNAs and recruits microRNAs to
inhibit apoptosis. Nature. 550:275–279. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen D, Hu S, Wu Z, Liu J and Li S: The
role of MiR-132 in regulating neural stem cell proliferation,
differentiation and neuronal maturation. Cell Physiol Biochem.
47:2319–2330. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang LX, Kang ZP, Yang ZC, Ma RX, Tan Y,
Peng XB, Dai RZ, Li J, Yu Y and Xu M: MicroRNA-135a inhibits
nasopharyngeal carcinoma cell proliferation through targeting
interleukin-17. Cell Physiol Biochem. 46:2232–2238. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bao L, You B, Shi S, Shan Y, Zhang Q, Yue
H, Zhang J, Zhang W, Shi Y, Liu Y, et al: Metastasis-associated
miR-23a from nasopharyngeal carcinoma-derived exosomes mediates
angiogenesis by repressing a novel target gene TSGA10. Oncogene.
37:2873–2889. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiang N, Jiang X, Chen Z, Song X, Wu L,
Zong D, Song D, Yin L, Wang D, Chen C, et al: MiR-203a-3p
suppresses cell proliferation and metastasis through inhibiting
LASP1 in nasopharyngeal carcinoma. J Exp Clin Cancer Res.
36:1382017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tang T, Huan L, Zhang S, Zhou H, Gu L,
Chen X and Zhang L: MicroRNA-212 functions as a tumor-suppressor in
human non-small cell lung cancer by targeting SOX4. Oncol Rep.
38:2243–2250. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li D, Li Z, Xiong J, Gong B, Zhang G, Cao
C, Jie Z, Liu Y, Cao Y, Yan Y, et al: MicroRNA-212 functions as an
epigenetic-silenced tumor suppressor involving in tumor metastasis
and invasion of gastric cancer through down-regulating PXN
expression. Am J Cancer Res. 5:2980–2997. 2015.PubMed/NCBI
|
|
15
|
Jiang C, Wang H, Zhou L, Jiang T, Xu Y and
Xia L: MicroRNA-212 inhibits the metastasis of nasopharyngeal
carcinoma by targeting SOX4. Oncol Rep. 38:82–88. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ma C, Nong K, Wu B, Dong B, Bai Y, Zhu H,
Wang W, Huang X, Yuan Z and Ai K: miR-212 promotes pancreatic
cancer cell growth and invasion by targeting the hedgehog signaling
pathway receptor patched-1. J Exp Clin Cancer Res. 33:542014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Oliver JR, Kushwah R and Hu J: Multiple
roles of the epithelium-specific ETS transcription factor, ESE-1,
in development and disease. Lab Invest. 92:320–330. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang JL, Chen ZF, Chen HM, Wang MY, Kong
X, Wang YC, Sun TT, Hong J, Zou W, Xu J and Fang JY: Elf3 drives
β-catenin transactivation and associates with poor prognosis in
colorectal cancer. Cell Death Dis. 5:e12632014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng L, Xu M, Xu J, Wu K, Fang Q, Liang
Y, Zhou S, Cen D, Ji L, Han W and Cai X: ELF3 promotes
epithelial-mesenchymal transition by protecting ZEB1 from
miR-141-3p-mediated silencing in hepatocellular carcinoma. Cell
Death Dis. 9:3872018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li L and Luo Z: Dysregulated miR-27a-3p
promotes nasopharyngeal carcinoma cell proliferation and migration
by targeting Mapk10. Oncol Rep. 37:2679–2687. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang Y and Xu Z: miR-93 enhances cell
proliferation by directly targeting CDKN1A in nasopharyngeal
carcinoma. Oncol Lett. 15:1723–1727. 2018.PubMed/NCBI
|
|
23
|
Li LN, Xiao T, Yi HM, Zheng Z, Qu JQ,
Huang W, Ye X, Yi H, Lu SS, Li XH and Xiao ZQ: MiR-125b increases
nasopharyngeal carcinoma radioresistance by targeting A20/NF-κB
signaling pathway. Mol Cancer Ther. 16:2094–2106. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cheng JZ, Chen JJ, Wang ZG and Yu D:
MicroRNA-185 inhibits cell proliferation while promoting apoptosis
and autophagy through negative regulation of TGF-β1/mTOR axis and
HOXC6 in nasopharyngeal carcinoma. Cancer Biomark. 23:107–123.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jia P, Wei G, Zhou C, Gao Q, Wu Y, Sun X
and Li X: Upregulation of MiR-212 inhibits migration and
tumorigenicity and inactivates Wnt/β-catenin signaling in human
hepatocellular carcinoma. Technol Cancer Res Treat.
17:15330346187652212018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hu B, Jin X and Wang J: MicroRNA-212
targets mitogen-activated protein kinase 1 to inhibit proliferation
and invasion of prostate cancer cells. Oncol Res. 26:1093–1102.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Song Y, Liu Y and Chen X: MiR-212
attenuates MPP+-induced neuronal damage by targeting
KLF4 in SH-SY5Y cells. Yonsei Med J. 59:416–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu Y, Zhang XL, Li XF, Tang YC and Zhao
X: miR-212-3p reduced proliferation, and promoted apoptosis of
fibroblast-like synoviocytes via down-regulating SOX5 in rheumatoid
arthritis. Eur Rev Med Pharmacol Sci. 22:461–471. 2018.PubMed/NCBI
|