Introduction
Ovarian cancer (OC) is one of the most frequent
causes of mortality associated with gynecologic malignancy, and the
fifth leading cause of health issues among women and
cancer-associated deaths worldwide during the past 2 decades
(1,2). A large number of patients (>50%) are
diagnosed at an advanced stage, mainly due to the asymptomatic
development of OC (3). Currently,
the therapeutic strategies for OC consist of radical surgical
resection, chemotherapy based on taxanes and platinum, and targeted
therapeutic management (4). Despite
the aforementioned treatments, the overall survival (OS) rate
remains at only ~30% (5), partially
due to drug resistance and a lack of specific biomarkers that can
be used to detect the disease. Therefore, it is urgent to identify
favorable prognostic factors of OC to improve the clinical outcomes
of patients.
Sirtuins (SIRTs) are a family of deacetylases that
comprises seven types in mammals (SIRT1-7), with different
subcellular localization patterns and enzymatic activities
(6). Since the discovery of SIRTs,
the seven members, activated by nicotinamide adenine dinucleotide,
have been closely associated with an extended life span by
counteracting oxidative damage (7).
Therefore, SIRTs could contribute greatly to aging (8). Among the seven identified SIRTs, SIRT1
is located in the nucleus; SIRT2 is located in the cytoplasm;
SIRT3, SIRT4 and SIRT5 are localized in the mitochondria; and SIRT6
and SIRT7 are present in the nucleus. Notably, due to the unique
ability of SIRTs to control the redox environment, accumulating
evidence has demonstrated that SIRTs are involved in the pathology
of various cancer types such as lung cancer, prostate cancer,
gastric cancer and breast cancer (9–13). More
specifically, previous studies have reported that SIRTs act as
independent prognostic factors of several carcinomas, including
colorectal cancer and non-small cell lung cancer (14,15). To
date, few individual SIRTs have been reported to be associated with
OC. Shuang et al (16) found
that SIRT1 could contribute to chemoresistance and the invasive
capacity of OC cells, thereby boosting the proliferation of OC.
Additionally, silencing of SIRT1 increases the protein expression
of estrogen receptor β, which is regarded as an effective inhibitor
of OC cells (17). On the other
hand, SIRT3 exerts an antitumor effect on the induction of
mitochondrial-dependent apoptosis via 5′ AMP-activated protein
kinase activation in OC cells (18).
Regarding SIRT6, its dual roles as a tumor oncogene and suppressor
in OC remain ambiguous (19,20). Furthermore, the prognostic values of
the SIRT family in OC remain to be elucidated. In the present
study, using the Kaplan-Meier (KM) plotter, the prognostic
significance of the SIRT transcription family was comprehensively
investigated in patients with OC.
Materials and methods
Acquisition of data and statistical
analysis
The prognostic values of individual SIRT mRNA levels
from 1,657 patients with OC were investigated using the online KM
plotter (http://kmplot.com/analysis) database.
Until now, 54,675 genes are included in the database and thus can
be examined to analyze the survival of patients with breast cancer
(21), lung cancer (22), OC (23) and gastric cancer. In the present
study, OS, progression-free survival (PFS) and post-progression
survival (PPS) of patients with primary epithelial OC were assessed
using the KM survival plot. Furthermore, clinical characteristics,
including two main primary epithelial OC histologies, stage, grade,
tumor protein p53 (TP53) mutation status and treatment choice were
analyzed. Generally, seven SIRT subtypes (SIRT1, SIRT2, SIRT3,
SIRT4, SIRT5, SIRT6 and SIRT7) were input into the database
(http://kmplot.com/analysis/index.php?p=service&cancer=ovar)
to generate KM survival plots. Individuals were divided into two
groups (high expression group and low expression group), according
to the median expression of the SIRT gene. The hazard ratios (HRs)
with 95% CIs and log-rank P values were illustrated with the Cox
proportional hazard model in the database. P<0.05 was considered
to indicate a statistically significant difference.
Tumor xenograft model
A2780 OC cells were purchased from American type
culture collection and cultured to establish a nude mice tumor
model. For culture, Dulbecco's modified Eagle's medium (DMEM)
(Thermo Fisher Scientific, Inc.) containing streptomycin (100
µg/ml), penicillin (100 U/ml) and 10% FBS (Thermo Fisher
Scientific, Inc.) were used. The medium was replaced every 2 days,
and the cells were incubated in a moist atmosphere containing 5%
CO2 at 37°C. Once adherent cells had grown to ~90%
confluence, the cells were digested with 0.25% trypsin-0.02% EDTA
for subculture and subsequent experimental treatment.
The athymic nude mice (BALB/C-nu/nu; age, 6 weeks;
male; weight, 18–22 g) were purchased from Shanghai SLAC Laboratory
Animal Co., Ltd. and bred in pathogen-free conditions under 22°C,
12 h light/12 h dark, with free access to sterile water and food,
in the Wenzhou Medical University Laboratory Animal Center
(Wenzhou, China). A total of 15 randomized nude mice were used for
the tumor xenograft model. The mice were anesthetized with, and
1×107 A2780 OC cells (in 100 µl PBS) were
injected subcutaneously into the armpit of each nude mouse. After 3
weeks, the mice were euthanized with 2% isoflurane excess carbon
dioxide with the flow rate of 3l/min. The animal study was approved
by the Wenzhou Medical University Ethics Committee (approval no.
wydw2019-0214).
Immunohistochemical staining
The tumor tissues were harvested for
immunohistochemistry. Tumor tissues were fixed in 4%
paraformaldehyde at 25°C overnight, dehydrated with different
concentrations of ethanol (75, 85, 95 and 100%) and 100%
dimethylbenzene separately, and embedded in paraffin. The specimens
were subsequently cut into 4-µm thick sections. The sections
were rehydrated by placing them in a descending alcohol series
(100, 90, 85 and 75%) and ddH20 for 5 min. Subsequently,
the sections were washed with PBS for 5 min. The washing step was
repeated twice more. Following that, slides were placed in 0.01 M
sodium citrate buffer (Merck KGaA) at 95°C for 5 min and then
cooled to room temperature. The slides were blocked with 10% bovine
serum albumin (BSA; Merck KGaA) for 1 h at 37°C. Tissue sections
were incubated with SIRT1 (1:200; cat. no. 13161-1-AP; Proteintech
Group, Inc.), SIRT3 (1:200; cat. no. ab217319; Abcam) and SIRT6
(1:200; cat. no. 13572-1-AP; ProteinTech Group, Inc.) primary
antibodies overnight at 4°C. Secondary antibody conjugated to
horseradish peroxidase (1:200; cat. no. PV-6001; Origene
Technologies, Inc.) was then added to the slides for 1 h at 37°C
and 3,3′-diaminobenzidine were subsequently added to the slices for
2 min. The sections were washed with PBS 3 times. The sections were
dryed and stained with hematoxylin for 5 min followed by staining
with differentiation solution for 1 min. The sections were washed
once with running water for 10 min and covered with a coverslip of
neutral resin. An inverted light microscope was used to observe the
expression of SIRT1, SIRT3 and SIRT6 in the tumor tissues at a
magnification of ×200.
Results
Multivariate analysis and survival
outcomes of patients with OC based on the expression of SIRTs
KM survival data on all seven SIRT members examined
in the present study can be acquired from www.kmplot.com. Firstly, the prognostic value of SIRT1
(Affymetrix ID, 218878_s_at) was evaluated. OS, PFS and PPS curves
were generated for all patients with OC (Fig. 1A). High SIRT1 expression was
significantly associated with worse OS (P=0.0029; HR, 1.22; 95% CI,
1.07–1.39) and PFS (P=0.016; HR, 1.17; 95% CI, 1.03–1.33). However,
there was no association identified between SIRT1 and PPS (P=0.2;
HR, 1.12; 95% CI, 0.94–1.34). In the present study, the two common
histological subtypes of ovarian cancer (endometrioid and serous
cancer) were used for subsequent analyses. Furthermore, the OS
curves of patients with different OC subtypes were plotted
(Fig. 1B). The results demonstrated
that the OS of patients with endometrioid cancer (P=0.053; HR,
4.94; 95% CI, 0.82–29.69) or serous cancer (P=0.074; HR, 1.15; 95%
CI, 0.99–1.34) was not associated with SIRT1 mRNA expression.
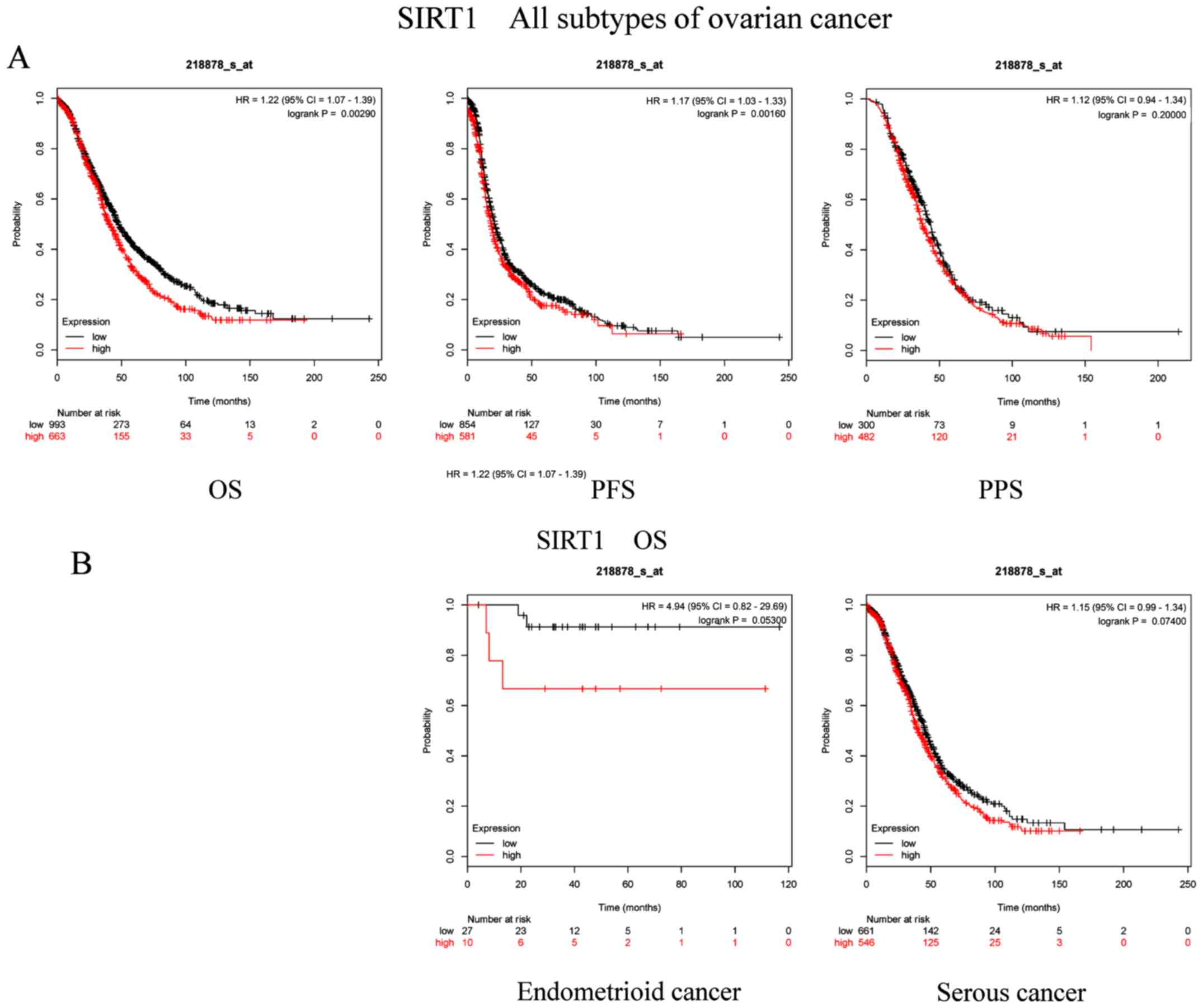 | Figure 1.Prognostic significance of SIRT1 mRNA
expression in OC patients. Prognostic significance of SIRT1 mRNA
expression (A) in all patients with OC and (B) in patients with
different subtypes of OC. All patients with OC, n=1,656; patients
with endometrioid cancer, n=37; patients with serous cancer,
n=1,207. OC, ovarian cancer; SIRT, sirtuin; OS, overall survival;
PFS, progression-free survival; PPS, post-progression survival; HR,
hazard ratio. |
Furthermore, the prognostic significance of SIRT2
mRNA expression (Affymetrix ID, 220605_s_at) was analyzed. Elevated
SIRT2 levels were significantly associated with PFS (P=0.025; HR,
0.85; 95% CI, 0.74–0.98) and PPS (P=0.011; HR, 1.27; 95% CI,
1.06–1.53) in patients with OC (Fig.
2A). By contrast, mRNA expression levels of SIRT2 in patients
with OC did not exhibit any association with OS (P=0.4; HR, 0.95;
95% CI, 0.83–1.08). Histological subtype outcomes indicated that
SIRT2 expression had no effect on the OS of patients with
endometrioid cancer (P=0.19; HR, 3.84; 95% CI, 0.43–34.41) or
serous cancer (P=0.17; HR, 1.13; 95% CI, 0.95–1.33; Fig. 2B).
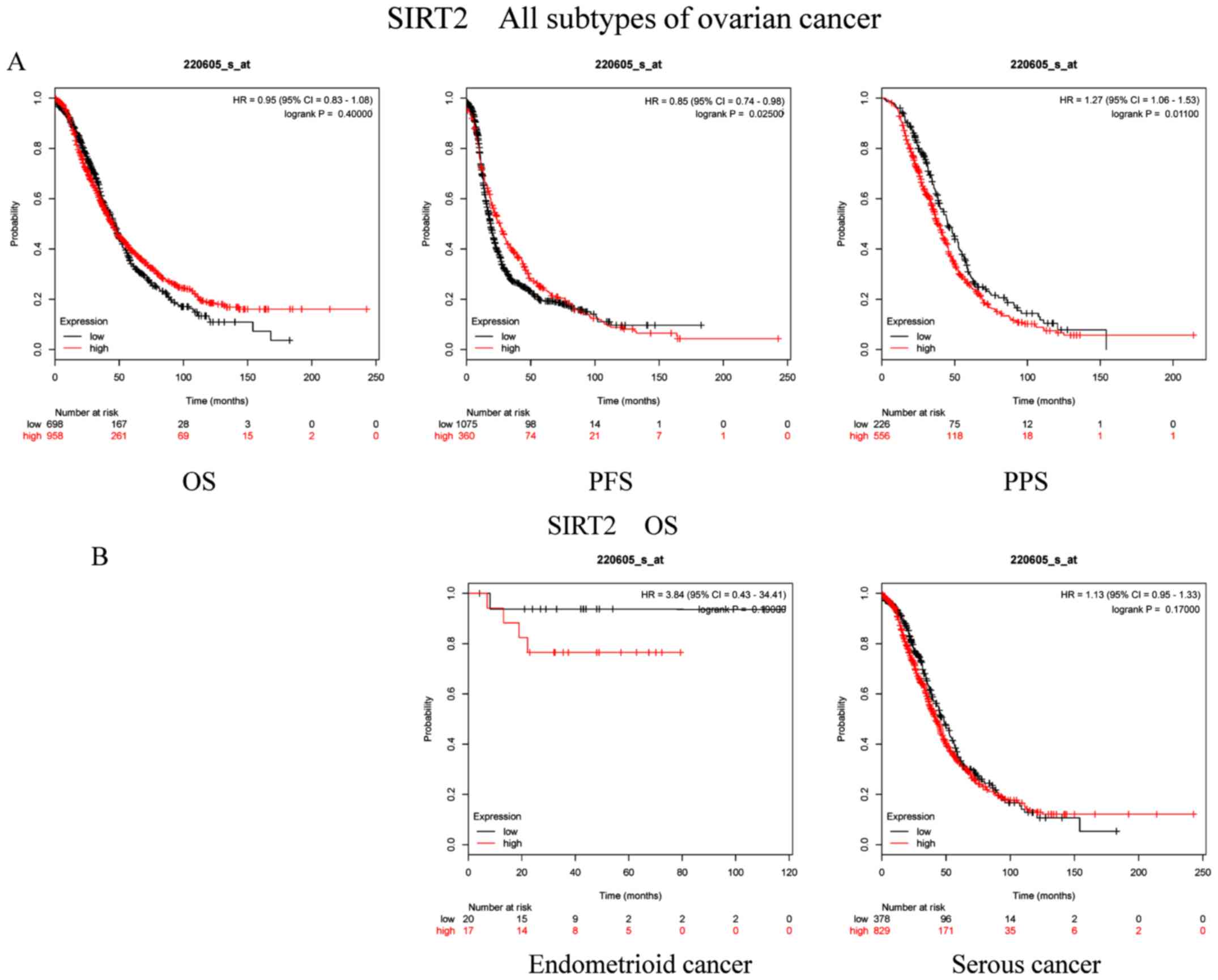 | Figure 2.Prognostic significance of SIRT2 mRNA
expression in OC patients. Prognostic significance of SIRT2 mRNA
expression (A) in all patients with OC and (B) in patients with
different subtypes of OC. All patients with OC, n=1,656; patients
with endometrioid cancer, n=37; patients with serous cancer,
n=1,207. OC, ovarian cancer; SIRT, sirtuin; OS, overall survival;
PFS, progression-free survival; PPS, post-progression survival; HR,
hazard ratio. |
Additionally, the prognostic role of SIRT3 mRNA
expression (Affymetrix ID, 221913_at) was examined (Fig. 3A and B). High expression of SIRT3 was
associated with favorable OS (P=0.00093; HR, 0.8; 95% CI, 0.7–0.91)
and PPS (P=0.045; HR, 0.84; 95% CI, 0.71–1.00) in patients with OC.
However, there was no significant association between PFS and SIRT3
mRNA levels in patients with OC (P=0.083; HR, 0.89; 95% CI,
0.77–1.02). With regard to serous cancer, high expression of SIRT3
exhibited an evident effect on OS among patients with OC
(P=0.00962; HR, 0.82; 95% CI, 0.7–0.95). Furthermore, there was no
significant association between the SIRT3 mRNA levels and the OS of
patients with endometrioid cancer (P=0.38; HR, 0.46; 95% CI,
0.08–2.75).
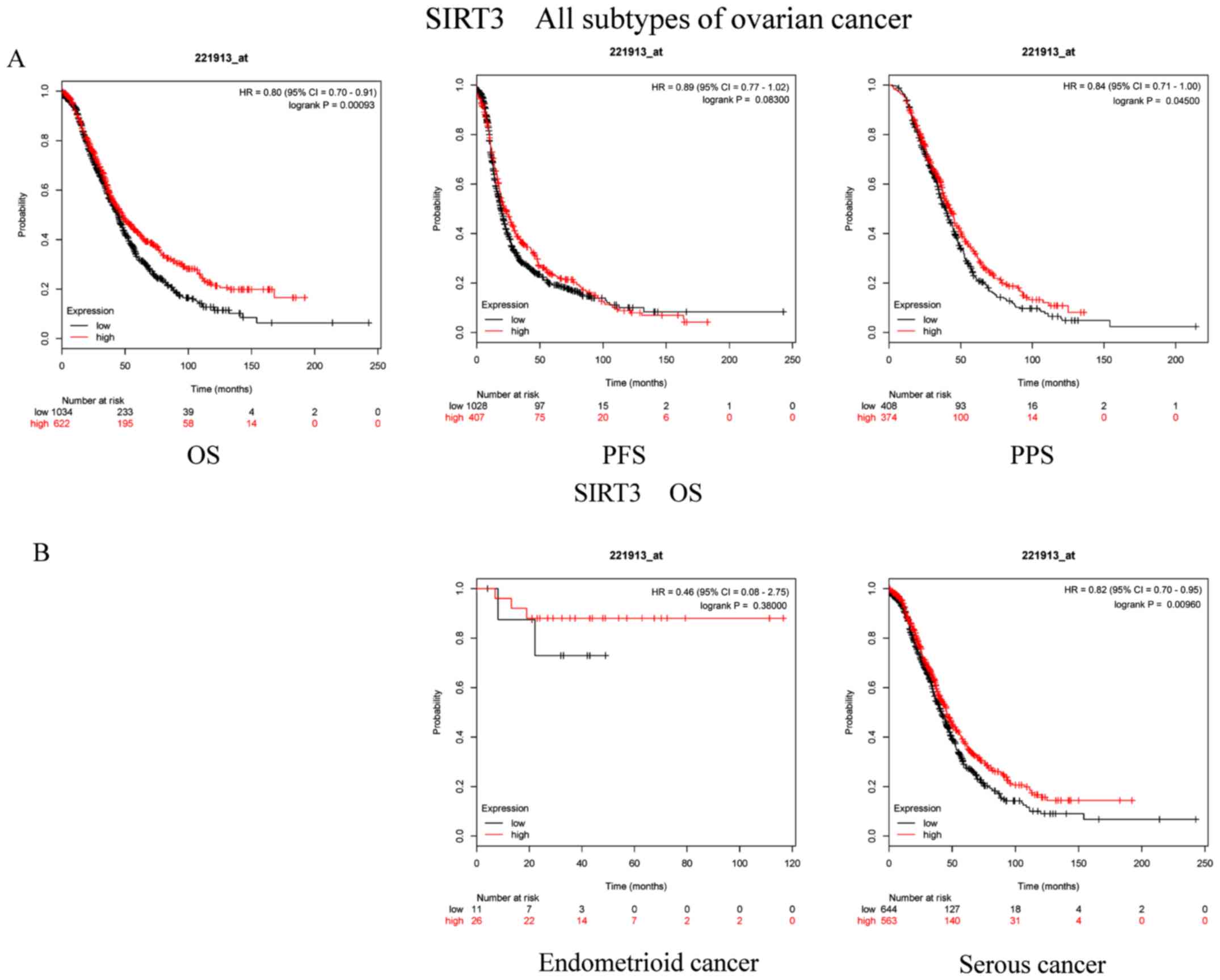 | Figure 3.Prognostic significance of SIRT3 mRNA
expression in OC patients. Prognostic significance of SIRT3 mRNA
expression (A) in all patients with OC and (B) in patients with
different subtypes of OC. All patients with OC, n=1,656; patients
with endometrioid cancer, n=37; patients with serous cancer,
n=1,207. OC, ovarian cancer; SIRT, sirtuin; OS, overall survival;
PFS, progression-free survival; PPS, post-progression survival; HR,
hazard ratio. |
Subsequently, the prognostic implications of SIRT4
mRNA expression (Affymetrix ID, 220047_at) were explored. High mRNA
expression levels of SIRT4 were significantly associated with
unfavorable OS (P=0.0012; HR, 1.24; 95% CI, 1.09–1.41), PFS
(P=0.00017; HR, 1.27; 95% CI, 1.12–1.44) and PPS (P=0.013; HR,
1.24; 95% CI, 1.05–1.47) in patients with OC (Fig. 4A). Additionally, increased SIRT4 mRNA
expression also indicated poor OS in patients with both
endometrioid cancer (P=0.016; HR, 9.36; CI, 1.04–84.6) and serous
cancer (P=0.011; HR, 1.22; 95% CI, 1.05–1.42; Fig. 4B).
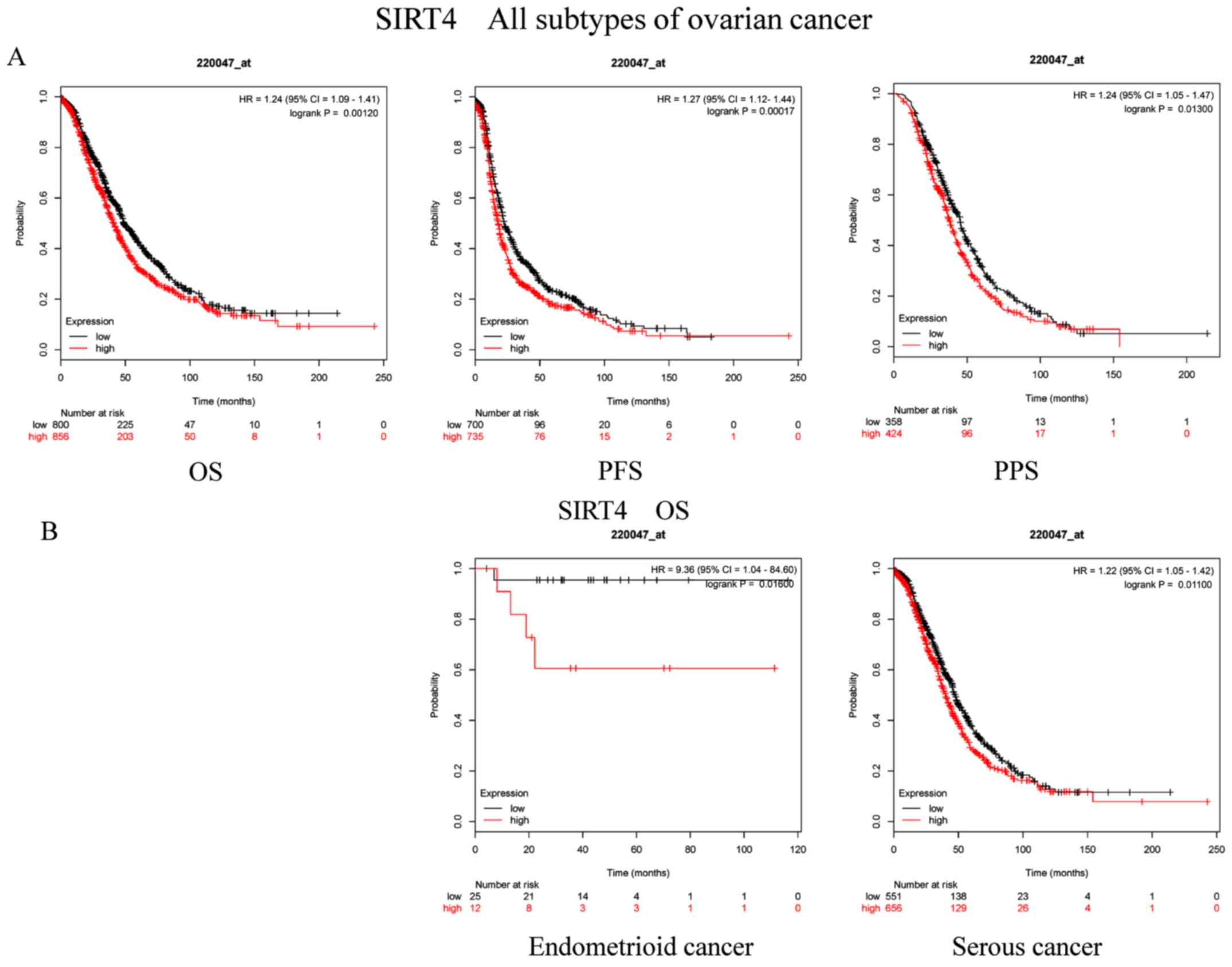 | Figure 4.Prognostic significance of SIRT4 mRNA
expression in OC patients. Prognostic significance of SIRT4 mRNA
expression (A) in all patients with OC and (B) in patients with
different subtypes of OC. All patients with OC, n=1,656; patients
with endometrioid cancer, n=37; patients with serous cancer,
n=1,207. OC, ovarian cancer; SIRT, sirtuin; OS, overall survival;
PFS, progression-free survival; PPS, post-progression survival; HR,
hazard ratio. |
The prognostic importance of SIRT5 mRNA expression
(Affymetrix ID, 229112_at) was subsequently examined.
Interestingly, enhanced SIRT5 mRNA levels were notably associated
with improved OS (P=0.048; HR, 0.81; 95% CI, 0.66–1.00) and PPS
(P=0.0011; HR, 0.66; 95% CI, 0.52–0.85), but with poor PFS
(P=0.0018; HR, 1.36; 95% CI, 1.12–1.65; Fig. 5A). Additionally, Fig. 5B reveals the association between
SIRT5 and the OC subtypes. It was clear that the upregulation of
SIRT5 mRNA expression was notably associated with favorable OS in
serous cancer (P=0.036; HR, 0.78; 95% CI, 0.62–0.98). However,
SIRT5 mRNA expression was not associated with OS in patients with
endometrioid cancer (P=0.32; HR, 3.01; 95% CI, 0.31–29).
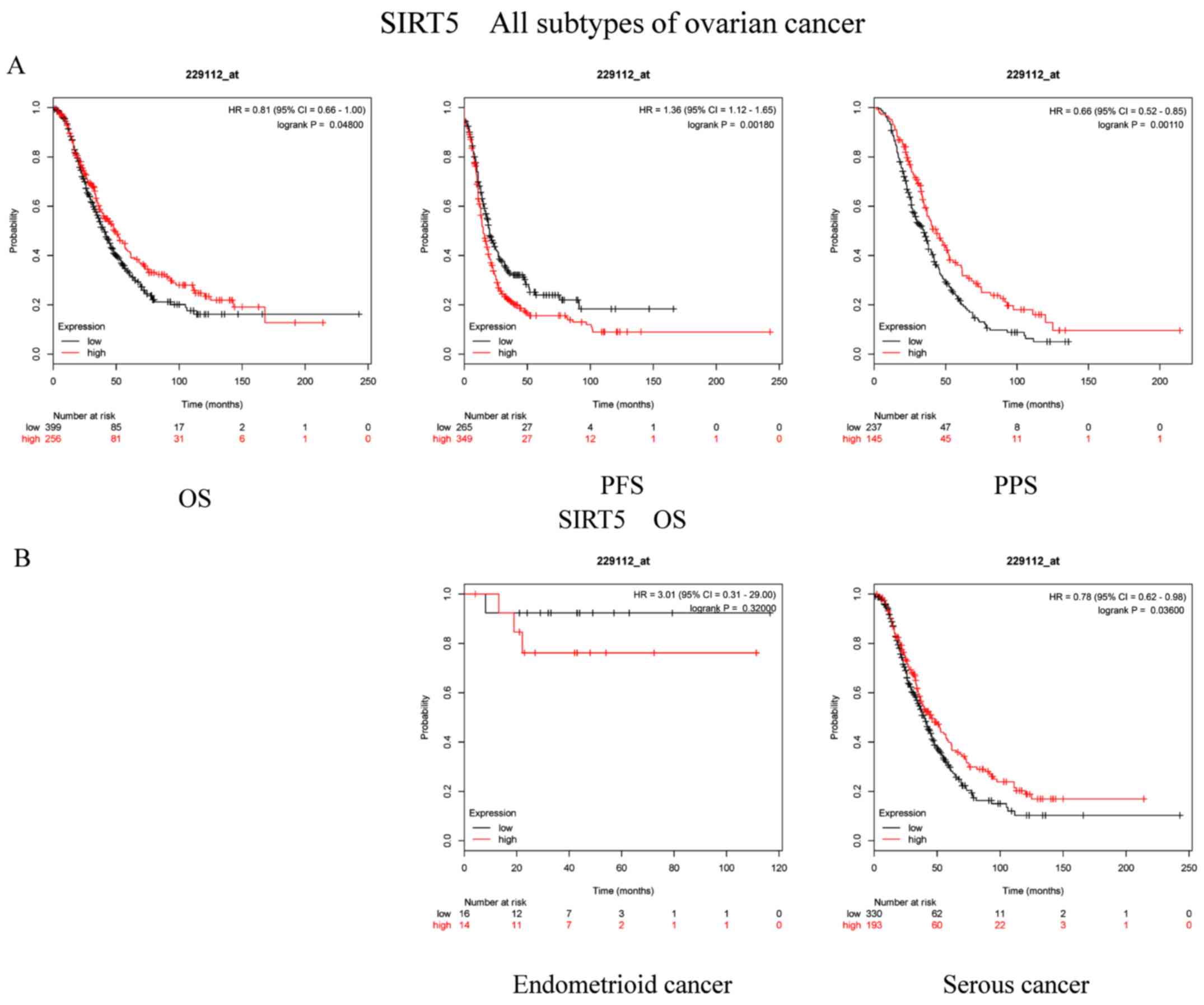 | Figure 5.Prognostic significance of SIRT5 mRNA
expression in OC patients. Prognostic significance of SIRT5 mRNA
expression (A) in all patients with OC and (B) in patients with
different subtypes of OC. All patients with OC, n=1,656; patients
with endometrioid cancer, n=37; patients with serous cancer,
n=1,207. OC, ovarian cancer; SIRT, sirtuin; OS, overall survival;
PFS, progression-free survival; PPS, post-progression survival; HR,
hazard ratio. |
Subsequently, the prognostic significance of SIRT6
mRNA expression levels (Affymetrix ID, 219613_s_at) was
investigated. High SIRT6 expression was significantly associated
with improved OS (P=0.0012; HR, 0.79; 95% CI, 0.69–0.91) and PFS
(P=0.00042; HR, 0.79; 95% CI, 0.69–0.90) in patients with OC.
Despite these results, SIRT6 exhibited no effect on PPS (P=0.29;
HR, 0.91; 95% CI, 0.76–1.09) in patients with OC (Fig. 6A). With respect to the histological
subtype of OC, elevated SIRT6 mRNA expression was associated with
good OS in patients with serous cancer (P=0.0062; HR, 0.81; 95% CI,
0.69–0.94), but not in patients with endometrioid cancer (P=0.069;
HR, 0.17; 95% CI, 0.02–1.50; Fig.
6B).
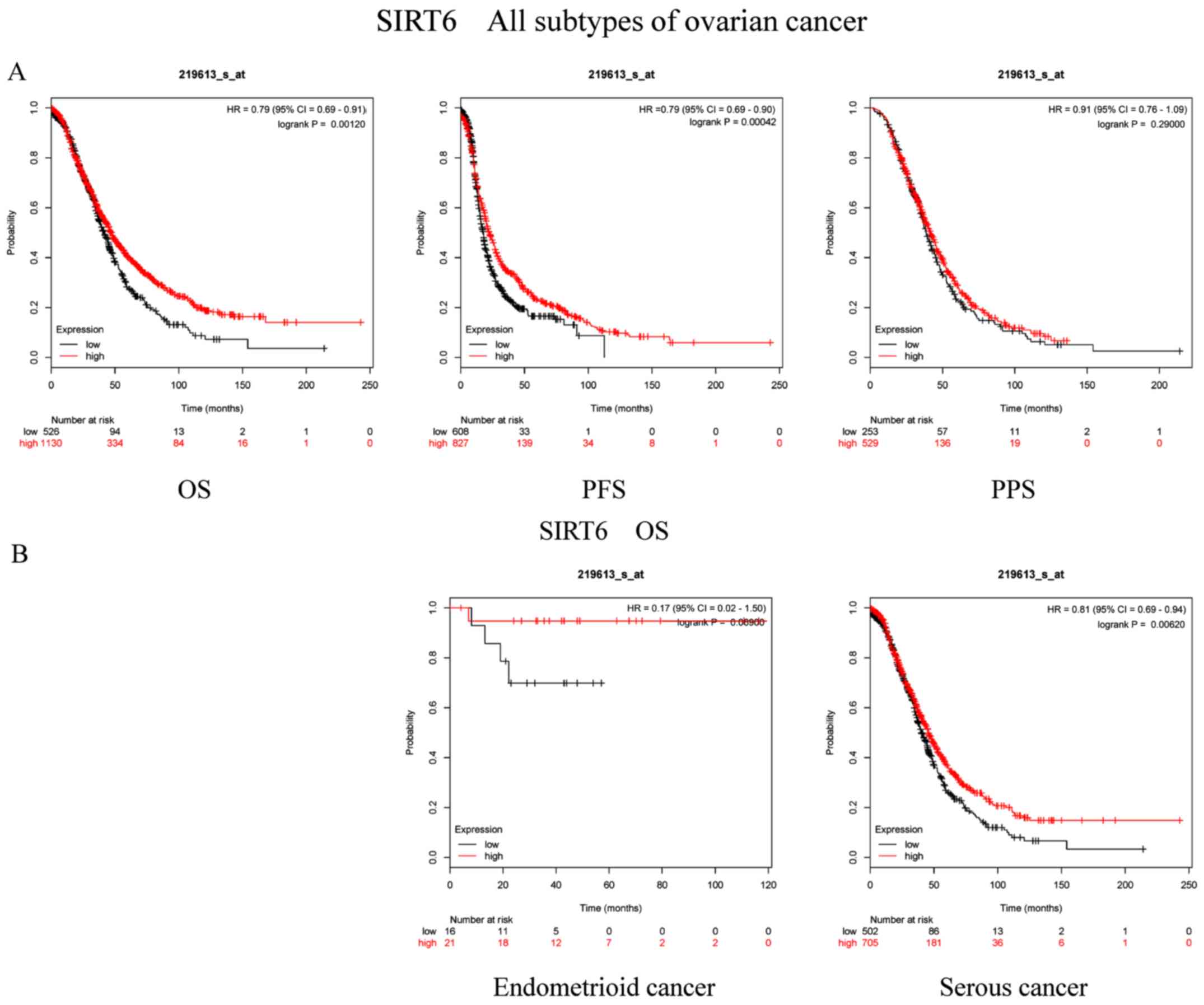 | Figure 6.Prognostic significance of SIRT6 mRNA
expression in OC patients. Prognostic significance of SIRT6 mRNA
expression (A) in all patients with OC and (B) in patients with
different subtypes of OC. All patients with OC, n=1,656; patients
with endometrioid cancer, n=37; patients with serous cancer,
n=1,207. OC, ovarian cancer; SIRT, sirtuin; OS, overall survival;
PFS, progression-free survival; PPS, post-progression survival; HR,
hazard ratio. |
Finally, the prognostic role of SIRT7 (Affymetrix
ID, 218797_s_at) was studied. High SIRT7 mRNA expression was
significantly associated with good OS (P=5.3×10−5; HR,
0.76; 95% CI, 0.67–0.87) and PFS (P=0.0022; HR, 0.81; 95% CI,
0.71–0.93) in all patients with OC. However, there was no
association between SIRT7 expression and PPS (P=0.2; HR, 0.88; 95%
CI, 0.73–1.07) in patients diagnosed with OC (Fig. 7A). Regarding the subtypes, increased
SIRT7 mRNA levels were significantly associated with improved OS in
patients with serous cancer (P=0.0044; HR, 0.8; 95% CI, 0.69–0.93),
but not in patients with endometrioid cancer [P=0.18; HR,
294,193,658.36 (0-inf); Fig.
7B].
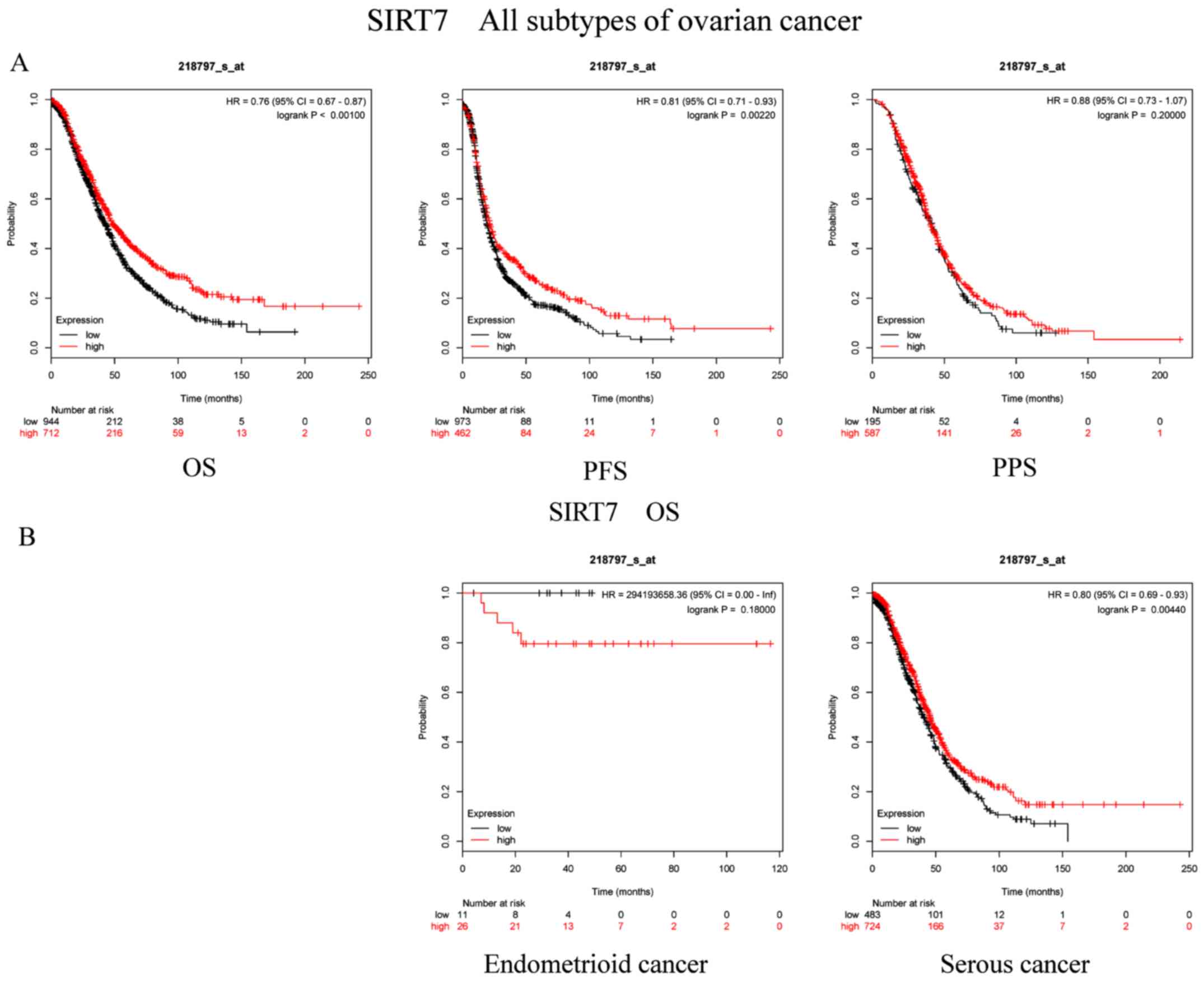 | Figure 7.Prognostic significance of SIRT7 mRNA
expression in OC patients. Prognostic significance of SIRT7 mRNA
expression in (A) all patients with OC and (B) in patients with
different subtypes of OC. All patients with OC, n=1,656; patients
with endometrioid cancer, n=37; patients with serous cancer,
n=1,207. OC, ovarian cancer; SIRT, sirtuin; OS, overall survival;
PFS, progression-free survival; PPS, post-progression survival; HR,
hazard ratio. |
Associations between high mRNA
expression levels of SIRT members and other clinicopathological
characteristics
Additionally, the associations of each SIRT family
member with pathological grade (Table
I), clinical stage (Table II),
TP53 mutation status (Table III)
and chemotherapy choice (Table IV)
were determined. Table II shows
that SIRT1 and SIRT4 were significantly associated with poor OS in
patients with stage III/IV OC, whereas SIRT3 and SIRT5 predicted
improved OS. Moreover, SIRT6 and SIRT7 were associated with
favorable OS in patients with stage I/II OC, as well as those with
stage III/IV. Consistent to the KM outcomes, SIRT2 exhibited no
association with OS in patients with stage I/II or III/IV. With
regard to the pathological grade in Table I, SIRT2 and SIRT4 indicated poor OS
in pathological grades I and III, respectively. However, SIRT3 and
SIRT7 were associated with significantly improved OS in patients
with OC of pathological grades II and III, respectively. Elevated
levels of SIRT6 exhibited a significant association with improved
OS in both pathological grades II and III. In terms of TP53
mutation (Table III), the results
demonstrated the significant associations of SIRT2 and SIRT5 with
OS in patients with OC that have a TP53 mutation. In addition,
SIRT3 was associated with favorable OS in patients with mutant and
wild-type TP53. With the exception of SIRT2 and SIRT5, the
associations between high mRNA expression levels of other SIRT
family members and chemotherapy agents were significant (Table IV).
 | Table I.Associations between elevated SIRT
mRNA expression and overall survival of all patients with ovarian
cancer, according to different pathological grades. |
Table I.
Associations between elevated SIRT
mRNA expression and overall survival of all patients with ovarian
cancer, according to different pathological grades.
| SIRT | Pathological
grade | Cases, n | HR (95% CI) | P-value |
|---|
| SIRT1 | I | 56 | 0.68
(0.25–1.68) | 0.4498 |
|
| II | 324 | 1.29
(0.95–1.77) | 0.1025 |
|
| III | 1,015 | 1.18
(0.99–1.41) | 0.0719 |
| SIRT2 | I | 56 | 3.04
(0.98–9.38) | 0.0425 |
|
| II | 324 | 0.82
(0.58–1.15) | 0.2536 |
|
| III | 1,015 | 1.10
(0.92–1.33) | 0.3024 |
| SIRT3 | I | 56 | 1.75
(0.67–4.55) | 0.2446 |
|
| II | 324 | 0.58
(0.43–0.79) | 0.0005 |
|
| III | 1,015 | 0.84
(0.70–1.00) | 0.0522 |
| SIRT4 | I | 56 | 0.71
(0.28–1.81) | 0.4768 |
|
| II | 324 | 1.24
(0.91–1.69) | 0.1805 |
|
| III | 1,015 | 1.32
(1.10–1.58) | 0.0026 |
| SIRT5 | I | 41 | 2.06
(0.71–6.96) | 0.1744 |
|
| II | 162 | 0.71
(0.44–1.13) | 0.1488 |
|
| III | 392 | 0.81
(0.61–1.08) | 0.1560 |
| SIRT6 | I | 56 | 0.43
(0.16–1.13) | 0.0792 |
|
| II | 324 | 0.61
(0.45–0.83) | 0.0013 |
|
| III | 1,015 | 0.82
(0.69–0.97) | 0.0183 |
| SIRT7 | I | 56 | 0.63
(0.24–1.64) | 0.3374 |
|
| II | 324 | 0.78
(0.57–1.05) | 0.1052 |
|
| III | 1,015 | 0.73
(0.61–0.88) | 0.0008 |
 | Table II.Associations between elevated SIRT
mRNA expression and overall survival of all patients with ovarian
cancer, according to different clinical stages. |
Table II.
Associations between elevated SIRT
mRNA expression and overall survival of all patients with ovarian
cancer, according to different clinical stages.
| SIRT | Clinical
stages | Cases, n | HR (95% CI) | P-value |
|---|
| SIRT1 | I+II | 135 | 1.96
(0.90–4.28) | 0.0862 |
|
| III+IV | 1,220 | 1.18
(1.02–1.37) | 0.0298 |
| SIRT2 | I+II | 135 | 0.50
(0.23–1.09) | 0.0763 |
|
| III+IV | 1,220 | 1.11
(0.94–1.31) | 0.2335 |
| SIRT3 | I+II | 135 | 0.51
(0.19–1.35) | 0.1682 |
|
| III+IV | 1,220 | 0.75
(0.64–0.89) | 0.0007 |
| SIRT4 | I+II | 135 | 0.52
(0.20–1.40) | 0.1883 |
|
| III+IV | 1,220 | 1.26
(1.07–1.49) | 0.0052 |
| SIRT5 | I+II | 83 | 1.93
(0.70–5.36) | 0.1976 |
|
| III+IV | 487 | 0.68
(0.54–0.87) | 0.0015 |
| SIRT6 | I+II | 135 | 0.23
(0.07–0.77) | 0.009 |
|
| III+IV | 1,220 | 0.77
(0.67–0.90) | 0.0008 |
| SIRT7 | I+II | 135 | 0.37
(0.17–0.81) | 0.0097 |
|
| III+IV | 1,220 | 0.83
(0.71–0.96) | 0.0130 |
 | Table III.Associations between elevated SIRT
mRNA expression and overall survival of all patients with ovarian
cancer, according to the TP53 mutation condition. |
Table III.
Associations between elevated SIRT
mRNA expression and overall survival of all patients with ovarian
cancer, according to the TP53 mutation condition.
| SIRT | TP53 mutation | Cases, n | HR (95% CI) | P-value |
|---|
| SIRT1 | Mutant | 506 | 1.21
(0.95–1.53) | 0.1256 |
|
| Wild | 94 | 1.53
(0.85–2.76) | 0.1545 |
| SIRT2 | Mutant | 506 | 1.55
(1.22–1.97) | 0.0003 |
|
| Wild | 94 | 0.64
(0.37–1.12) | 0.1153 |
| SIRT3 | Mutant | 506 | 0.51
(0.19–1.35) | 0.0072 |
|
| Wild | 94 | 0.54
(0.29–0.99) | 0.0432 |
| SIRT4 | Mutant | 506 | 1.19
(0.94–1.49) | 0.1407 |
|
| Wild | 94 | 1.69
(0.95–3.02) | 0.0720 |
| SIRT5 | Mutant | 124 | 0.50
(0.32–0.77) | 0.0015 |
|
| Wild | 19 | 0.51
(0.16–1.64) | 0.2497 |
| SIRT6 | Mutant | 506 | 1.17
(0.92–1.49) | 0.2102 |
|
| Wild | 94 | 0.67
(0.38–1.18) | 0.1677 |
| SIRT7 | Mutant | 506 | 1.19
(0.94–1.50) | 0.1515 |
|
| Wild | 94 | 1.30
(0.72–2.34) | 0.3808 |
 | Table IV.Associations between elevated SIRT
mRNA expression and overall survival of all patients with ovarian
cancer, according to different chemotherapy methods. |
Table IV.
Associations between elevated SIRT
mRNA expression and overall survival of all patients with ovarian
cancer, according to different chemotherapy methods.
| SIRT | Chemotherapy
method | Cases, n | HR (95% CI) | P-value |
|---|
| SIRT1 | Contains
Platinum | 1,049 | 1.21
(1.05–1.40) | 0.0092 |
|
| Contains Taxol | 793 | 1.25
(1.03–1.51) | 0.0207 |
|
| Contains
Taxol+Platinum | 776 | 1.28
(1.06–1.55) | 0.0113 |
| SIRT2 | Contains
Platinum | 1,049 | 0.90
(0.76–1.05) | 0.1792 |
|
| Contains Taxol | 793 | 0.88
(0.73–1.07) | 0.1980 |
|
| Contains
Taxol+Platinum | 776 | 0.88
(0.73–1.07) | 0.2007 |
| SIRT3 | Contains
Platinum | 1,049 | 0.83
(0.70–0.97) | 0.0199 |
|
| Contains Taxol | 793 | 0.77
(0.64–0.93) | 0.0069 |
|
| Contains
Taxol+Platinum | 776 | 0.81
(0.67–0.98) | 0.0289 |
| SIRT4 | Contains
Platinum | 1,049 | 1.26
(1.09–1.44) | 0.0014 |
|
| Contains Taxol | 793 | 1.31
(1.07–1.60) | 0.0090 |
|
| Contains
Taxol+Platinum | 776 | 1.30
(1.06–1.60) | 0.0111 |
| SIRT5 | Contains
Platinum | 478 | 0.79
(0.62–1.00) | 0.0540 |
|
| Contains Taxol | 357 | 0.82
(0.61–1.10) | 0.1903 |
|
| Contains
Taxol+Platinum | 356 | 0.82
(0.61–1.11) | 0.2017 |
| SIRT6 | Contains
Platinum | 1,049 | 0.81
(0.70–0.93) | 0.0031 |
|
| Contains Taxol | 793 | 0.77
(0.63–0.94) | 0.0094 |
|
| Contains
Taxol+Platinum | 776 | 0.75
(0.61–0.92) | 0.0057 |
| SIRT7 | Contains
Platinum | 1,049 | 0.79
(0.68–0.90) | 0.0008 |
|
| Contains Taxol | 793 | 0.74
(0.61–0.89) | 0.0014 |
|
| Contains
Taxol+Platinum | 776 | 0.74
(0.60–0.91) | 0.0043 |
Tumor xenograft and IHC results
Fig. S1 represents
excised tumors with scale bars and tumor volume over time
statistical graph (Fig. S1B and
S1C). In addition, using IHC
staining it was demonstrated that SIRT1, SIRT3, SIRT6 were
expressed in various degrees in the tumor tissues (Fig. S1A).
Discussion
OC is one of the most lethal types of gynecological
malignancies, which affects the health condition of female patients
worldwide. Despite the development of medical technology, the
incidence and OS rates remain unsatisfactory, due to the unique
biological characteristics of OC (24). Thus, identifying a novel biomarker
for the prognostic prediction of OC is necessary. Previously, the
SIRT family has been reported to serve a critical part in the
process of tumorigenesis (25–27).
Each individual SIRT may act either as a tumor suppressor or an
oncogene in different types of malignancies, potentially via
tumor-associated signaling pathways or mitosis regulation (13,28,29).
However, the specific association between SIRTs and OC remains
controversial, and remains to be further clarified.
In the present study, the prognostic values of seven
individual SIRTs were comprehensively assessed in OC via the KM
plotter online database. According to the analysis, most of the
SIRT members act as either tumor promoters or inhibitors in OC. As
a consequence, the high mRNA expression levels of SIRT1 and SIRT4
were associated with an unfavorable prognosis in patients with OC.
Similarly, previous studies have also verified the roles of the
aforementioned two SIRTs in other carcinomas, such as lung cancer
(30,31), breast cancer (32–34),
gastric cancer (35,36), and prostate cancer (37). Specifically, Shin et al
(30) reported that high SIRT1
expression was identified in non-small cell lung cancer, which was
highly associated with drug resistance via the
SIRT1-mitogen-activated protein kinase signaling pathway.
Additionally, SIRT1 could aggravate invasion capacity and
metastasis in prostate cancer via the induction of
epithelial-to-mesenchymal transition (37). Consistently, the present study
demonstrated that elevated mRNA levels of SIRT1 were associated
with poor OS in patients with OC, notably those at a high clinical
stage (III+IV) and those that received chemotherapy. SIRT4 has been
demonstrated to be a tumor suppressor as it causes inhibition of
cell proliferation and metastasis (38). To the best of our knowledge, no study
has reported the prognostic value of SIRT4 in OC. The present study
demonstrated that SIRT4 expression was inversely associated with
prognosis in OC. In patients with OC of a high clinical stage
(III+IV), poor differentiation (pathological grade III) and who
received chemotherapy, high SIRT4 expression was associated with
unfavorable OS. This observation may provide a novel insight into
the effect of SIRT4 on the regulation of OC. Above all, these
results support the use of SIRT1 and SIRT4 as potential biomarkers
for prognostic prediction in OC.
The tumor suppressive effects of SIRT3, SIRT5, SIRT6
and SIRT7 in OC were also determined in the present study. To the
best of our knowledge, mitochondrial localized SIRT3 (39), with its unique characteristic, serves
as a crucial regulator of the Warburg effect (40) in breast cancer via the inhibition of
reactive oxygen species and the destabilization of HIF-1α. Previous
studies have demonstrated that SIRT3 can suppress the metastasis
capacity and induce apoptosis in OC (18,41).
Consistently, the KM plotter results of the present study
demonstrated that high SIRT3 expression may be associated with a
favorable prognosis in patients. Additionally, patients with
pathological grade II, a high clinical stage (III+IV), a TP53
mutation and those who received chemotherapy experienced improved
OS, further indicating that increased SIRT3 expression predicts
prolonged OS. SIRT5 has been reported to possess dual roles in the
regulation of various types of carcinoma, suggesting that SIRT5
acts as a tumor suppressor in hepatic cancer by inhibiting acyl-CoA
oxidase 1 (42). However, SIRT5 has
also been demonstrated to function as an oncogene in the
carcinogenesis of colorectal cancer (CRC), potentially by
stimulating glutamine metabolism through the activation of
dehydrogenase 1 during the tricarboxylic-acid cycle in CRC cells
(43). The fact that SIRT5 exhibits
dual effects on the regression of different tumors may depend on
the dominant factor in the microenvironment and the signaling
pathway. In the present study, SIRT5 overexpression was associated
with a favorable prognosis in OC, and elevated SIRT5 levels in
patients with clinical stage III+IV and TP53 mutations were
associated with improved OS, implying that SIRT5 is a potential
biomarker of OC. Previous studies referring to SIRT6 have
investigated its role as a tumor suppressor, and its decreased
expression is associated with poor OS in patients with pancreatic
cancer (44), hepatic carcinoma
(45) and colon cancer (46). The present study demonstrated that
SIRT6 overexpression was associated with good prognosis in OC. An
increased SIRT6 level also predicted favorable OS in patients with
poor differentiation (pathological grades II+III), in all clinical
stages and in patients who received chemotherapy. However, Bae
et al (19) have demonstrated
that SIRT6 expression is notably associated with a poor prognosis
in OC. This difference could be attributed to the patient
selection, sample size, study design, statistical method and
detection means (the present study focused on mRNA expression,
whereas the former study focused on the protein degrees). According
to the SIRT7 mRNA analysis, the present study revealed that
increased levels of SIRT7 predicted a good prognosis in patients
with OC, notably those with serous cancer and pathological grade II
and in all clinical stages. Additionally, SIRT7 overexpression was
associated with improved OS in patients who received any type of
chemotherapy. The data in the present study differed from those
described in previous studies, in that SIRT7 acted as an oncogene
in tumor progression (14,47,48),
which provides a novel basis for predicting the potential of SIRT7
in OC.
Furthermore, the analysis using the KM plotter of
each of the SIRT members revealed contradictory associations with
PFS, PPS and OS. Theoretically, OS may better represent patient
prognosis (in contrast to PFS and PPS); however, because PFS and
PPS compose OS, OS is in turn affected by these two factors
(49). These conflicting
associations may partially result from differences in the
chemotherapy choice, sample size, sensitivity to diagnosis and
treatment between two SIRT expression groups (50,51).
Therefore, the PFS and PPS data may provide another direction for
further studies investigating the role of the SIRT family in the
prognosis of OC.
In conclusion, the present study suggested that the
mRNA expression levels of SIRTs (SIRT1, SIRT3, SIRT4, SIRT5, SIRT6
and SIRT7) are associated with prognosis in patients with OC.
Additionally, it was revealed that the SIRT family members
demonstrated potential prediction capability for other
clinicopathological features, including the histological type,
pathological grade, clinical stage, TP53 mutation status and
mainstream chemotherapy choice. However, these results are limited
to the mRNA expression levels of SIRTs. Therefore, analysis of the
protein expression levels is required in future studies. Overall,
the present study provides a novel prospect for future studies on
specific signaling pathways through which SIRTs may participate in
the tumorigenesis, progression and metastasis of OC.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Jie Zhang
(Department of Hepatobiliary Surgery, The First Affiliated Hospital
of Wenzhou Medical University) for their technical help.
Funding
The present study was supported by the Natural
Science Foundation of China (grant no. 81470874).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LW and QH conceived and designed the present study.
KC and PG analyzed the data. QH, RY and ND wrote the manuscript. RY
performed the animal experiments and histological examinations. ND
revised the manuscript for intellectual content and analyzed the
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The animal study was approved by the Wenzhou Medical
University Ethics Committee (approval no. wydw2019-0214).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
SIRT
|
sirtuin
|
|
OC
|
ovarian cancer
|
|
KM plotter
|
Kaplan-Meier plotter
|
|
OS
|
overall survival
|
|
PFS
|
progression-free survival
|
|
PPS
|
post-progression survival
|
|
HR
|
hazard ratio
|
|
CI
|
confidence interval
|
|
TP53
|
tumor protein p53
|
|
CRC
|
colorectal cancer
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen X, Xu Z, Zeng S, Wang X, Liu W, Qian
L, Wei J, Yang X, Shen Q, Gong Z and Yan Y: SIRT5 downregulation is
associated with poor prognosis in glioblastoma. Cancer Biomark.
24:449–459. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jayson GC, Kohn EC, Kitchener HC and
Ledermann JA: Ovarian cancer. Lancet. 384:1376–1388. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Coleman MP, Forman D, Bryant H, Butler J,
Rachet B, Maringe C, Nur U, Tracey E, Coory M, Hatcher J, et al:
Cancer survival in Australia, Canada, Denmark, Norway, Sweden, and
the UK, 1995–2007 (the International Cancer Benchmarking
Partnership): An analysis of population-based cancer registry data.
Lancet. 377:127–138. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kane AE and Sinclair DA: Sirtuins and
NAD+ in the development and treatment of metabolic and
cardiovascular diseases. Circ Res. 123:868–885. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Singh CK, Chhabra G, Ndiaye MA,
Garcia-Peterson LM, Mack NJ and Ahmad N: The role of sirtuins in
antioxidant and redox signaling. Antioxid Redox Signal. 28:643–661.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
O'Callaghan C and Vassilopoulos A:
Sirtuins at the crossroads of stemness, aging, and cancer. Aging
Cell. 16:1208–1218. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Karbasforooshan H, Roohbakhsh A and Karimi
G: SIRT1 and microRNAs: The role in breast, lung and prostate
cancers. Exp Cell Res. 367:1–6. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu Y, Lin J, Lin Y, Chen X, Zhu G and
Huang G: Overexpression of SIRT4 inhibits the proliferation of
gastric cancer cells through cell cycle arrest. Oncol Lett.
17:2171–2176. 2019.PubMed/NCBI
|
|
11
|
Park EY, Woo Y, Kim SJ, Kim DH, Lee EK, De
U, Kim KS, Lee J, Jung JH, Ha KT, et al: Anticancer effects of a
new SIRT inhibitor, MHY2256, against human breast cancer MCF-7
cells via regulation of MDM2-p53 binding. Int J Biol Sci.
12:1555–1567. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang G, Cui F, Yu F, Lu H, Zhang M, Tang
H and Peng Z: Sirtuin-4 (SIRT4) is downregulated and associated
with some clinicopathological features in gastric adenocarcinoma.
Biomed Pharmacother. 72:135–139. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim HS, Vassilopoulos A, Wang RH, Lahusen
T, Xiao Z, Xu X, Li C, Veenstra TD, Li B, Yu H, et al: SIRT2
maintains genome integrity and suppresses tumorigenesis through
regulating APC/C activity. Cancer Cell. 20:487–499. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu H, Ye W, Wu J, Meng X, Liu RY, Ying X,
Zhou Y, Wang H, Pan C and Huang W: Overexpression of sirt7 exhibits
oncogenic property and serves as a prognostic factor in colorectal
cancer. Clin Cancer Res. 20:3434–3445. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Azuma Y, Yokobori T, Mogi A, Altan B,
Yajima T, Kosaka T, Onozato R, Yamaki E, Asao T, Nishiyama M and
Kuwano H: SIRT6 expression is associated with poor prognosis and
chemosensitivity in patients with non-small cell lung cancer. J
Surg Oncol. 112:231–237. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shuang T, Wang M, Zhou Y and Shi C:
Over-expression of Sirt1 contributes to chemoresistance and
indicates poor prognosis in serous epithelial ovarian cancer (EOC).
Med Oncol. 32:2602015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pinton G, Nilsson S and Moro L: Targeting
estrogen receptor beta (ERβ) for treatment of ovarian cancer:
Importance of KDM6B and SIRT1 for ERβ expression and functionality.
Oncogenesis. 7:152018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu Y, Gao WN, Xue YN, Zhang LC, Zhang JJ,
Lu SY, Yan XY, Yu HM, Su J and Sun LK: SIRT3 aggravates
metformin-induced energy stress and apoptosis in ovarian cancer
cells. Exp Cell Res. 367:137–149. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bae JS, Noh SJ, Kim KM, Park SH, Hussein
UK, Park HS, Park BH, Ha SH, Lee H, Chung MJ, et al: SIRT6 is
involved in the progression of ovarian carcinomas via
β-catenin-mediated epithelial to mesenchymal transition. Front
Oncol. 8:5382018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang J, Yin XJ, Xu CJ, Ning YX, Chen M,
Zhang H, Chen SF and Yao LQ: The histone deacetylase SIRT6 inhibits
ovarian cancer cell proliferation via down-regulation of Notch 3
expression. Eur Rev Med Pharmacol Sci. 19:818–824. 2015.PubMed/NCBI
|
|
21
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Györffy B, Surowiak P, Budczies J and
Lánczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gyorffy B, Lánczky A and Szállási Z:
Implementing an online tool for genome-wide validation of
survival-associated biomarkers in ovarian-cancer using microarray
data from 1287 patients. Endocr Relat Cancer. 19:197–208. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Qin L, Qiu H, Zhang M, Zhang F, Yang H,
Yang L, Jia L, Qin K, Jia L, Dou X, et al: Soluble CD40 ligands
sensitize the epithelial ovarian cancer cells to cisplatin
treatment. Biomed Pharmacother. 79:166–175. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Giannopoulou AF, Velentzas AD,
Konstantakou EG, Avgeris M, Katarachia SA, Papandreou NC, Kalavros
NI, Mpakou VE, Iconomidou V, Anastasiadou E, et al: Revisiting
histone deacetylases in human tumorigenesis: The paradigm of
urothelial bladder cancer. Int J Mol Sci. 20:2019. View Article : Google Scholar
|
|
26
|
Huang G and Zhu G: Sirtuin-4 (SIRT4), a
therapeutic target with oncogenic and tumor-suppressive activity in
cancer. Onco Targets Ther. 11:3395–3400. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Geng CH, Zhang CL, Zhang JY, Gao P, He M
and Li YL: Overexpression of Sirt6 is a novel biomarker of
malignant human colon carcinoma. J Cell Biochem. 119:3957–3967.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Serrano L, Martínez-Redondo P,
Marazuela-Duque A, Vazquez BN, Dooley SJ, Voigt P, Beck DB,
Kane-Goldsmith N, Tong Q, Rabanal RM, et al: The tumor suppressor
SirT2 regulates cell cycle progression and genome stability by
modulating the mitotic deposition of H4K20 methylation. Genes Dev.
27:639–653. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee BB, Kim Y, Kim D, Cho EY, Han J, Kim
HK, Shim YM and Kim DH: Metformin and tenovin-6 synergistically
induces apoptosis through LKB1-independent SIRT1 down-regulation in
non-small cell lung cancer cells. J Cell Mol Med. 23:2872–2889.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shin DH, Choi YJ and Park JW: SIRT1 and
AMPK mediate hypoxia-induced resistance of non-small cell lung
cancers to cisplatin and doxorubicin. Cancer Res. 74:298–308. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fu L, Dong Q, He J, Wang X, Xing J, Wang
E, Qiu X and Li Q: SIRT4 inhibits malignancy progression of NSCLCs,
through mitochondrial dynamics mediated by the ERK-Drp1 pathway.
Oncogene. 36:2724–2736. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee H, Kim KR, Noh SJ, Park HS, Kwon KS,
Park BH, Jung SH, Youn HJ, Lee BK, Chung MJ, et al: Expression of
DBC1 and SIRT1 is associated with poor prognosis for breast
carcinoma. Human Pathol. 42:204–213. 2011. View Article : Google Scholar
|
|
33
|
Jin X, Wei Y, Xu F, Zhao M, Dai K, Shen R,
Yang S and Zhang N: SIRT1 promotes formation of breast cancer
through modulating Akt activity. J Cancer. 9:2012–2023. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jeong SM, Xiao C, Finley LW, Lahusen T,
Souza AL, Pierce K, Li YH, Wang X, Laurent G, German NJ, et al:
SIRT4 has tumor-suppressive activity and regulates the cellular
metabolic response to DNA damage by inhibiting mitochondrial
glutamine metabolism. Cancer Cell. 23:450–463. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cha EJ, Noh SJ, Kwon KS, Kim CY, Park BH,
Park HS, Lee H, Chung MJ, Kang MJ, Lee DG, et al: Expression of
DBC1 and SIRT1 is associated with poor prognosis of gastric
carcinoma. Clin Cancer Res. 15:4453–4459. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sun H, Huang D, Liu G, Jian F, Zhu J and
Zhang L: SIRT4 acts as a tumor suppressor in gastric cancer by
inhibiting cell proliferation, migration, and invasion. Onco
Targets Ther. 11:3959–3968. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Byles V, Zhu L, Lovaas JD, Chmilewski LK,
Wang J, Faller DV and Dai Y: SIRT1 induces EMT by cooperating with
EMT transcription factors and enhances prostate cancer cell
migration and metastasis. Oncogene. 31:4619–4629. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Csibi A, Fendt SM, Li C, Poulogiannis G,
Choo AY, Chapski DJ, Jeong SM, Dempsey JM, Parkhitko A, Morrison T,
et al: The mTORC1 pathway stimulates glutamine metabolism and cell
proliferation by repressing SIRT4. Cell. 153:840–854. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pryds K, Nielsen RR, Jorsal A, Hansen MS,
Ringgaard S, Refsgaard J, Kim WY, Petersen AK, Bøtker HE and
Schmidt MR: Effect of long-term remote ischemic conditioning in
patients with chronic ischemic heart failure. Basic Res Cardiol.
112:672017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dong XC, Jing LM, Wang WX and Gao YX:
Down-regulation of SIRT3 promotes ovarian carcinoma metastasis.
Biochem Biophys Res Commun. 475:245–250. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen XF, Tian MX, Sun RQ, Zhang ML, Zhou
LS, Jin L, Chen LL, Zhou WJ, Duan KL, Chen YJ, et al: SIRT5
inhibits peroxisomal ACOX1 to prevent oxidative damage and is
downregulated in liver cancer. EMBO Rep. 19:2018. View Article : Google Scholar
|
|
43
|
Wang YQ, Wang HL, Xu J, Tan J, Fu LN, Wang
JL, Zou TH, Sun DF, Gao QY, Chen YX and Fang JY: Sirtuin5
contributes to colorectal carcinogenesis by enhancing
glutaminolysis in a deglutarylation-dependent manner. Nat Commun.
9:5452018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kugel S, Sebastián C, Fitamant J, Ross KN,
Saha SK, Jain E, Gladden A, Arora KS, Kato Y, Rivera MN, et al:
SIRT6 suppresses pancreatic cancer through control of Lin28b. Cell.
165:1401–1415. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Marquardt JU, Fischer K, Baus K, Kashyap
A, Ma S, Krupp M, Linke M, Teufel A, Zechner U, Strand D, et al:
Sirtuin-6-dependent genetic and epigenetic alterations are
associated with poor clinical outcome in hepatocellular carcinoma
patients. Hepatology. 58:1054–1064. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li N, Mao D, Cao Y, Li H, Ren F and Li K:
Downregulation of SIRT6 by miR-34c-5p is associated with poor
prognosis and promotes colon cancer proliferation through
inhibiting apoptosis via the JAK2/STAT3 signaling pathway. Int J
Oncol. 2018.
|
|
47
|
Wei W, Jing ZX, Ke Z and Yi P: Sirtuin 7
plays an oncogenic role in human osteosarcoma via downregulating
CDC4 expression. Am J Cancer Res. 7:1788–1803. 2017.PubMed/NCBI
|
|
48
|
Li H, Tian Z, Qu Y, Yang Q, Guan H, Shi B,
Ji M and Hou P: SIRT7 promotes thyroid tumorigenesis through
phosphorylation and activation of Akt and p70S6K1 via DBC1/SIRT1
axis. Oncogene. 38:345–359. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
He WZ and Xia LP: RE: Detecting overall
survival benefit derived from survival postprogression rather than
progression-free survival. J Natl Cancer Inst. Oct 16–2015.(Epub
ahead of print). doi: 10.1093/jnci/djv311.
|
|
50
|
Sundar S, Wu J, Hillaby K, Yap J and
Lilford R: A systematic review evaluating the relationship between
progression free survival and post progression survival in advanced
ovarian cancer. Gynecol Oncol. 125:493–499. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Shimokawa M, Ohki M and Kaku T:
Correlation of progression-free and post-progression survival with
overall survival in phase III trials of first-line chemotherapy for
advanced epithelial ovarian cancer. Eur J Gynaecol Oncol.
36:370–375. 2015.PubMed/NCBI
|
|
52
|
Telloni SM: Tumor staging and grading: A
primer. Methods Mol Biol. 1606:1–17. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Šišovská I, Minář L, Felsinger M, Anton M,
Bednaříková M, Hausnerová J, Jandáková E and Weinberger V: Current
FIGO staging classification for cancer of ovary, fallopian tube and
peritoneum. Ceska Gynekol. 82:230–236. 2017.(In Czech). PubMed/NCBI
|





















