Introduction
Lung cancer is the most commonly diagnosed cancer
and the leading cause of cancer-associated mortality in China and
globally (1). Low-dose computed
tomography has been recommended as the primary method for screening
lung cancer (1). Multiple in
vitro diagnostic methods for lung cancer screening and early
detection, including next-generation sequencing and blood-based
circulating tumor DNA (ctDNA) assays, are currently under
development with promising application prospects (2–7).
However, there are few effective blood-based tests for assessing
the therapeutic response or predicting the prognosis of patients
with lung cancer (8–10). The sensitivity of clinically used
protein markers, such as carcinoma antigen 125 (CA125),
carcinoembryonic antigen (CEA), cytokeratin 19-fragments
(cyfra21-1) and neuron-specific enolase (NSE) is not high enough
for effective screening (2–10). In addition, patients with negative
test results prior to treatment are unable to be assessed using
protein markers, which are generally more sensitive to advanced
lung cancer than early-stage lung cancer (8–10).
Therefore, protein markers are more suitable to be used as
recurrence indicators, rather than response monitoring markers.
However, CT imaging, despite being a widely-used, non-invasive
method for therapeutic response assessment, is unable to be used
routinely due to radiation, hence renders the immediate detection
of tumors changes impossible. Currently, effective methods for
response monitoring and prognosis prediction in lung cancer therapy
are lacking.
Methylated prostaglandin E2 receptor EP4 subtype
(mPTGER4) was recently reported as a methylation marker for the
early detection of lung cancer. To the best of our knowledge, the
only two studies investigating the application of mPTGER4 as a
marker involve the use of this marker in the differential diagnosis
of benign and malignant lung diseases (11,12).
Other diagnostic and therapeutic applications of mPTGER4 have not
yet been reported. In the aforementioned two reports, mPTGER4 was
used in conjunction with methylated short stature homeobox 2
(mSHOX2) to differentiate between benign and malignant lung
diseases, with an enhanced sensitivity and specificity of 67 and
90%, respectively compared with mPTGER4 or mSHOX2 alone (11,12).
Methylation markers generally exhibit stage-dependent detection
sensitivity, which is typically higher in advanced cases. In
early-stage patients, however, the detection sensitivity is
determined by the actual performance of markers in early detection
and differential diagnosis (11–16). For
patients with advanced disease, methylation markers can potentially
be used for therapeutic response monitoring, due to their high
positive detection rate (PDR) in advanced stage cancers, as well as
their sensitivity to changes in tumor burden following treatment
(15,16). Conversely, protein markers are not as
potent as methylation markers, due to their unsatisfactory PDR even
in advanced cancer, thus limiting their use in monitoring
techniques (8–10,17).
However, the specificity of protein markers is generally
satisfactory (18,19), therefore, they are often better
indicators of recurrence instead.
In the present study, the efficacy of methylated
PTGER4 was systematically assessed and compared with four
clinically- used protein markers (CA125, CEA, Cyfra211 and NSE) in
therapeutic response monitoring and survival prediction in patients
with stage IV lung cancer. Blood samples were analyzed before and
after two cycles of treatment, and the effectiveness of these
markers in monitoring the therapeutic response was assessed.
Concurrently, follow-up was performed on the patients for up to 891
days, and the efficacy of the markers in predicting survival based
on pre- and post-therapeutic blood levels was compared, as well as
the relevant percentage changes. It was revealed that methylated
PTGER4 was better than protein markers in therapeutic response
monitoring, while methylated PTGER4, CA125, CEA and NSE were useful
for predicting the overall survival rate. The present study
validated methylated PRGER4 as a potential marker for future
continuous response monitoring and prognosis prediction in patients
with stage IV lung cancer.
Materials and methods
Ethics
The protocol of the present clinical study was
approved by the ethics committees of the affiliated hospital of
Jiangnan university and the eighth medical center of the Chinese
People's Liberation Army general hospital prior to sample
collection. Written informed consent was obtained from all
subjects, and information on the intended usage of plasma and test
results was provided to all subjects.
Study design, patients and
therapy
The current study was designed and implemented in
the Affiliated Hospital of Jiangnan University (Jiangsu, China) and
The 8th Medical Center of the Chinese PLA General Hospital
(Beijing, China). A methylated PTGER4 assay was used as previously
reported by Weiss et al (11). Clinical status was determined prior
to blood collection for the methylated PTGER4 assay, and blood
samples were obtained from all subjects who met the selection
criteria. The main inclusion criteria: adults >18 years old with
complete clinicopathological information and confirmed diagnosis of
lung cancer by imaging examination (including MRI, CT, etc.) and/or
subsequent pathological examination. The main exclusion criteria
include: pregnancy, history of any cancer, or history of therapy
for any cancer. Subjects with incomplete information were also
excluded. All technicians were blinded to the clinical information
of subjects, and patients' test results and corresponding clinical
information were only revealed after all tests were finished. All
patients were ≥18 years old with no previous history of cancer and
both male and female patients were included. All lung cancer
subtypes, including small-cell lung cancer (SCLC) and non-small
cell lung cancer (NSCLC) were included. Adenocarcinoma (ADC) and
SCLC were the most prevalent subtypes in the present study
(Table I). A total of 146 subjects
were recruited in the current study, and all patients had primary
lung cancer without any history of treatment. The population
comprised 30 patients at stage I, 29 at stage II, 36 at stage III
and 51 at stage IV (Table I). Tumors
were staged in accordance with the NCCN guidelines (13). Determination of stage was achieved by
at least two independent pathologists via pathological examination
of biopsies from needle aspiration. All patients at stage IV were
diagnosed using imaging and pathological examinations, in which ≥1
primary lung cancer lesion and ≥1 confirmed distal metastasis were
discovered. First-line standard chemotherapy, combined radio- and
chemotherapy or tyrosine kinase inhibitor (TKI)-based targeted
therapy was used to treat stage IV patients. The diameter of the
primary tumors was assessed based on RECIST 1.1 criteria, and
correlated with the level of mPTGER4 (20).
 | Table I.Number of enrolled subjects and
demographic characteristics by diagnosis group. |
Table I.
Number of enrolled subjects and
demographic characteristics by diagnosis group.
|
| Sex, n | Age, years |
|---|
|
|
|
|
|---|
| Diagnosis
group | Total | Male | Female | <50 | 50–59 | 60–69 | ≥70 |
|---|
| Overall | 146 | 95 | 51 | 20 | 51 | 48 | 27 |
| Stage |
|
|
|
|
|
|
|
| I | 30 | 21 | 9 | 2 | 11 | 12 | 5 |
| II | 29 | 17 | 12 | 4 | 9 | 10 | 6 |
|
III | 36 | 25 | 11 | 9 | 14 | 8 | 5 |
| IV | 51 | 32 | 19 | 5 | 17 | 18 | 11 |
| Pathological type
(stage IV) |
|
|
|
|
|
|
|
|
SCLC | 12 | 8 | 4 | 1 | 4 | 4 | 3 |
|
ADC | 35 | 21 | 14 | 4 | 11 | 12 | 8 |
| SC | 3 | 2 | 1 | 0 | 1 | 2 | 0 |
| LC | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
Population size estimation
The equation for known positive detection rate was
used for Population size estimation:
n=Z2x[p(1-p)]/E2. Z is a statistical
parameter (Z=1.96 for 95% CI) and E represented the error (10% was
selected in the present study), and p represented the putative
positive detection rate. The E value is generally 0.05–0.1,
depending on the error that is allowed in a study. Since the
effectiveness of the mPTGER4 test has been validated in previous
studies (11,12) and a previous pilot study, 0.1(10%)
was used in the present study. The p value represented the known
sensitivity for the assay on lung cancer (11), and was obtained from a previous pilot
study (data not shown). If the known sensitivity for stage IV lung
cancer equals to 0.85, an estimated 49 lung cancer cases were
required. A total of 51 patients (32 male and 19 female; age range,
43–82 year; median age, 63) with stage IV lung cancer were included
in the current study (Table I); and
this total comprised 12 patients with SCLC and 39 with NSCLC. Of
the patients with NSCLC, 14 exhibited epidermal growth factor
receptor (EGFR)-sensitive mutations (EGFR M+) and underwent
TKI-based first-line therapy and 25 patients did not have
EGFR-sensitive mutations (EGFR M-) and underwent
either standard chemotherapy or combined chemo- and
radiotherapy.
Sample collection and storage
A 10 ml peripheral blood sample was collected from
lung cancer patients of the two participating hospitals using 10 ml
K2EDTA anticoagulant tubes (BD Biosciences). Sample
storage and transportation followed the methods previously reported
by Weiss et al (11). The
first blood sample was collected prior to initiation of any
therapy, and the second blood sample was collected following two
cycles of therapy. One cycle of chemotherapy lasts 21 days, while
the TKI therapy requires patients to take medicine daily.
Therefore, the blood collection point was on the 42nd day following
treatment initiation for all patients, prior to the beginning of
the third cycle of therapy. The sample information was recorded in
sample collection forms. Plasma samples from all participating
hospitals were prepared in the individual hospitals and stored at
−20°C prior to delivery to the laboratory, and all assays were
performed in the same laboratory ≤3 weeks from the sample
collection date. The sample quality was examined when the samples
arrived at the medical laboratory. Samples with plasma volume
<3.5 ml, or with apparent hemolysis, high bilirubin, chylemia or
visible particles/pellets were not tested and repeated blood draw
was requested.
DNA extraction, qualitative PCR
analysis of PTGER4 and measurement of protein marker levels
A commercial kit, Epi proLung (cat. no. M6-02-002,
Epigenomics AG) was used in this study. The experiments were
carried out according to the manufacturer's instructions (21) (https://www.epigenomics.com/wp-content/uploads/2018/08/IFU_0018_GB_
rev4_Instructions_for_Use_Epi_proLung.pdf). In brief, DNA
extraction and bisulfite conversion were performed manually from
plasma circulating DNA following the methods reported by Weiss
et al (11). The
bisulfite-converted DNA was assayed using an ABI7500 Fast Dx Real
Time PCR device (Thermo Fisher Scientific, Inc.). PCR was performed
in triplicate with 15 µl template DNA per well, and run for 45
cycles. The validity of each sample batch was determined on the
basis of methylated PTGER4 and β-actin (ACTB) threshold
count (Ct) values for the positive and negative controls.
ACTB was used as an internal reference to assess the
integrity of each sample. The sequences of the primers and probes
for PTGER4 are listed in the corresponding patents (patent numbers,
EP2143807A1, EP3234184B1, AU2008207110B2 and US20090203011A1). The
sequence of primers for β-actin detection used in PCR amplification
were as follows: forward primer, 5′-GTGATGGAGGAGGTTTAGTAAGTT-3′ and
reverse primer, 5′-CCAATAAAACCTACTCCTCCCTTAA-3′ and probe,
5′-ACCACCACCCAACACACAATAACAAACACA-3′. The blood levels of protein
markers, including CA125, CEA, Cyfra21-1 and NSE, were measured by
trained technicians in the corresponding hospitals, according to
the in-house procedures.
Data analysis and interpretation
Test data of the mPRGER4 assay were analyzed
by calculating the ΔΔCt values using the Ct values from samples,
ACTB internal controls and the positive controls (14). Statistical analysis was performed and
figures were plotted using GraphPad Prism software (version 5.0;
GraphPad Software, Inc.). For each sample, a relative methylation
value was determined using the ΔΔCt method adapted for DNA
methylation analyses as previously described (14). In brief, ΔΔCt values were calculated
as below: ΔΔCtSample=ΔCtSample-
ΔCtCalibrator, where ΔCtSample=CtACTB of
sample-CtPTGER4 of sample and
ΔCtCalibrator=CtACTB of
calibrator-CtPTGER4 of calibrator.
The unpaired Student's t-test was used to compare
two groups. The χ2 test and Fisher tests were performed
when rate or percentage was compared for significance. *P<0.05
represented significant changes, **P<0.01 represented highly
significant changes and ***P<0.001 represented very highly
significant changes. Kaplan-Meier curves were plotted and analyzed
using Graphpad Prism software (version 5.0; GraphPad Software,
Inc.) and the log-rank (Mantel-Cox) test was performed to compare
the survival curves. Patients were divided into two groups in the
survival analysis by the median levels of mPTGER4, CA125, CEA,
Cyfra211 and NSE in the blood. As the total number of stage IV lung
cancer patients was 51 in the present study, each group had 25 or
26 subjects depending on the exact median value. In the survival
analysis of NSCLC patients the total number of patients was 39 and
each group had 19 or 20 subjects depending on the exact median
value. P<0.05 was considered to indicate a statistically
significant difference between survival curves.
Results
Methylated PTGER4 exhibits better
performance as a marker in therapeutic response assessment compared
with CA125, CEA, Cyfra211 and NSE
In order to investigate the performance of
methylated PTGER4, CA125, CEA, Cyfra21-1 and NSE as indicators of
therapeutic efficacy in patients with stage IV lung cancer, the
positive detection rate (PDR) of each molecule was individually
calculated. It was revealed that the PDR of methylated PTGER4
increased as the cancer stage progressed, with a high PDR reaching
78.0% in stage IV lung cancer (Fig.
1A). Meanwhile, the PDRs of CA125, CEA, Cyfra21-1 and NSE were
revealed to be 46.2, 48.0, 52.0 and 58.3%, respectively, which were
significantly lower compared with methylated PTGER4 (Fig. 1B). A higher PDR value signifies that
a higher ratio of patients may benefit from the assessment, and
methylated PTGER4 exhibited a significantly higher coverage of
patients with stage IV lung cancer than the protein markers
measured.
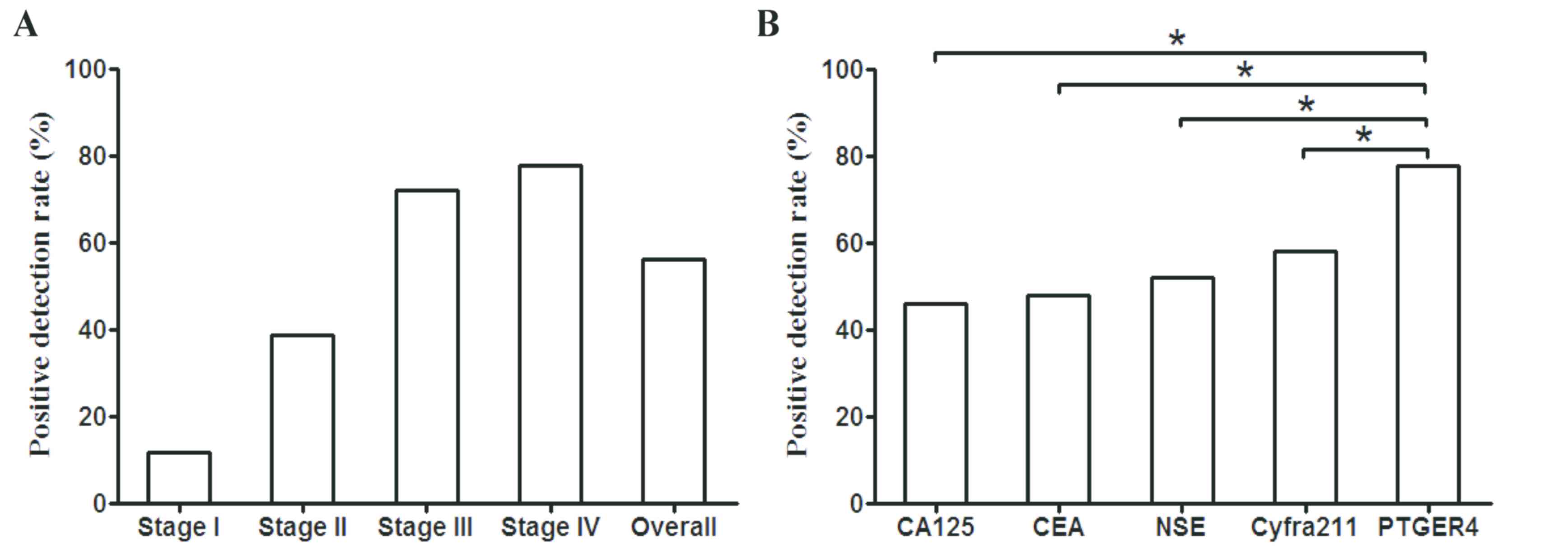 | Figure 1.Positive detection rate of methylated
PTGER4 in all stages of lung cancer and a comparison with CA125,
CEA, Cyfra211 and NSE in stage IV lung cancer. (A) The
stage-dependent PDR and overall PDR of methylated PTGER4 in lung
cancer detection. (B) Comparison of PDR (sensitivity) between the
pre- and post-therapeutic groups of CA125, CEA, NSE, Cyfra211 with
mPRGER4 when the specificity was set to 90%. χ2 test has
been performed to compare the PDR and a significant difference
(*P<0.05) was found between PTGERS and the other four markers.
PTGER4, prostaglandin E receptor 4; CA125, carcinoma antigen 125;
CEA, carcinoembryonic antigen; cyfra21-1, cytokeratin 19-fragment;
NSE, neuron-specific enolase; PDR positive detection rate. |
Blood level changes of the five markers were also
measured pre- and post-treatment. Patients were divided into a PR
group and SD group based on RECIST1.1 criteria (20), as detailed in Fig. 2. In the PR groups, while the levels
of methylated PTGER4 and NSE significantly decreased following
treatment, there were no significant difference observed between
CA125, CEA and Cyfra21-1 levels. In the SD group, there was no
significant difference in blood levels before and after treatment
for all markers. It is worth noting that the plasma level of
methylated PTGER4 decreased significantly. The average ΔΔCt value
(mean ± SD) decreased 85.10-fold from −2.634±1.486 before treatment
to −9.045±1.139 after treatment
(2−2.634-(−9.045)=26.411=85.1), which was a
fold-change much larger than any protein marker tested. The present
results indicate that methylated PTGER4 and NSE accurately
reflected the therapeutic response of patients with stage IV cancer
in the PR group, while other markers were not effective in
reflecting such a response.
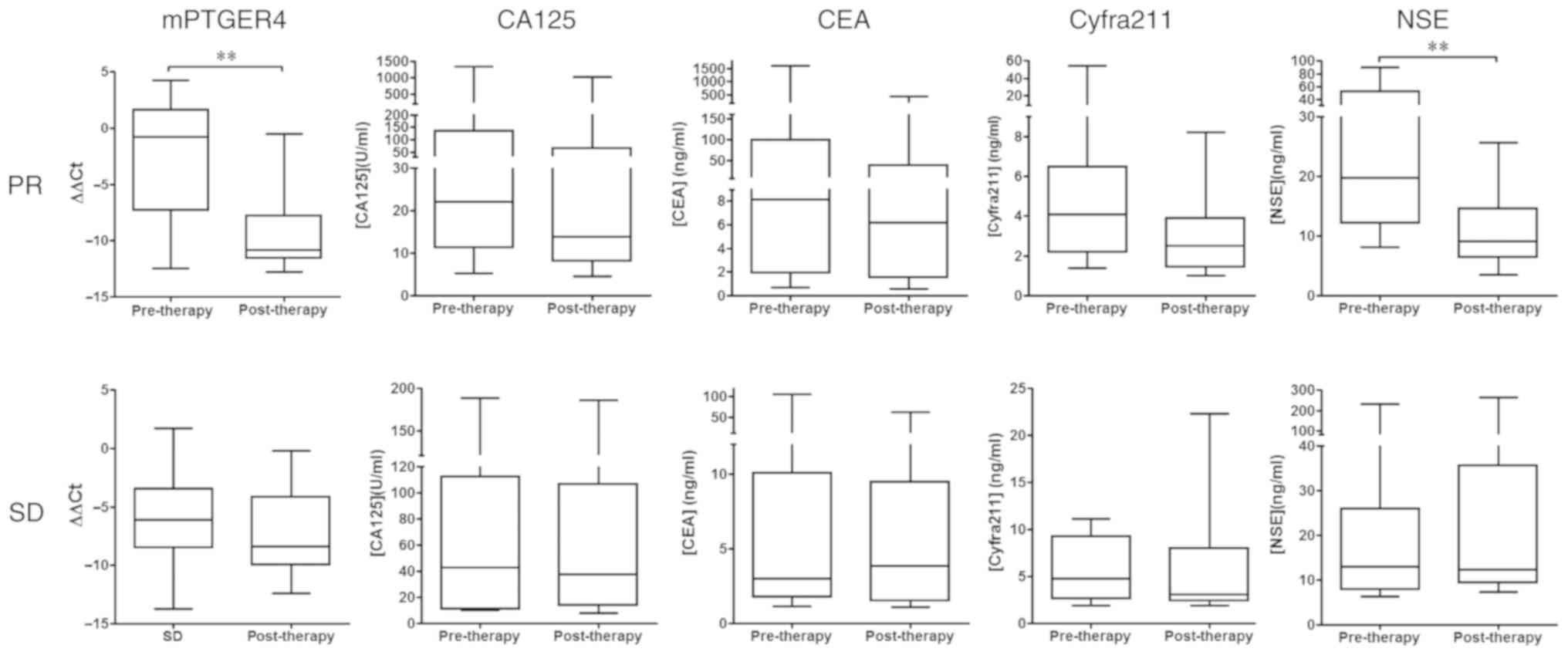 | Figure 2.Box plot for the levels of methylated
PTGER4, CA125, CEA, Cyfra211 and NSE before and after therapy in
patients with stage IV lung cancer. Absolute values of marker blood
levels before and after therapy are shown for each marker based on
therapeutic responses (PR or SD). The biomarker levels of the pre-
and post-therapeutic groups are compared in each panel.
**P<0.01. PTGER4, prostaglandin E receptor 4; CA125, carcinoma
antigen 125; CEA, carcinoembryonic antigen; cyfra21-1, cytokeratin
19-fragment; NSE, neuron-specific enolase; PR, partial response;
SD, stable disease. |
Since the level changes of the markers shown in
Fig. 2 only indicates the overall
change in this specific population, the normalized percentage
changes in the PR and SD group were also calculated. Fig. 3 depicts a comparison of the
percentage change of each marker in the PR and SD groups,
normalized against the pre-therapeutic marker level. Significant
differences in percentage between the PR and SD group in mPRGER4,
CA125, CEA and NSE were observed, reflecting the response
differences in patients with stage IV lung cancer patients.
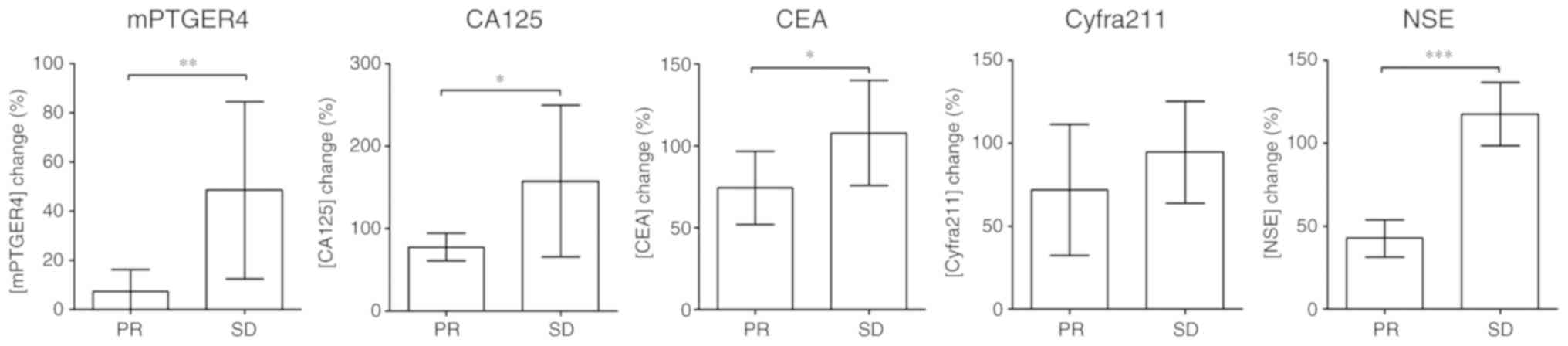 | Figure 3.Comparison of level change between PR
and SD groups for methylated PTGER4, CA125, CEA, Cyfra211 and NSE.
Post-therapeutic marker levels were normalized to pre-therapeutic
level for each patient and grouped by therapeutic response (PR or
SD). The percentage change of the PR and SD groups are compared in
each panel. *P<0.05; **P<0.01; ***P<0.001. PTGER4,
prostaglandin E receptor 4; CA125, carcinoma antigen 125; CEA,
carcinoembryonic antigen; cyfra21-1, cytokeratin 19-fragment; NSE,
neuron-specific enolase; PR, partial response; SD, stable disease;
HR, hazard ratio. |
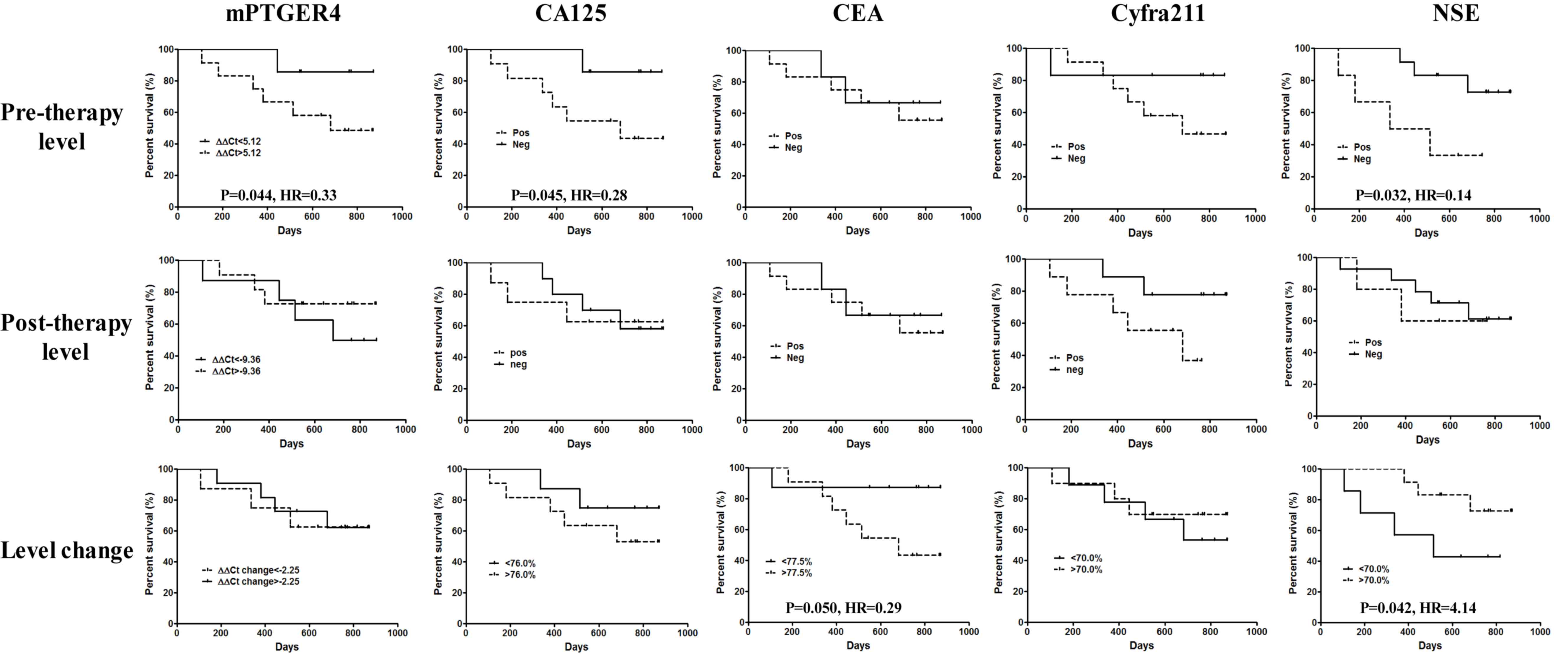
Methylated PTGER4 is capable of
predicting the overall survival rate of patients with stage IV
NSCLC
To investigate the efficacy of the five markers in
predicting long-term prognosis, patients were followed up for ≤891
days and the association between pre- and post-treatment levels,
level percentage changes and overall survival rate were calculated.
The survival of all patients with lung cancer by predicting the
efficacy of all markers, as detailed in Fig. 4. The results indicated that if
patients were grouped by the detection threshold of each marker,
NSE was the only marker able to predict the overall survival rate
at pre-therapy level (P=0.026; HR, 0.27), and patients with
undetectable pre-therapy NSE levels exhibited a better overall
survival rate. Nevertheless, all markers failed to predict the
overall survival rate at the post-therapy level. If the median of
percentage change was used as the grouping threshold, CA125
(P=0.045; HR=0.35), CEA (P=0.048; HR, 0.31) and NSE (P=0.049; HR,
3.01) were all able to predict the overall survival rate. Notably,
a decrease in marker blood level of CA125 (<76.0%) and CEA
(<77.5%) was associated with a greater survival benefit, while a
decrease in NSE level (<70.0%) was associated with a shorter
survival time. This indicates that a higher NSE expression is
associated with a longer survival time.
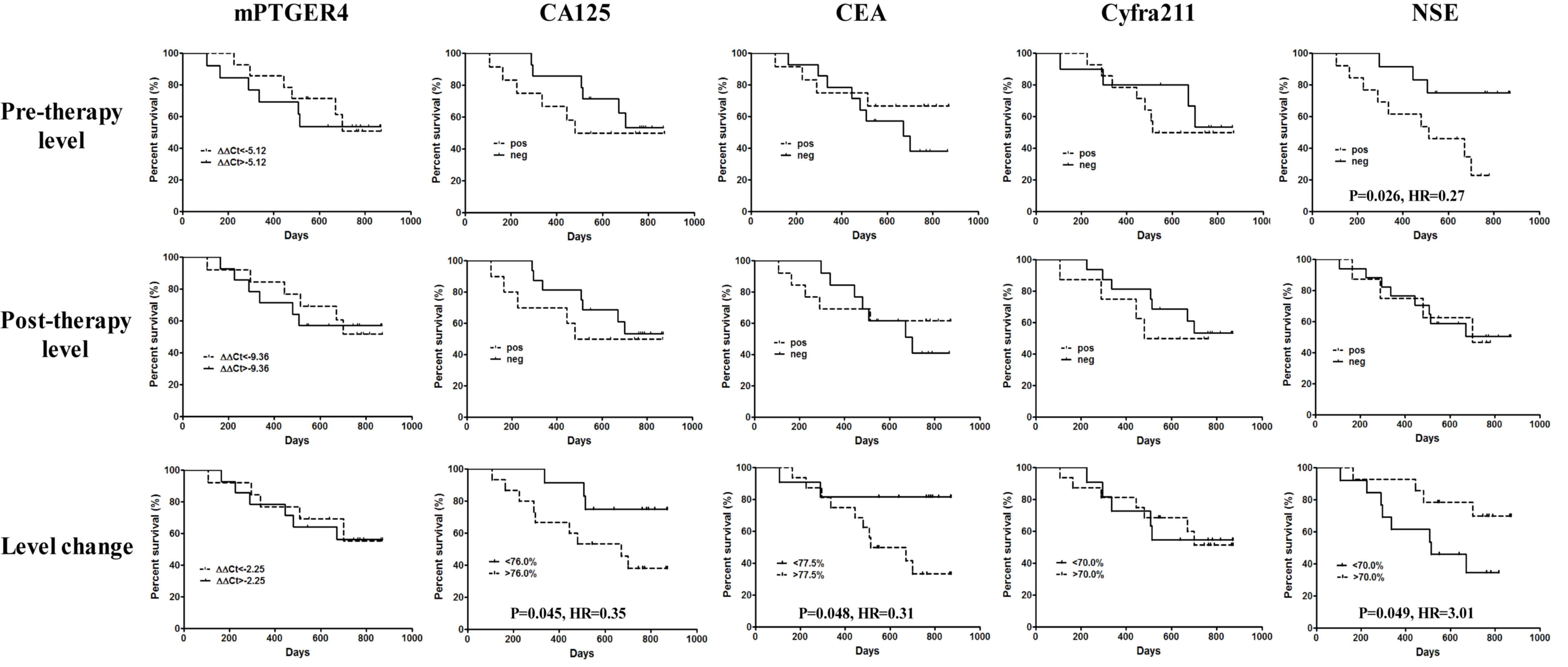 | Figure 4.Efficacy of methylated PTGER4, CA125,
CEA, Cyfra211 and NSE in predicting the overall survival rate of
all patients with stage IV lung cancer in the present study.
Pre-therapeutic level, post-therapeutic level and level changes in
percentage were used in the prediction for each marker. Comparison
has been performed between the two survival curves in each panel.
P<0.05 was regarded as a significant difference. PTGER4,
prostaglandin E receptor 4; CA125, carcinoma antigen 125; CEA,
carcinoembryonic antigen; cyfra21-1, cytokeratin 19-fragment; NSE,
neuron-specific enolase; HR, hazard ratio. |
A total of 76.5% (39/51) of patients in the present
study had stage IV non-small cell lung carcinoma (NSCLC), which can
be detected using CA125, CEA and Cyfra211 with a high level of
sensitivity, hence, the predictive performance of each marker for
the overall survival rate time of patients with NSCLC was analyzed.
As indicated in Fig. 5, the blood
level of pre-therapeutic mPRGER4 (P=0.044; HR, 0.33), CA125
(P=0.045; HR, 0.28) and NSE (P=0.032; HR, 0.14) were all capable of
predicting the overall survival rate time of patients with NSCLC,
and patients with the three markers exhibited improved survival
times. Nevertheless, none of the markers were able to predict the
post-therapy overall survival rate time. However, if the median of
the percentage change was used as the grouping threshold, the level
change of CEA (P=0.050; HR, 0.29) and NSE (P=0.042; HR, 4.14) were
able to predict overall survival rate times in the present
population. Notably, patients with a greater decrease in CEA
(<77.5%) exhibited longer overall survival rate, while
conversely patients with a greater decrease in NSE levels
(<70.0%) exhibited shorter overall survival rate, supporting the
results exhibited in Fig. 4. The
changes in the level of Cyfra211 before and after therapy also
showed a potential efficacy of prediction but the results were not
significant.
Discussion
Performance comparison between
methylation and protein markers in therapeutic response
assessment
Blood protein markers have long been used as a tool
for cancer detection. Protein markers were first discovered to have
a strong correlation with the occurrence of tumors, but later
studies revealed that the majority of protein markers are unable to
detect early-stage tumors (17–19).
Protein markers are particularly elevated during tumor recurrence
and are generally associated with tumors in more advanced stages
(17). Hence, protein tumor markers
are predominantly used for recurrence monitoring. An increased
level of certain blood protein markers is associated with tumor
progression, while a decreased level is an indication of tumor
remission (17–19). However, significant changes in blood
protein level may not be observed in a group of patients due to
variation between individuals. In the present study, no significant
difference in CA125, CEA and Cyfra211 levels was observed in the PR
group before and after treatment, indicating that protein markers
may not be accurate in monitoring therapeutic response. Conversely,
methylation markers are more sensitive to tumor burden change
compared with protein markers (15,22,23), as
they are sensitive to the changes of ctDNA levels in plasma, which
reflects the release of cellular DNA during apoptosis or necrosis
(24). In the present study, the
change in methylated PTGER4 level from pre- to post-treatment was
greater than that of the protein markers, indicating that
methylated PTGER4 represents a more sensitive marker for
therapeutic response assessment. Notably, previous studies have
reported the application of methylation markers in monitoring tumor
response to therapy (15,16,22–25). The
methylation markers chosen for response monitoring share certain
common features. Firstly, the pre-treatment sensitivity of the
markers is generally satisfactory with a wide coverage of patients.
Secondly, whilst having a good sensitivity to tumor burden change,
their plasma level changes have a strong association with either
disease progression or remission. Thirdly, the level of change is
quantifiable and may therefore be able to inform clinical practices
(15,16,22–25).
Methylation markers are applicable not only in
response monitoring, but also for early diagnosis. Previous studies
have suggested that the overall performance of methylation markers
in both these applications is an improvement compared with protein
markers (15,16,22–25).
However, protein markers also have certain advantages. For
instance, their specificity for cancer is generally better with a
low false-positive rate in healthy subjects, suggesting that the
potential of measuring levels of protein markers in the blood
requires further investigation (17–19). By
contrast, the positive detection rate of methylation markers is
generally higher in older patients, with an increased false
positive rate in healthy elderly subjects (26,27).
Therefore, a combination of methylation and protein markers may be
beneficial in enhancing both the sensitivity and specificity of
cancer diagnosis and therapeutic monitoring.
Characteristics of markers for
predicting long-term survival
Both methylation and protein markers may be useful
for predicting long-term survival, but not all of them are
effective in predicting overall survival rate. Markers capable of
predicting long-term overall survival rate must exhibit the
following characteristics: i) The blood level of markers
post-treatment must exhibit a significant decrease in patients with
PR, and an insignificant change in patients with SD, or a
significant increase associated with disease progression; ii) an
identifiable association between the marker blood level and the
change in tumor size; and iii) the short- or mid-term
post-treatment remission of a tumor must have a strong association
with the increased long-term survival of patients, while the short-
or mid-term tumor progression must also be strongly correlated with
the decreased long-term survival of the patients. In the present
study, NSE met the above criteria, and its predictive efficacy for
survival time was demonstrated in the patients with stage IV NSCLC.
The pre-therapeutic level of methylated PTGER4 also exhibited
similar efficacy in predicting the overall survival rate of
patients with NSCLC. However, the level of methylated PTGER4
decreased in the SD group by ~2.71 times (the mean ΔΔCt decreased
from −5.96 to −7.4). This subsequently affected the discrimination
of level change between PR and SD patients, interfering with the
predictive performance of long-term survival in all stage IV
patients. It appeared that SCLC may be a contributing factor that
interfered with the predictive ability of mPRGER4. Other previously
reported methylation markers, including mSHOX2 (15,16,20) and
methylated septin 9 (SEPT9) (22–25),
were revealed to be effective in survival prediction in patients
with advanced lung or colorectal cancer.
Meanwhile, CA125 during pre-treatment exhibited an
ability to predict overall survival rate time in patients with
NSCLC. It was also able to predict overall survival rate time of
all included patients with stage IV lung cancer. For CEA, its level
change had the ability to predict overall survival rate time for
all stage IV subtypes and NSCLC specifically. In general, if
long-term survival is able to be predicted before treatment, more
options and earlier interventions will be available to patients. In
the current study, the level of mPRGER4, CA125 and NSE during
pre-treatment exhibited clear predictive capability and were
therefore more suitable than other markers.
Previous studies have revealed that protein markers
that are specific to certain lung cancer types can be used to
predict survival (28–34). For instance, lung adenocarcinoma
survival can be predicted by measuring CA125 (29,30) and
CEA (31) levels, while the survival
of patients with squamous cell carcinoma (SC) and SCLC can be
predicted by Cyfra211 (32) and NSE
levels (33,34), respectively. In the present study, it
was revealed that these markers were specific to each cancer type,
suggesting the potential application of these markers for the
survival prediction of different pathological subtypes accordingly.
Furthermore, NSE expression was associated with the survival of
patients with NSCLC. Notably, a longer overall survival time was
predicted by a higher post-treatment NSE level and the opposite was
true of CEA and CA125 levels, which warrants further
investigation.
Application of monitoring and
predictive biomarkers in clinical settings
Generally, there are two therapeutic options for
patients with advanced NSCLC, which depends on the patients'
susceptibility to a drug given TKI is normally used as the
first-line therapy for patients with TKI-associated drug-sensitive
mutations; otherwise, first-line chemotherapy or radiochemotherapy
is recommended (13). The
second-line treatment is often determined by the patients'
condition. Patients who have received first- or second-generation
TKI therapy will proceed with third-generation TKI if they have
developed resistance induced by specific mutations; otherwise,
chemotherapy or immunotherapy are considered (13). Patients with first-line chemotherapy
are generally treated with second-line chemotherapy or
immunotherapy upon the onset of resistance (13). For patients with SCLC, chemotherapy
is currently used as the first-line therapy, with second-line
chemotherapy or immunotherapy also considered viable options
(35). Furthermore, the FDA has
approved the use of immunotherapy as the first-line treatment of
NSCLC and SCLC (35). Notably, the
treatment of SC is distinct from that of the ADC due to the
differences in tumor biology, symptoms, targeting population and
molecular characteristics. SC generally exhibits a much lower rate
of mutations in key driver genes, such as EGFR or ALK (36), making chemotherapy as a more suitable
first-line therapy compared with TKI-based target therapy in the
majority of patients with ADC (13).
Second-line chemotherapy and/or immunotherapy are typically
suitable for patients with SC who have developed resistance to
first-line chemotherapy. Recently, first-line immunotherapy
(37) or first-line chemo/immuno
combined therapy (38) have been
suggested as an improved therapeutic options for patients with
late-stage SC.
Further development of therapeutic
strategies will provide more options in the treatment of late-stage
lung cancer
Regardless of whether the first or multiple lines of
therapies were given to patients with ADC, SC or SCLC, current
therapies on late-stage lung cancer require intensive real-time
monitoring of the patients' condition, as it is necessary to
investigate the patients' response before deciding whether to
maintain the current treatment or switch to alternative therapies.
In the present study, the effects of targeted therapy and
chemotherapy were evaluated simultaneously, although they are two
different types of therapies. This was because the current study
focused on the association between therapeutic response and the
mPTGER4 level, and the results of the present study support
previous reports (12,13,15)
which indicate that the level of methylation markers is closely
associated with therapeutic response, but not treatment regime.
Different treatments exhibit distinct responses that may be
reflected by methylation markers; however, treatment type was not
the focus of the present study and therefore all therapies were
assessed together.
Currently, CT examination is still the most commonly
used method, but it is radiative and hence not suitable for
repeated use in a short time period. The findings from CT images
only reflect a condition that has already happened instead of
continuously monitoring the real-time condition, resulting in
patients missing out on the best opportunity for treatment.
Therefore, response monitoring with blood markers may be an ideal
method, but it requires high sensitivity markers that exhibit a
good correlation with changes in tumor burden. Previous studies
have revealed that both methylation and protein markers can be used
for response monitoring, but not all changes in blood marker levels
were correlated with tumor burden, and there is a large variation
between individuals (17–19). These issues warrant further
investigation to characterize the performance of individual
markers. In addition, the positive detection rate of methylation
markers prior to treatment is typically higher than that of protein
markers, making them more suitable for response monitoring.
At present, the performance of a monitoring marker
can be assessed according to three aspects: i) Whether it can
accurately and timely reflect the real-time changes of a disease;
ii) whether it can predict the change or outcome of the disease;
and iii) whether it can predict long-term survival and prognosis of
a patient. Ideally, one marker or a panel of markers can meet all
three aspects simultaneously. The results of disease monitoring are
comparable, and a patient's response can be quantified. However,
such markers are rare and may not be effective for all patients,
therefore, the combination of multiple markers may still be
necessary to be sensitive to as many patients as possible.
Future in vivo study and validation on
PTGER4 protein expression
It is difficult to build up an in vivo
methylation model so far as methylation status varies greatly
across different genes and individuals. Since methylation is
dynamic and reversible and may be affected by many in vivo
factors, its level is hard to evaluate under experimental
conditions. Notably, in vivo methylation models targeting
certain genes are even more difficult to achieve. Therefore, the
majority of methylation studies now use in vitro models,
such as cell lines from patients or animals with confirmed
methylation status at certain genes. However, the majority of these
models represent an ideal situation where the methylation of a
certain gene is relatively stable, and the effect of intervention
can be assessed and quantified. Examples of studies using these
cell line models include a colorectal cancer cell line studying the
methylation marker SEPT9 (39), cell
lines with abnormal mutL homolog 1 methylation in colorectal cancer
and lung cancer (40) and cell lines
with abnormal PR/SET domain 2/5/16 gene methylation in lung cancer
(41).
There are several limitations in the present study.
The number of patients in the present study was small especially
when survival analysis was performed on NSCLC patients. Future
large-scale studies are needed to verify the conclusions of this
study. Furthermore, the methylation change at the individual
patient level may not accurately reflect the patient condition,
since blood methylation level is affected by many factors.
Therefore, decision of therapeutic strategies based on combined
information from multiple examinations including methylation,
protein markers and imaging methods should be applied to patients.
There was a small proportion of patients who cannot be assessed by
PTGER4, due to it's sensitivity prior to therapy not being 100%.
The therapeutic effect in these patients must be assessed by other
methods, such as the use of protein markers or imaging methods
(e.g. CT and MRI).
In the present study, the blood methylation level of
PTGER4 gene was tested, which was developed as a new marker for
discriminating benign from malignant lung nodules (11), while the validation of PTGER4 at
protein level has not yet been performed. At present, there is no
systematic study comparing the expression level of PTGER4 in lung
cancer tissues, para-carcinoma tissues or normal tissues.
Theoretically, hypermethylation at the promoter region of PTGER4 in
lung cancer may result in decreased expression of PTGER4 protein,
but the mechanism is yet to be elucidated. Moreover, the expression
level of PTGER4 may also be influenced by changes in other pathways
in lung cancer. Therefore, it is also worth exploring the potential
affecting factors of PTGER4 expression. Investigation of PTGER4
expression may help to validate the PTGER4 methylation assay and
potentially indicate PTGER4 as a new protein marker.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Special Funds
for Strategic Emerging Industry Development of Shenzhen (grant no.
20170922151538732) and the Science and Technology Project of
Shenzhen (grant no. JSGG20180703164202084). Funding bodies did not
influence the study design, study implementation, data collection,
data analysis, data interpretation and manuscript writing of the
present study.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FZ and LS designed the study. YZ and JH performed
the statistical analysis and wrote the manuscript. YZ, JH, QZ, JC,
KY, QF, DQ, JW and EB collected the samples, clinical information,
diagnostic information and performed the patient follow-up. YZ and
JH analyzed the data. LS proof read the manuscript and submitted
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Affiliated Hospital of Jiangnan University and the
eighth medical center of the Chinese People's Liberation Army
general hospital. Prospective samples were collected and written
informed consent to participate was acquired from the patients.
Patient consent for publication
Consent for publication was obtained from the
participants before the start of the study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
National Lung Screening Trial Research
Team, ; Church TR, Black WC, Aberle DR, Berg CD, Clingan KL, Duan
F, Fagerstrom RM, Gareen IF, Gierada DS, et al: Results of initial
low-dose computed tomographic screening for lung cancer. N Engl J
Med. 368:1980–1991. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Belloni E, Veronesi G, Rotta L, Volorio S,
Sardella D, Bernard L, Pece S, Di Fiore PP, Fumagalli C, Barberis
M, et al: Whole exome sequencing identifies driver mutations in
asymptomatic computed tomography-detected lung cancers with normal
karyotype. Cancer Genet. 208:152–155. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Uchida J, Kato K, Kukita Y, Kumagai T,
Nishino K, Daga H, Nagatomo I, Inoue T, Kimura M, Oba S, et al:
Diagnostic accuracy of noninvasive genotyping of EGFR in lung
cancer patients by deep sequencing of plasma cell-free DNA. Clin
Chem. 61:1191–1196. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jin X, Chen Y, Chen H, Fei S, Chen D, Cai
X, Liu L, Lin B, Su H, Zhao L, et al: Evaluation of tumor-derived
exosomal miRNA as potential diagnostic biomarkers for early-stage
non-small cell lung cancer using next-generation sequencing. Clin
Cancer Res. 23:5311–5319. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Leng Q, Lin Y and Jiang F, Lee CJ, Zhan M,
Fang H, Wang Y and Jiang F: A plasma miRNA signature for lung
cancer early detection. Oncotarget. 8:111902–111911. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ye M, Li S, Huang W, Wang C, Liu L, Liu J,
Liu J, Pan H, Deng Q, Tang H, et al: Comprehensive targeted
super-deep next generation sequencing enhances differential
diagnosis of solitary pulmonary nodules. J Thorac Dis. 10 (Suppl
7):S820–S829. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Feng Y, Feng G, Lu X, Qian W, Ye J,
Manrique CA, Ma C and Lu Y; written on behalf of the AME Lung
Cancer Collaborative Group, : Exploratory analysis of introducing
next-generation sequencing-based method to treatment-naive lung
cancer patients. J Thorac Dis. 10:5904–5912. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Oellerich M, Schütz E, Beck J and Walson
PD: Circulating cell-free DNA-diagnostic and prognostic
applications in personalized cancer therapy. Ther Drug Monit.
41:115–120. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lim M, Kim CJ, Sunkara V, Kim MH and Cho
YK: Liquid biopsy in lung cancer: Clinical applications of
circulating biomarkers (CTCs and ctDNA). Micromachines (Basel).
9:E1002018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mayo-de-Las-Casas C, Garzón Ibáñez M,
Jordana-Ariza N, García-Peláez B, Balada-Bel A, Villatoro S,
Malapelle U, Karachaliou N, Troncone G, Rosell R and Molina-Vila
MA: An update on liquid biopsy analysis for diagnostic and
monitoring applications in non-small cell lung cancer. Expert Rev
Mol Diagn. 18:35–45. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Weiss G, Schlegel A, Kottwitz D, König T
and Tetzner R: Validation of the SHOX2/PTGER4 DNA methylation
marker panel for plasma-based discrimination between patients with
malignant and nonmalignant lung disease. J Thorac Oncol. 12:77–84.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chu X, Zhao W and Wang B: DNA methylation
detection of SHOX2 and PTGER4 in plasma contributes to differential
diagnosis of pulmonary nodule patients. Xi Bao Yu Fen Zi Mian Yi
Xue Za Zhi. 35:357–361. 2019.(In Chinese). PubMed/NCBI
|
|
13
|
Ettinger DS, Wood DE, Aisner DL, Akerley
W, Bauman J, Chirieac LR, D'Amico TA, DeCamp MM, Dilling TJ,
Dobelbower M, et al: Non-small cell lung cancer, version 5.2017,
NCCN clinical practice guidelines in oncology. J Natl Compr Canc
Netw. 15:504–535. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schmidt B, Beyer J, Dietrich D, Bork I,
Liebenberg V and Fleischhacker M: Quantification of cell-free
mSHOX2 Plasma DNA for therapy monitoring in advanced stage
non-small cell (NSCLC) and small-cell lung cancer (SCLC) patients.
PLoS One. 10:e01181952015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peng XM, Liu XL, Xu L, Li Y, Wang H, Song
L and Xiao W: The mSHOX2 is capable of assessing the therapeutic
effect and predicting the prognosis of stage IV lung cancer. J Thor
Dis. 11:2458–2469. 2019. View Article : Google Scholar
|
|
17
|
Duffy MJ and O'Byrne K: Tissue and blood
biomarkers in lung cancer: A review. Adv Clin Chem. 86:1–21. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang B, Li X, Ren T and Yin Y:
Autoantibodies as diagnostic biomarkers for lung cancer: A
systematic review. Cell Death Discov. 5:1262019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qin J, Zeng N, Yang T, Wan C, Chen L, Shen
Y and Wen F: Diagnostic value of autoantibodies in lung cancer: A
systematic review and meta-analysis. Cell Physiol Biochem.
51:2631–2646. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Epi proLung. Instruction for use.
Epigenomics AG; Berlin: 2018, https://www.epigenomics.com/wp-content/uploads/2018/08/IFU_0018_GB_rev4_Instructions_for_Use_Epi_proLung.pdfAugust
6–2018
|
|
22
|
Song L, Guo S, Wang J, Peng X, Jia J, Gong
Y, Yang B, Xiao W, Dong C, Liu H and Li Y: The blood mSEPT9 is
capable of assessing the surgical therapeutic effect and the
prognosis of colorectal cancer. Biomark Med. 12:961–973. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Song L, Yu H, Jia J and Li Y: A systematic
review of the performance of the SEPT9 gene methylation assay in
colorectal cancer screening, monitoring, diagnosis and prognosis.
Cancer Biomark. 18:425–432. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jahr S, Hentze H, Englisch S, Hardt D,
Fackelmayer FO, Hesch RD and Knippers R: DNA fragments in the blood
plasma of cancer patients: Quantitations and evidence for their
origin from apoptotic and necrotic cells. Cancer Res. 61:1659–1665.
2001.PubMed/NCBI
|
|
25
|
Dietrich D, Hasinger O, Liebenberg V,
Field JK, Kristiansen G and Soltermann A: DNA methylation of the
homeobox genes PITX2 and SHOX2 predicts outcome in non-small-cell
lung cancer patients. Diagn Mol Pathol. 21:93–104. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bergheim J, Semaan A, Gevensleben H,
Groening S, Knoblich A, Dietrich J, Weber J, Kalff JC, Bootz F,
Kristiansen G and Dietrich D: Potential of quantitative SEPT9 and
SHOX2 methylation in plasmatic circulating cell-free DNA as
auxiliary staging parameter in colorectal cancer: A prospective
observational cohort study. Br J Cancer. 118:1217–1228. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Song L, Jia J, Yu H, Peng X, Xiao W, Gong
Y, Zhou G, Han X and Li Y: The performance of the mSEPT9 assay is
influenced by algorithm, cancer stage and age, but not sex and
cancer location. J Cancer Res Clin Oncol. 143:1093–1101. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Song L, Jia J, Peng X, Xiao W and Li Y:
The performance of the SEPT9 gene methylation assay and a
comparison with other CRC screening tests: A meta-analysis. Sci
Rep. 7:30322017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang H, Shen L, Geng J, Wu Y, Xiao H,
Zhang F and Si H: Prognostic value of cancer antigen −125 for lung
adenocarcinoma patients with brain metastasis: A random survival
forest prognostic model. Sci Rep. 8:56702018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Isaksson S, Jönsson P, Monsef N,
Brunnström H, Bendahl PO, Jönsson M, Staaf J and Planck M: CA 19-9
and CA 125 as potential predictors of disease recurrence in
resectable lung adenocarcinoma. PLoS One. 12:e01862842017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Grunnet M and Sorensen JB:
Carcinoembryonic antigen (CEA) as tumor marker in lung cancer. Lung
Cancer. 76:138–143. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu Z, Zhang G, Yang M, Zhang S, Zhao B,
Shen G and Chai Y: Systematic review of CYFRA 21-1 as a prognostic
indicator and its predictive correlation with clinicopathological
features in non-small cell lung cancer: A meta-analysis.
Oncotarget. 8:4043–4050. 2017.PubMed/NCBI
|
|
33
|
Xue F, Zhu L, Wang L and Wang Q: Serum
neuron specific enolase levels correlate with patient prognosis for
advanced lung cancer. Int J Clin Exp Med. 8:9498–9504.
2015.PubMed/NCBI
|
|
34
|
Liu X, Zhang W, Yin W, Xiao Y, Zhou C, Hu
Y and Geng S: The prognostic value of the serum neuron specific
enolase and lactate dehydrogenase in small cell lung cancer
patients receiving first-line platinum-based chemotherapy. Medicine
(Baltimore). 96:e82582017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kalemkerian GP, Loo BW, Akerley W, Attia
A, Bassetti M, Boumber Y, Decker R, Dobelbower MC, Dowlati A,
Downey RJ, et al: NCCN guidelines insights: Small cell lung cancer,
version 2.2018. J Natl Compr Canc Netw. 16:1171–1182. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Roviello G: The distinctive nature of
adenocarcinoma of the lung. Onco Targets Ther. 8:2399–2406. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Reck M, Rodríguez-Abreu D, Robinson AG,
Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe
S, et al: Updated analysis of KEYNOTE-024: Pembrolizumab versus
platinum-based chemotherapy for advanced non-small-cell lung cancer
with PD-L1 tumor proportion score of 50% or greater. J Clin Oncol.
37:537–546. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Paz-Ares L, Luft A, Vicente D, Tafreshi A,
Gümüş M, Mazières J, Hermes B, Çay Şenler F, Csőszi T, Fülöp A, et
al: Pembrolizumab plus chemotherapy for squamous non-small-cell
lung cancer. N Engl J Med. 379:2040–2051. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ravegnini G, Zolezzi Moraga JM, Maffei F,
Musti M, Zenesini C, Simeon V, Sammarini G, Festi D, Hrelia P and
Angelini S: Simultaneous analysis of SEPT9 promoter methylation
status, micronuclei frequency, and folate-related gene
polymorphisms: The potential for a novel blood-based colorectal
cancer biomarker. Int J Mol Sci. 16:28486–28497. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ma Y, Chen Y and Petersen I: Expression
and promoter DNA methylation of MLH1 in colorectal cancer and lung
cancer. Pathol Res Pract. 213:333–338. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tan SX, Hu RC, Liu JJ, Tan YL and Liu WE:
Methylation of PRDM2, PRDM5 and PRDM16 genes in lung cancer cells.
Int J Clin Exp Pathol. 7:2305–2311. 2014.PubMed/NCBI
|