Introduction
Renal cell carcinoma (RCC) is the most common type
of kidney cancer (1). The incidence
of RCC has gradually increased worldwide in the last decades
(2). In 2015, 12,547 new cases of
RCC were registered in the UK, with an Age-Standardized Incidence
Rate of 20.8 (2). As RCC has
acquired chemoresistance, its prognosis remains low (3,4). In
addition, only 10% of patients with RCC and metastases survive
longer than five years (5,6). Furthermore, the underlying mechanisms
of RCC progression are poorly understood. A previous study reported
that molecular-targeted therapies have significant therapeutic
efficacy in RCC (7). It is therefore
crucial to identify novel diagnostic biomarkers in order to allow
the development of potential therapeutic interventions in RCC.
MicroRNAs (miRNAs) are small non-coding RNAs (~22
nucleotides in length) that regulate gene expression on a
post-transcriptional level through base-pairing with complementary
sequences of the 3′untranslated region (UTR) of mRNAs (8). Recent studies demonstrated that miR-720
and eight other miRNAs serve crucial roles in RCC (9,10). For
example, miR-92a-3p has been reported to serve a crucial role in
the progression of various types of cancer, including lung cancer,
esophageal squamous cell carcinoma, colorectal, breast, ovarian,
and cervical cancer (11–16). However, the functional significance
and molecular mechanism of miR-92a-3p in RCC remain unclear.
F-box and WD repeat domain containing 7 (FBXW7) has
been reported to act as a general tumor suppressor and to be one of
the most frequently dysregulated ubiquitin-proteasome system
proteins in human cancer (17). It
has been reported that FBXW7 is a substrate specifying subunit of
the evolutionarily conserved SKP1-CUL1-F-box ubiquitin ligase
complex, and that FBXW7 functions as a cell cycle regulatory gene
which protein products that regulate the stability of numerous
oncoprotein substrates, including cyclin E, c-Myc, Notch, c-Jun,
mammalian target of rapamycin and MCL1 (18,19). The
dysregulation of FBXW7 expression therefore serves critical roles
in cancer development. Furthermore, FBXW7 was demonstrated to be a
functional target of numerous miRNAs involved in the regulation of
cancer progression (20,21). However, the significance of miRNAs
and FBXW7 association in RCC requires further investigation
The present study investigated the effects of
miR-92a-3p in RCC cells. In addition, the association between
miR-92a-3p and its direct target gene FBXW7 was determined in order
to highlight the underlying mechanisms of miR-92a-3p in the
progression of RCC.
Materials and methods
Cell culture and tumor tissues
The human proximal tubule epithelial (HK-2) and
human renal cancer (ACHN and SN12PM6) cell lines were purchased
from the American Type Culture Collection. Cells were cultured in
Dulbecco's Modified Eagle's medium (Thermo Fisher Scientific, Inc)
supplemented with 10% (v/v) fetal bovine serum (Thermo Fisher
Scientific, Inc.) and placed at 37°C in a humidified incubator
containing 5% CO2.
A total of 16 pairs of RCC tissues and adjacent
non-tumor tissues were obtained from the tissue bank of the Hubei
University of Chinese Medicine. The inclusion criteria were as
follows: i) Provision of prior nephrectomy; ii) histological
confirmation of clear-cell RCC with metastases; iii) Eastern
Cooperative Oncology Group performance status of 0 or 1; and iv)
adequate hematologic, hepatic, renal and cardiac functions.
Patients were excluded if they had cardiac disease,
antibiotic-requiring systemic infections, coagulation disorders,
second malignancies, organ allografts, corticosteroid dependence,
and infection with human immunodeficiency virus or hepatitis. The
age of patients ranged from 51 to 82 years (median age 67 years)
and 68% of patients were men. The mean tumor size was 4.7 cm and
histologic necrosis was present in 25.1% of cases. Tissues were
collected between January 2015 and November 2017, immediately
minced on ice, frozen in liquid nitrogen and stored at −80°C until
further analysis. Patients were staged using radiographic reports
and postoperative pathological data according to the 2010 American
Joint Committee on Cancer Tumor-Node-Metastasis classification
(22). Patients with N1- or M1-stage
tumors were considered to have metastatic disease and were excluded
from this study. The present study was approved by the Ethics
Committee of Hubei University of Chinese Medicine, and informed
consent was obtained from each patient.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
A mirVana™ miRNA Isolation Kit (Ambion; Thermo
Fisher Scientific, Inc.) was used to extract total RNA from tissue
samples and cells according to the manufacturer's instructions.
Subsequently, RNAs were reverse transcribed into cDNA using a High
Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific, Inc.) following
removal of residual DNA by DNase I (Invitrogen; Thermo Fisher
Scientific, Inc.). FBXW7 gene expression was detected by TaqMan™
RT-qPCR in a QuantStudio 6 Flex System (Thermo Fisher Scientific,
Inc.). β-actin was used as an internal control. The relative
expression level of FBXW7 was presented as the fold difference
relative to β-actin. The relative expression level of FBXW7 was
normalized to endogenous controls and was expressed as
2−ΔΔCq (23).
To quantify the expression of mature miRNAs, total
RNA was reverse-transcribed using a TaqMan™ Advanced miRNA cDNA
Synthesis Kit, according to the manufacturer's recommended
protocols (Applied Biosystems; Thermo Fisher Scientific, Inc.). U6
small nuclear RNA (snRNA) was used as the internal control and
reverse-transcribed using a TaqMan™ microRNA Reverse Transcription
Kit, according to the manufacturer's protocol. The relative
expression level of miR-92a-3p was calculated as the fold
difference relative to U6. All TaqMan probes were purchased from
Thermo Fisher Scientific, Inc. (FBXW7, cat. no. Hs00217794;
β-actin, cat. no. Hs99999903; miR-92a-3p, cat no. 477827; U6, cat
no. 001973). The thermocycling conditions were as follows: Hold
stage, 95°C for 20 sec; PCR stage, 95°C for 1 sec, 60°C for 20 sec
for 1 cycle, 40 cycles total.
Western blotting
Tissues and ACHN and SN12PM6 cell lines were lysed
in ice cold RIPA Lysis and Extraction buffer (Thermo Fisher
Scientific, Inc.). Briefly, 0.5 or 1 ml RIPA buffer was added to
5×106 cells in suspension or 0.1 g tissue, respectively.
Protein concentration was determined using a DC Protein Assay kit
(Bio-Rad Laboratories, Inc.). Proteins (15 µg) were separated on
4–15% precast gels (Bio-Rad Laboratories, Inc.) and transferred
onto nitrocellulose membranes (Bio-Rad Laboratories, Inc.).
Membranes were blocked with 5% skimmed milk in TBST for 1 h at room
temperature, and incubated with primary antibodies against FBXW7
(1:1,000; cat. no. ab109617; Abcam), CDC42 (1:1,000; cat. no.
ab187643; Abcam) and β-actin (1:500; cat. no. sc-47778; Santa Cruz
Biotechnology, Inc.) at 4°C overnight. Membranes were washed three
times with TBST and incubated with the goat anti-rabbit horseradish
peroxidase (HRP)-conjugated secondary antibody (1:2,000; cat. no.
STAR124P; Bio-Rad Laboratories, Inc.) or goat anti-mouse
HRP-conjugated secondary antibody (1:5,000; cat. no. STAR207P;
Bio-Rad Laboratories, Inc.) for 2 h at room temperature. Bands were
detected using the Enhanced Chemiluminescence Kit (Pierce; Thermo
Fisher Scientific, Inc.). Each experiment was repeated three times.
In order to avoid possible problems related to incomplete
stripping, all results were taken from separate blots. The relative
protein expression was normalized to endogenous control β-actin
using ImageJ software version 1.50i (National Institutes of
Health).
Cell transfection
The miR-92a-3p lentivirus expression vector and
inhibitor hsa-miR-92b-3p lentivector (anti-miR-92a-3p) were
purchased from Applied Biological Materials Inc. and were used to
stably overexpress or knock down miR-92a-3p, respectively, in ACHN
and SN12PM6 cell lines. Non-relevant sequence inserts acted as
negative controls for miR-92a-3p and miR-92a-3p inhibitor, named
miR-control and anti-miR-control, respectively (Applied Biological
Materials Inc.). The lentiviral construct expressing human FBXW7
short hairpin RNA (sh-FBXW7) that was used to knock down FBXW7 and
the negative control (sh-control) were generated using pLVTHM-GFP
lentiviral RNAi expression system (Addgene, Inc.) as previously
described (24). Furthermore, the
lentivirus-covered coding region FBXW7 cDNA sequence (FBXW7-OE) was
used to overexpress FBXW7, whereas control-OE was used as negative
control lentivirus (both from GeneChem, Inc.). ACHN and SN12PM6
cells (6×105 cells) were infected with 2.5 µg of
lentiviral particles containing the aforementioned vectors. Media
containing 2 µg/ml puromycin (Sigma-Aldrich; Merck KGaA) was used
to select infected cells after 48 h. The successfully infected
cells were maintained in complete medium containing 0.5 µg/ml
puromycin.
All transfection procedures were performed using 5
µg/ml polybrene (GeneChem, Inc.) according to the manufacturer's
instructions. Cells were collected for subsequent measurements
after 48 h transfection.
Target gene analysis of
miR-92a-3p
The available databases of TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org/ and http://www.mirbase.org/) were used to search for
candidate targets of miR-92a-3p. The term ‘miR-92a-3p’ was typed in
the search box and the candidate genes were then provided.
3′ UTR dual luciferase assay
For reporter assays, the 3′-UTR of FBXW7 within the
predicted target sites was cloned into the pRL-TK vector (Promega
Corporation) for luciferase expression, as previously described
(25,26). Cells were seeded into 96-well plates
at a density of 2×104 cells/well. Cells were
co-transfected with 120 ng miR-92a-3p or miR-control and 30 ng of
the wild-type 3′-UTR of FBXW7 mRNA. Following 48 h transfection,
the cells were collected and gene expression was assessed using the
dual luciferase assay system (Promega Corporation), according to
the manufacturer's instructions. The pRL-TK expressing Renilla
luciferase was co-transfected as an internal control to reduce
variation from different transfection and harvest efficiencies
(27).
Cell proliferation assay
ACHN and SN12PM6 cells that were transfected with
different vectors were seeded into 96-well plates at a density of
5×103 cells/well and incubated for 48 h. The Cell
Counting Kit-8 (CCK-8) assay (Invitrogen; Thermo Fisher Scientific,
Inc.) was used to measure cell proliferation, according to the
manufacturer's protocol. Cell proliferation was detected at 0, 24,
48, 72, 96 and 120 h. Briefly, 10 µl CCK-8 solution was added to
each well and incubated for 2 h at 37°C. Optical density was
measured at 450 nm with an automatic microplate reader (28) and data were expressed as the means ±
standard error (SE).
Soft agar assay
Anchorage-independent growth is the ability of
transformed cells to grow independently on a solid surface and is
considered as a hallmark of carcinogenesis (29). The anchorage-independent growth of
ACHN and SN12PM6 cells that were transfected with different vectors
was examined by colony formation in soft agarose. Some 6-well
plates were coated with 0.5% (v/v) agarose diluted in cell media at
42°C for 1 h. Cells (5,000/well) were resuspended in heated medium
containing 0.4% (v/v) low-melt agarose and incubated in the 6-well
plate for 1–4 weeks. Colonies were stained with crystal violet
(0.5% v/v; EMD Millipore) for 15 min at room temperature and
counted manually by light microscopy.
Statistical analysis
Each experiment was set up in triplicate and
repeated three times. A two-tailed unpaired Student's t-test was
used to assess significance between two conditions. One-way ANOVA
followed by Tukey's post-hoc test was used to compare significant
differences between more than two conditions. Means and SEs were
calculated from numerical data. The SPSS software package (version
20.0; IBM Corp.) was used to perform statistical analyses.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-92a-3p is upregulated in RCC
tissues and cells
According to preliminary miRNA microarray data for
differentially expressed miRNAs in RCC tissues and adjacent
non-tumor tissues, miR-92a-3p is one of the upregulated miRNAs in
RCC tissues (data not shown). To verify these preliminary results,
16 pairs of RCC tissues and matched adjacent non-tumor tissues were
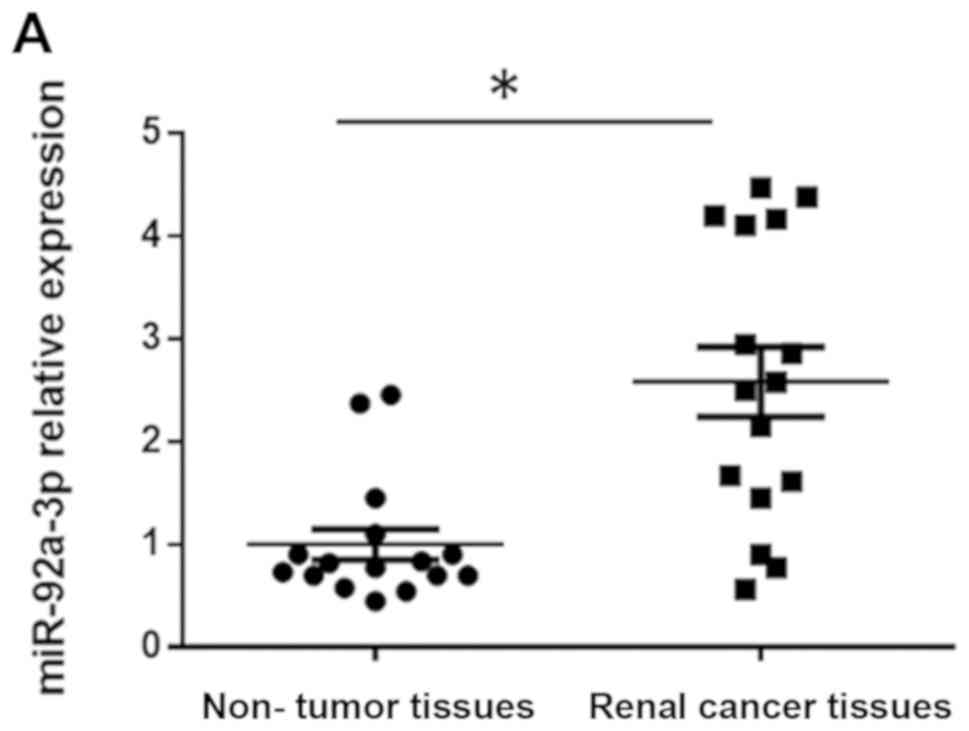
tested for miR-92a-3p expression level. The results demonstrated
that the mean level of miR-92a-3p expression was significantly
upregulated by 2.6-fold (P<0.05) in RCC tissues compared with
matched adjacent non-tumor tissues (Fig.
1A). Subsequently, miR-92a-3p expression level was determined
by RT-qPCR in the RCC cell lines ACHN and SN12PM6 and the
non-tumorigenic renal cell line HK-2. The results demonstrated that
miR-92a-3p relative expression level was significantly upregulated
in ACHN and SN12PM6 cells compared with HK-2 cells (Fig. 1B).
Effects of miR-92a-3p on RCC cell
proliferation
To determine the biological function of miR-92a-3p,
the miR-92a-3p and anti-miR-92a-3p were stably expressed in ACHN
and SN12PM6 cell lines, and the endogenous expression level of
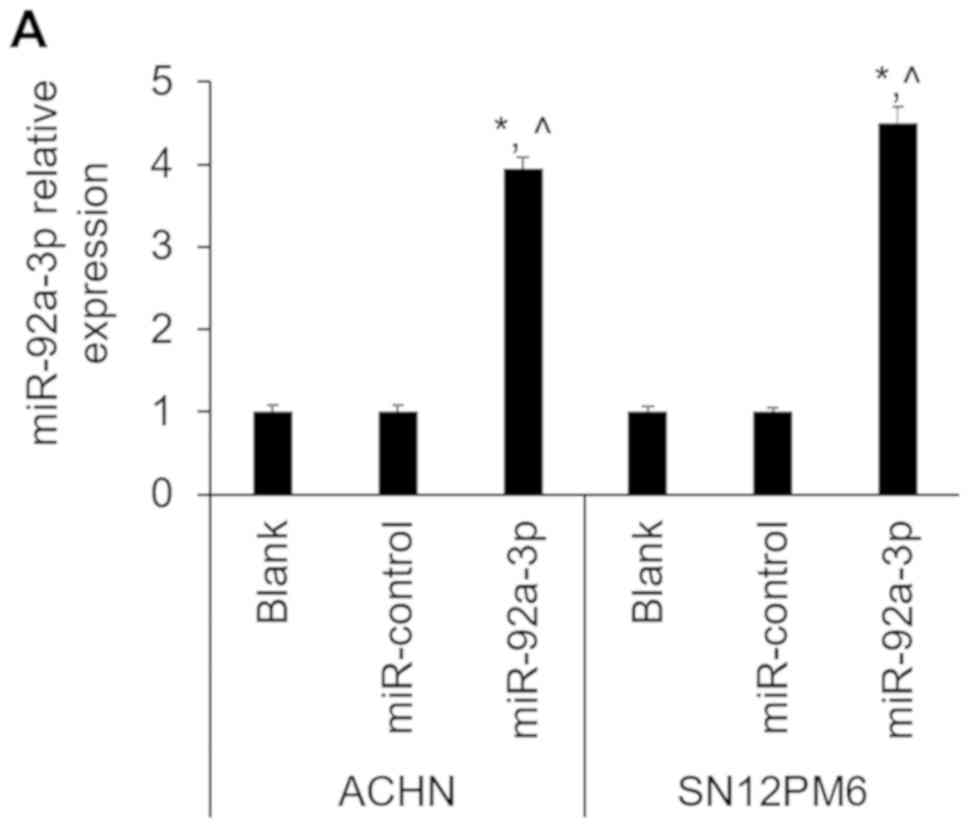
miR-92a-3p in ACHN and SN12PM6 cells was determined by RT-qPCR. The
results demonstrated that miR-92a-3p expression level was
significantly increased in miR-92a-3p-overexpressing cells compared
with miR-control and untransfected cells (blank) (Fig. 2A). Conversely, miR-92a-3p expression
level was significantly decreased following anti-miR-92a-3p cell
transfection compared with anti-miR-control and untransfected cells
(blank) (Fig. 2B). Furthermore,
miR-92a-3p-overexpressing cells exhibited significant increased
cell proliferation compared with miR-control and untransfected
cells (Fig. 2C). In addition, the
number of colonies formed under anchorage-independent conditions
was significantly increased (P<0.05) in
miR-92a-3p-overexpressing cells compared with untransfected ACHN
and SN12PM6 cells (Fig. 2D and E).
Conversely, miR-92a-3p downregulation inhibited cell proliferation
compared with anti-miR-control and untransfected cells (Fig. 2F). In particular, the number of
colonies formed under anchorage-independent conditions was also
decreased (P<0.05) following miR-92a-3p knockdown (Fig. 2D and E) compared with untransfected
ACHN and SN12PM6 cells. In addition, the protein expression of the
cell proliferation-associated marker Cdc42, was increased in cells
overexpressing miR-92a-3p compared with miR-control and
untransfected cells (Fig. 2G);
however, Cdc42 protein expression was decreased in cells following
miR-92a-3p knockdown compared with anti-miR-control and
untransfected cells (Fig. 2H).
FBXW7 is downregulated in RCC tissues
and cells
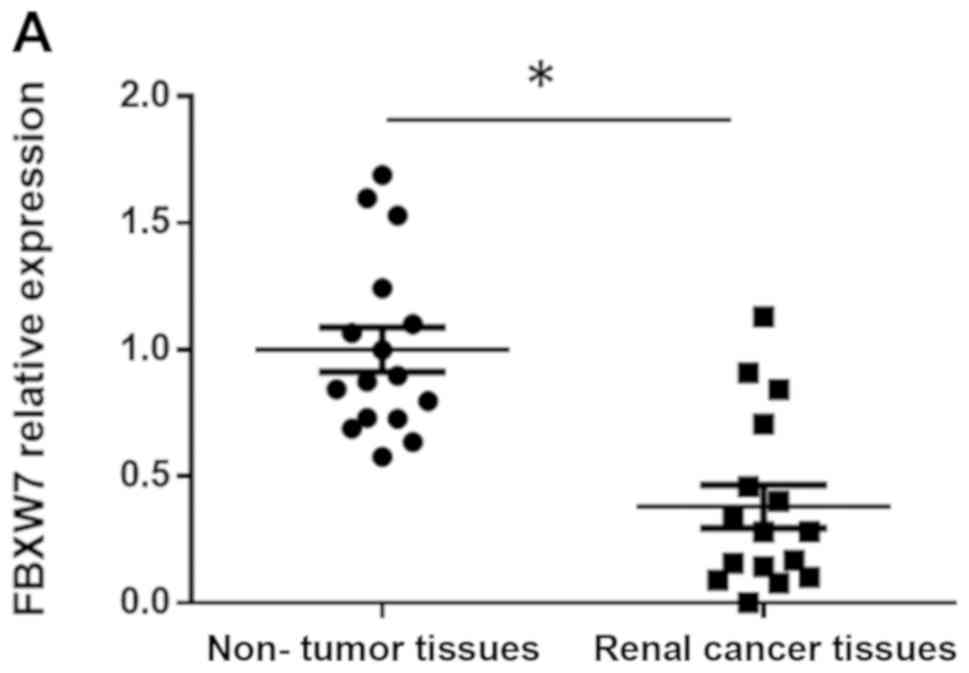
FBXW7 expression level was determined in 16 RCC
tissue samples by RT-qPCR and western blotting. The results
demonstrated that FBXW7 mRNA expression levels was significantly
lower in RCC tissues compared with in matched adjacent non-tumor
tissues (P<0.05; Fig. 3A).
Furthermore, FBXW7 protein expression was also lower in the RCC
tissues compared with paired non-tumor tissues (Fig. 3B). Subsequently, FBXW7 mRNA and
protein levels were determined in HK-2, ACHN and SN12PM6 cells by
RT-qPCR and western blotting. The results demonstrated that FBXW7
mRNA and protein expression levels were significantly downregulated
in ACHN and SN12PM6 cells compared with HK-2 cells (Fig. 3C-E).
FBXW7 is the direct target of
miR-92a-3p
The complimentary sequence of miR-92a-3p has already
been verified in the 3′UTR of FBXW7 mRNA in human hepatocellular
carcinoma (26). In the present
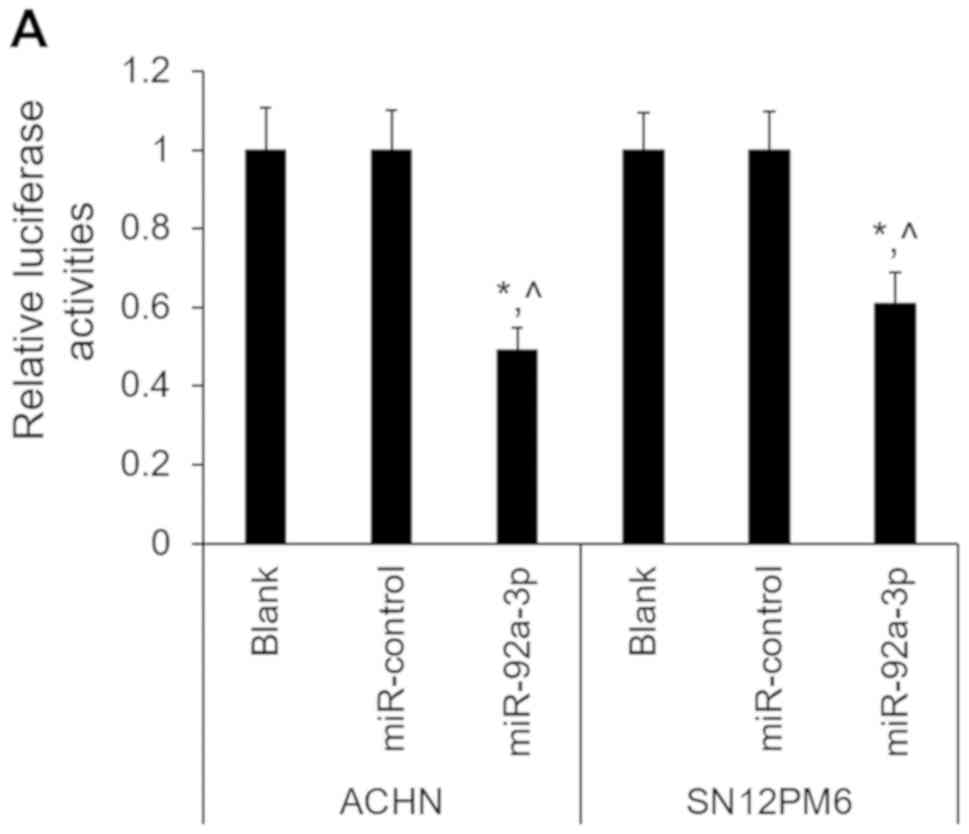
study, the results from the 3′-UTR luciferase assay confirmed that
FBXW7 was the direct target of miR-92a-3p in ACHN and SN12PM6
cells. Furthermore, miR-92a-3p significantly suppressed the
luciferase activity of FBXW7, which was not the case with
miR-control (Fig. 4A). In addition,
FBXW7 mRNA and protein expression were significantly decreased
following miR-92a-3p overexpression in ACHN and SN12PM6 cells
(Fig. 4B-D). Conversely, FBXW7 mRNA
and protein expression were significantly increased following
miR-92a-3p knockdown (Fig. 4E-G).
These results indicated that FBXW7 was directly suppressed by
miR-92a-3p in RCC cells.
Alterations in FBXW7 expression mimic
the effects of miR-92a-3p on RCC cells
To further confirm that FBXW7 may be a downstream
functional target of miR-92a-3p, FBXW7 was knocked down or
overexpressed by FBXW7 shRNA or FBXW-OE lentivirus, respectively,
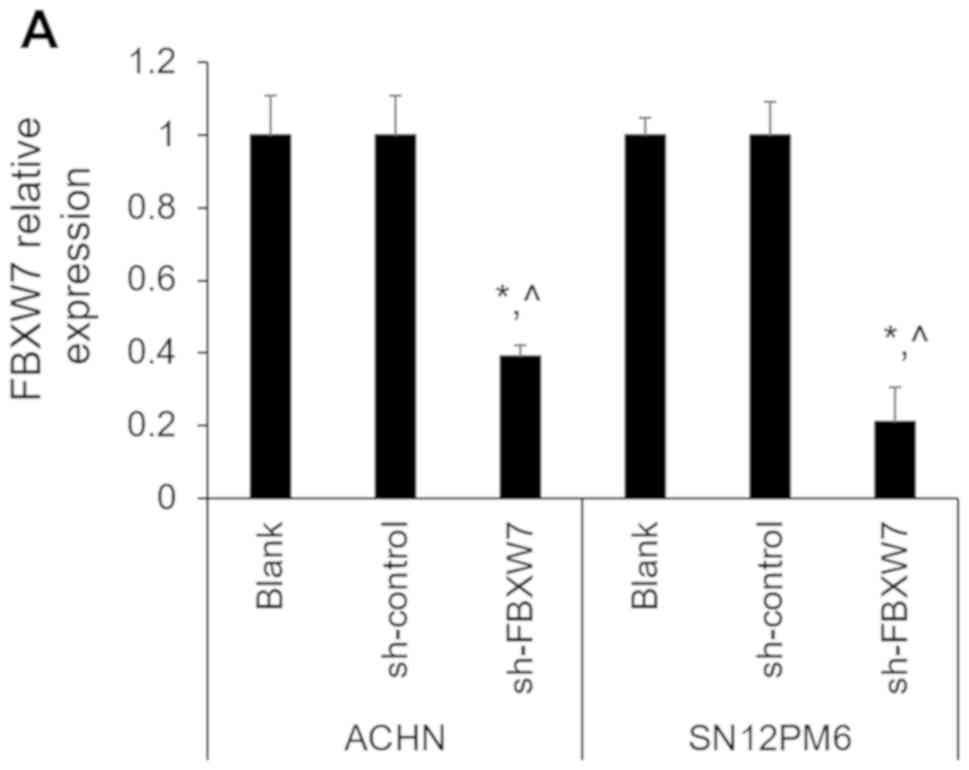
in ACHN and SN12PM6 cell lines. The efficiency of FBXW7 knockdown
or overexpression was verified by RT-qPCR and western blotting. As
presented in Fig. 5A and B, FBXW7
shRNA significantly reduced the mRNA expression level of FBXW7
compared with untransfected cells and sh-control, whereas FBXW-OE
significantly upregulated FBXW7 mRNA expression compared with
untransfected cells and control-OE. To further confirm these
results, western blotting was used to determine FBXW7 protein level
following sh-FBXW7 or FBXW7-OE-transfection of ACHN and SN12PM6
cells. The results demonstrated that FBXW7 protein level was
significantly decreased after sh-FBXW7 transfection compared with
sh-control and untransfected cells (Fig.
5C and D). Conversely, FBXW7 protein level was significantly
upregulated following FBXW7 overexpression compared with control-OE
and untransfected cells (Fig. 5E and
F). In addition, FBXW7 knockdown significantly stimulated cell
proliferation (Fig. 5G) and
increased the number of colonies formed under anchorage-independent
conditions compared with untransfected cells (Fig. 2D), which was similar to the results
observed following miR-92a-3p overexpression. Conversely, FBXW7
overexpression significantly inhibited cell proliferation (Fig. 5H) and decreased the number of
colonies formed under anchorage-independent conditions compared
with untransfected cells (Fig.
2D).
Discussion
At present, miRNAs dysregulation is an area of great
interest when investigating the molecular mechanisms of renal
tumorigenesis (30). Numerous
studies reported that miRNAs serve crucial roles in the regulation
of various cellular processes, including cell cycle control,
proliferation, migration and apoptosis (31,32).
Furthermore, aberrant miRNA expression has been associated with a
wide range of human diseases, including cancer (33–35).
Among these miRNAs, miR-92a-3p has been frequently reported to be
dysregulated in various types of cancer, including human acute
megakaryoblastic leukemia, glioma and Wilms tumor (36–38). For
example, miR-92a can mediate AZD6244-induced apoptosis and G1-phase
arrest of lymphoma cells by targeting Bim (39). Previous study reported that
miR-92a-3p inhibits proliferation, migration and invasion of Wilms
tumor cells by targeting notch receptor 1 (NOTCH1) (38). In addition, it has been demonstrated
that miR-92a-3p is upregulated in cervical cancer and promotes cell
proliferation and invasion by targeting FBXW7 (16). However, the biological function and
underlying mechanism of miR-92a-3p in RCC remain unknown. In the
present study, miR-92a-3p expression level was significantly
increased in RCC tissues compared with paired non-tumorous tissues.
Furthermore, miR-92a-3p expression was upregulated in ACHN and
SN12PM6 cells compared with HK-2 cells. In addition, miR-92a-3p
overexpression promoted RCC cell proliferation and stimulated the
number of colonies formed under anchorage-independent conditions.
Furthermore, miR-92a-3p downregulation inhibited cell proliferation
and reduced colonie number under anchorage-independent conditions.
These results suggested that miR-92a-3p may regulate RCC cell
proliferation.
The results from the present study suggested that
alteration of FBXW7 expression could interfere with the enhancing
effects of miR-92a-3p on tumor cell proliferation. In addition,
FBXW7 was confirmed to be a direct target of miR-92a-3p in human
hepatocellular carcinoma (26).
Furthermore, FBXW7 is associated with knockdown of family with
sequence similarity 83 member D, and could regulate the
proliferation, migration and invasion of colorectal cancer cells
(40). In addition, it has been
reported that FBXW7 functions as a tumor suppressor, inhibits
breast cancer growth and promotes cell apoptosis by targeting
metadherin for degradation (41). A
previous study reported that melanoma-associated antigen 1
interacts with FBXW7 to regulate the ubiquitin ligase-mediated
turnover of Notch1 intracellular domain 1 in breast and ovarian
cancer cells (42). FBXW7
upregulation also suppresses metastasis and epithelial mesenchymal
transition in RCC (43). However,
the biological function and underlying mechanism of FBXW7 on miRNAs
remain unknown in RCC. The results from the present study
demonstrated that FBXW7 was downregulated in RCC tissues compared
with adjacent non-tumorous tissues. Furthermore, miR-92a-3p
inversely regulated FBXW7 abundance in RCC cell lines. Besides,
miR-92a-3p directly suppressed FBXW7 using a 3′UTR luciferase assay
and western blotting. In addition, FBXW7 knockdown significantly
promoted cell proliferation and increased the number of colonies
formed under anchorage-independent conditions. Conversely, FBXW7
overexpression significantly suppressed cell proliferation and
decreased the number of colonies formed under anchorage-independent
conditions. These results strongly suggested that FBXW7 may be
considered a functional mediator of miR-92a-3p in RCC and that
miR-92a-3p may stimulate RCC cell proliferation by regulating
FBXW7; however, whether the downstream of FBXW7 is also
dysregulated in RCC remains to be investigated.
The results from this study indicated that
miR-92a-3p was upregulated in RCC tissues and cells. In addition,
miR-92a-3p could be considered as a novel miRNA promoting
proliferation and colony formation in RCC cells. The
tumor-promoting effect of miR-92a-3p may be associated with FBXW7.
In conclusion, the tight regulation of the newly discovered
miR-92a-3p/FBXW7 pathway may represent a potential novel strategy
and theoretical basis for improving the clinical treatment of
RCC.
Acknowledgements
Not applicable.
Funding
No funding was received
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Author's contributions
JL and RZ designed the experiments and wrote the
manuscript. RZ, JH and YS performed the in vitro study and
analyzed the data. All the authors read and approve the final
version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Hubei University of Chinese Medicine, and informed
consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kabaria R, Klaassen Z and Terris MK: Renal
cell carcinoma: Links and risks. Int J Nephrol Renovasc Dis.
9:45–52. 2016.PubMed/NCBI
|
|
2
|
Capitanio U, Bensalah K, Bex A, Boorjian
SA, Bray F, Coleman J, Gore JL, Sun M, Wood C and Russo P:
Epidemiology of renal cell carcinoma. Eur Urol. 75:74–84. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hsieh JJ, Purdue MP, Signoretti S, Swanton
C, Albiges L, Schmidinger M, Heng DY, Larkin J and Ficarra V: Renal
cell carcinoma. Nat Rev Dis Primers. 3:170092017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou J, Yun EJ, Chen W, Ding Y, Wu K, Wang
B, Ding C, Hernandez E, Santoyo J, Pong RC, et al: Targeting
3-phosphoinositide-dependent protein kinase 1 associated with
drug-resistant renal cell carcinoma using new oridonin analogs.
Cell Death Dis. 8:e27012017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Campbell SC, Flanigan RC and Clark JI:
Nephrectomy in metastatic renal cell carcinoma. Curr Treat Options
Oncol. 4:363–372. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Doberstein K, Steinmeyer N, Hartmetz AK,
Eberhardt W, Mittelbronn M, Harter PN, Juengel E, Blaheta R,
Pfeilschifter J and Gutwein P: MicroRNA-145 targets the
metalloprotease ADAM17 and is suppressed in renal cell carcinoma
patients. Neoplasia. 15:218–230. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Conti A, Santoni M, Amantini C, Burattini
L, Berardi R, Santoni G, Cascinu S and Muzzonigro G: Progress of
molecular targeted therapies for advanced renal cell carcinoma.
Biomed Res Int. 2013:4191762013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guo H, Ingolia NT, Weissman JS and Bartel
DP: Mammalian microRNAs predominantly act to decrease target mRNA
levels. Nature. 466:835–840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bhat NS, Colden M, Dar AA, Saini S, Arora
P, Shahryari V, Yamamura S, Tanaka Y, Kato T, Majid S and Dahiya R:
MicroRNA-720 regulates E-cadherin-αE-catenin complex and promotes
renal cell carcinoma. Mol Cancer Ther. 16:2840–2848. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ying G, Wu R, Xia M, Fei X, He QE, Zha C
and Wu F: Identification of eight key miRNAs associated with renal
cell carcinoma: A meta-analysis. Oncol Lett. 16:5847–5855.
2018.PubMed/NCBI
|
|
11
|
Chen ZL, Zhao XH, Wang JW, Li BZ, Wang Z,
Sun J, Tan FW, Ding DP, Xu XH, Zhou F, et al: microRNA-92a promotes
lymph node metastasis of human esophageal squamous cell carcinoma
via E-cadherin. J Biol Chem. 286:10725–10734. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nilsson S, Möller C, Jirström K, Lee A,
Busch S, Lamb R and Landberg G: Downregulation of miR-92a is
associated with aggressive breast cancer features and increased
tumour macrophage infiltration. PLoS One. 7:e360512012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ohyagi-Hara C, Sawada K, Kamiura S, Tomita
Y, Isobe A, Hashimoto K, Kinose Y, Mabuchi S, Hisamatsu T,
Takahashi T, et al: miR-92a inhibits peritoneal dissemination of
ovarian cancer cells by inhibiting integrin alpha5 expression. Am J
Pathol. 182:1876–1889. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ranade AR, Cherba D, Sridhar S, Richardson
P, Webb C, Paripati A, Bowles B and Weiss GJ: MicroRNA 92a-2*: A
biomarker predictive for chemoresistance and prognostic for
survival in patients with small cell lung cancer. J Thorac Oncol.
5:1273–1278. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu CW, Ng SS, Dong YJ, Ng SC, Leung WW,
Lee CW, Wong YN, Chan FK, Yu J and Sung JJ: Detection of miR-92a
and miR-21 in stool samples as potential screening biomarkers for
colorectal cancer and polyps. Gut. 61:739–745. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou C, Shen L, Mao L, Wang B, Li Y and Yu
H: miR-92a is upregulated in cervical cancer and promotes cell
proliferation and invasion by targeting FBXW7. Biochem Biophys Res
Commun. 458:63–69. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yeh CH, Bellon M and Nicot C: FBXW7: A
critical tumor suppressor of human cancers. Mol Cancer. 17:1152018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Inuzuka H, Shaik S, Onoyama I, Gao D,
Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, et al:
SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for
ubiquitylation and destruction. Nature. 471:104–109. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wertz IE, Kusam S, Lam C, Okamoto T,
Sandoval W, Anderson DJ, Helgason E, Ernst JA, Eby M, Liu J, et al:
Sensitivity to antitubulin chemotherapeutics is regulated by MCL1
and FBW7. Nature. 471:110–114. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chang H, Liu YH, Wang LL, Wang J, Zhao ZH,
Qu JF and Wang SF: MiR-182 promotes cell proliferation by
suppressing FBXW7 and FBXW11 in non-small cell lung cancer. Am J
Transl Res. 10:1131–1142. 2018.PubMed/NCBI
|
|
21
|
Hua J, Ding T and Yang L: Dysfunction of
microRNA-32 regulates ubiquitin ligase FBXW7 in multiple myeloma
disease. Onco Targets Ther. 9:6573–6579. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim SP, Alt AL, Weight CJ, Costello BA,
Cheville JC, Lohse C, Allmer C and Leibovich BC: Independent
validation of the 2010 American joint committee on cancer TNM
classification for renal cell carcinoma: Results from a large,
single institution cohort. J Urol. 185:2035–2039. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xiao Y, Yin C, Wang Y, Lv H, Wang W, Huang
Y, Perez-Losada J, Snijders AM, Mao JH and Zhang P: FBXW7 deletion
contributes to lung tumor development and confers resistance to
gefitinib therapy. Mol Oncol. 12:883–895. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu Y, Sengupta T, Kukreja L and Minella
AC: MicroRNA-223 regulates cyclin E activity by modulating
expression of F-box and WD-40 domain protein 7. J Biol Chem.
285:34439–34446. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang W, Dou C, Wang Y, Jia Y, Li C, Zheng
X and Tu K: MicroRNA-92a contributes to tumor growth of human
hepatocellular carcinoma by targeting FBXW7. Oncol Rep.
34:2576–2584. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Z, Tu K and Liu Q: Effects of
microRNA-30a on migration, invasion and prognosis of hepatocellular
carcinoma. FEBS Lett. 588:3089–3097. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qiu M, Xu Y, Wang J, Zhang E, Sun M, Zheng
Y, Li M, Xia W, Feng D, Yin R and Xu L: A novel lncRNA, LUADT1,
promotes lung adenocarcinoma proliferation via the epigenetic
suppression of p27. Cell Death Dis. 6:e18582015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Borowicz S, Van Scoyk M, Avasarala S,
Karuppusamy Rathinam MK, Tauler J, Bikkavilli RK and Winn RA: The
soft agar colony formation assay. J Vis Exp. e51998:2014.
|
|
30
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu Z, Sun J, Liu B, Zhao M, Xing E and
Dang C: miRNA222 promotes liver cancer cell proliferation,
migration and invasion and inhibits apoptosis by targeting BBC3.
Int J Mol Med. 42:141–148. 2018.PubMed/NCBI
|
|
32
|
Pan BL, Wu L, Pan L, Yang YX, Li HH, Dai
YJ, He ZQ, Tan L, Huang YG, Tong ZW and Liao JL: Up-regulation of
microRNA-340 promotes osteosarcoma cell apoptosis while suppressing
proliferation, migration, and invasion by inactivating the
CTNNB1-mediated Notch signaling pathway. Biosci Rep.
38:BSR201716152018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jafri MA, Al-Qahtani MH and Shay JW: Role
of miRNAs in human cancer metastasis: Implications for therapeutic
intervention. Semin Cancer Biol. 44:117–131. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Oliveto S, Mancino M, Manfrini N and Biffo
S: Role of microRNAs in translation regulation and cancer. World J
Biol Chem. 8:45–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Venkatadri R, Muni T, Iyer AK, Yakisich JS
and Azad N: Role of apoptosis-related miRNAs in resveratrol-induced
breast cancer cell death. Cell Death Dis. 7:e21042016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sharifi M and Salehi R: Blockage of
miR-92a-3p with locked nucleic acid induces apoptosis and prevents
cell proliferation in human acute megakaryoblastic leukemia. Cancer
Gene Ther. 23:29–35. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang Q, Teng Y, Wang R, Deng D, You Y,
Peng Y, Shao N and Zhi F: The long non-coding RNA SNHG14 inhibits
cell proliferation and invasion and promotes apoptosis by sponging
miR-92a-3p in glioma. Oncotarget. 9:12112–12124. 2018.PubMed/NCBI
|
|
38
|
Zhu S, Zhang L, Zhao Z, Fu W, Fu K, Liu G
and Jia W: MicroRNA92a3p inhibits the cell proliferation, migration
and invasion of Wilms tumor by targeting NOTCH1. Oncol Rep.
40:571–578. 2018.PubMed/NCBI
|
|
39
|
Lv XB, Zhang X, Deng L, Jiang L, Meng W,
Lu Z and Wang X: MiR-92a mediates AZD6244 induced apoptosis and
G1-phase arrest of lymphoma cells by targeting Bim. Cell Biol Int.
38:435–443. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mu Y, Zou H, Chen B, Fan Y and Luo S:
FAM83D knockdown regulates proliferation, migration and invasion of
colorectal cancer through inhibiting FBXW7/Notch-1 signalling
pathway. Biomed Pharmacother. 90:548–554. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen X, Li XY, Long M, Wang X, Gao ZW, Cui
Y, Ren J, Zhang Z, Liu C, Dong K and Zhang H: The FBXW7 tumor
suppressor inhibits breast cancer proliferation and promotes
apoptosis by targeting MTDH for degradation. Neoplasma. 65:201–209.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhao J, Wang Y, Mu C, Xu Y and Sang J:
MAGEA1 interacts with FBXW7 and regulates ubiquitin ligase-mediated
turnover of NICD1 in breast and ovarian cancer cells. Oncogene.
36:5023–5034. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cai Y, Zhang M, Qiu X, Wang B, Fu Y, Zeng
J, Bai J and Yang G: Upregulation of FBXW7 suppresses renal cancer
metastasis and epithelial mesenchymal transition. Dis Markers.
2017:82769392017. View Article : Google Scholar : PubMed/NCBI
|