Exosomes, ranging from 30–150 nm in diameter, are
nanoscale extracellular phospholipid bilayer vesicles, which
originate from endosomes and are stored within multivesicular
bodies (MVBs) (1). When MVBs fuse
with the cytomembrane, exosomes are released into the extracellular
environment or fuse with target cells, resulting in a series of
phenotypic changes (2,3). Exosomes encapsulate and transfer a
myriad of functional molecular cargoes, including proteins
(4), lipids, metabolites, DNA,
messenger RNA (mRNA), ribosomal RNA, microRNA (miRNA), transfer RNA
(tRNA), long non-coding RNA (lncRNA) and circular RNA (circRNA)
(5,6). As exosomes are released into the
external environment, circRNA begins cycling and binds miRNA or
proteins to exert various biological functions. Numerous studies
have confirmed that exosomes act as crucial mediators of
intercellular communication (7–9). In
addition, they function in numerous physiological processes, such
as blood coagulation (10), tissue
repair, skin regeneration (11) and
immune modulation (12); therefore,
exosomes play roles in cancer and other pathological processes such
as in cancer, cerebral ischemia and obesity (13–15).
circRNA belongs to the endogenous non-coding RNA
(ncRNA) family. Unlike linear RNA, circRNA is a closed circular
molecule that has a covalently closed loop structure, lacking a
poly A tail or 5′-3′ polarity (16).
Although circRNA was initially considered as a non-functional
by-product of aberrant RNA splicing, numerous studies have
investigated this molecule class (17–19), and
the development of bioinformatics approaches and next-generation
deep sequencing technology have contributed to the discovery and
identification of an increasing number of circRNAs with regulatory
functions (19). circRNAs are
molecules that display high enrichment and relative stability,
diversity and evolutionary conservation; they also exhibit
tissue-specific and developmental phase-specific expression
(20). These characteristics
indicate that circRNAs have distinct properties and diverse
cellular regulatory functions, including the regulation of cellular
processes, such as proliferation and apoptosis (21–23).
Furthermore, it has been revealed that circRNAs have crucial
regulatory functions in various aspects of cancer, such as
tumorigenesis (24), proliferation,
migration, invasion (25),
metastasis (26), apoptosis, and
chemotherapy and radiation resistance (27,28), as
well as in cancer prognosis (29).
Previous studies have demonstrated that circRNAs are
stable in cells and within exosomes (6). It has been revealed that exosomal
circRNAs may have important regulatory functions, and due to their
unique structure and high specificity, the use of combinations of
exosomes and circRNAs may increase the potential clinical
applications of these molecules as markers in the diagnosis and
prognosis of cancer (30,31). For example, high expression of plasma
exosomal circ-PDE8A is associated with lymphatic invasion and
advanced tumour stage, as well as poor survival in patients with
pancreatic ductal adenocarcinoma (PDAC) (32). Therefore, exosomal circ-PDE8A may be
used as a marker for determining diagnosis or progression of the
disease in patients with PDAC (32).
Notably, regarding the role of exosomal circRNA in cancer, exosomes
containing circRNAs may function to promote or inhibit cancer
progression. Exosomal circRNAs promote the progression
(proliferation and invasiveness) of cancer, the generation of
premetastatic niches and the occurrence of metastasis (33,34). By
contrast, exosomal circRNAs also regulate tumour immunity and
immunotherapy and play a role in cancer treatment (35,36).
In previous years, numerous studies on exosomal
lncRNAs and miRNAs in cancer have been conducted, but relatively
little attention has been devoted to exosomal circRNAs. In the
current review, an overview of exosomal circRNAs is provided; the
biogenesis and biological functions of circRNAs are discussed, the
abundance of circRNAs in exosomes and their possible sorting
mechanisms are analysed and their potential emerging roles in
promoting or inhibiting cancer progression are examined.
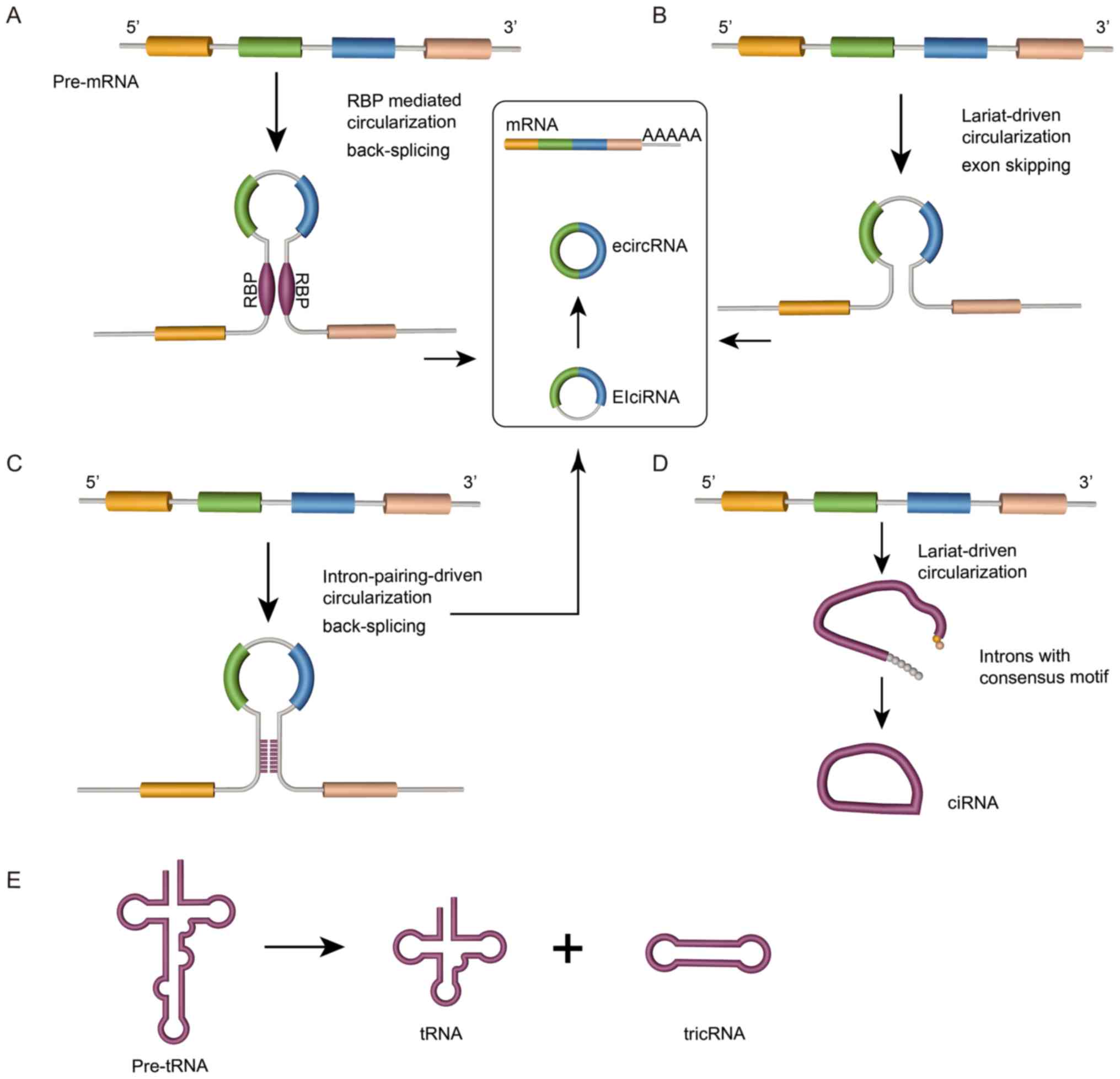
Unlike linear RNAs, circRNAs are generated by
back-splicing or exon skipping of pre-mRNAs, with direct
back-splicing occurring more frequently than exon skipping
(37). circRNAs are cyclized into a
continuous closed-loop structure that lacks a polyadenylation (poly
A) tail and 5′ cap (38). Based on
their composition, circRNAs are divided into four categories:
Exonic circRNA (ecircRNAs), exon-intron circRNAs (EIciRNAs),
circular intronic RNAs (ciRNAs) and tRNA intronic circular RNAs
(tricRNAs). Currently, three mechanisms for circRNA biogenesis are
widely accepted (Fig. 1):
RNA-binding protein (RBP)-mediated circularization, intron
pairing-driven circularization and lariat-driven circularization.
First, the RBP-mediated and intron pairing-driven circularization
mechanisms of circRNA biogenesis occur via the direct back-splicing
pathway (18). RBPs have an
important function in promoting circRNA biogenesis by regulating
adjacent splice sites (Fig. 1A). For
instance, the splicing factors Muscleblind (39), Quaking (40), adenosine deaminase RNA specific
(41) and DExH-box helicase 9
(42) are all reported to
participate in the formation of circRNA. Furthermore,
circularization can also occur by complementary pairing of flanking
introns that contain inverted complementary sequences (Fig. 1B) (43). Finally, lariat-driven circularization
is facilitated by an exon-skipping event (Fig. 1C). Internal splicing facilitates the
removal of the flanking intronic sequence, allowing the production
of ecircRNAs (44); if these
flanking sequences are retained, the constructs are called EIciRNAs
(45). Additionally, ciRNAs are
generated via lariat-driven circularization, which is facilitated
by a ciRNA-specific consensus motif (Fig. 1D) (46). TricRNAs are generated by a
combination of the released intron terminal ends, which come from
spliced pre-tRNAs via the tRNA splicing endonuclease complex
(Fig. 1E) (47).
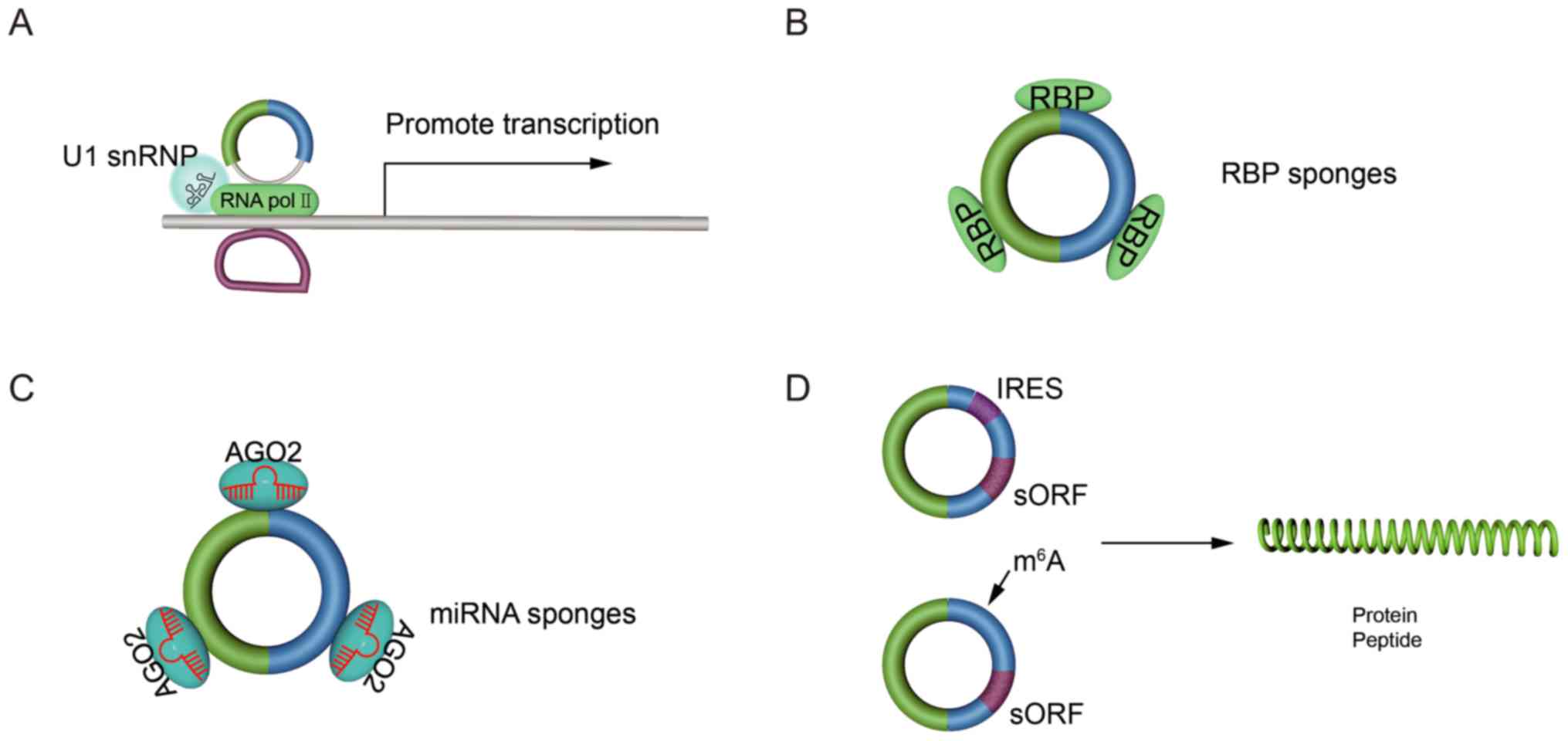
circRNAs are abundant in the cytoplasm,
evolutionarily conserved among species and relatively stable
compared with their linear counterparts (20). These features provide circRNAs with a
number of potential functions. The main functions of circRNAs are
regulating the expression of parental genes, acting as miRNA or RBP
sponges and being translated into peptides or proteins.
EIciRNAs interact with U1 small nuclear
ribonucleoproteins, and they increase host gene transcription by
binding to RNA polymerase II (RNA pol II) (45). Certain ciRNAs and the RNA pol II
complex directly interact to regulate parental gene transcription
(Fig. 2A) (46). circRNAs also function as miRNA
sponges (Fig. 2B). These circRNAs
contain miRNA response elements, which facilitate the binding of
circRNAs with miRNAs, thereby sequestering the miRNAs away from
target mRNA molecules (48). For
example, circ_0001730 is upregulated in glioblastoma and promotes
glioblastoma cell proliferation and invasion by serving as a ceRNA
for miR-326 to regulate the Wnt7B/β-catenin pathway (49). Moreover, certain circRNAs are rich in
binding sites for RBPs, so they inhibit the biological function of
RBPs by functioning as protein sponges (Fig. 2C) (50). For instance, circ-Sirt1 interacts
with NF-κB p65 in the cytoplasm, thereby sequestering the nuclear
translocation of p65 (induced by inflammation), and inhibiting the
expression of inflammatory factors (51). circRNAs are considered to serve as
protein scaffolds by harbouring binding sites for the assembly of
two or more proteins, such as enzymes and their substrates, which
may subsequently form large protein complexes (37). For example, circ-forkhead box O3
(Foxo3) may generate a circ-Foxo3-p21-CDK2 ternary complex by
interacting with CDK2 and p21, which functions to inhibit CDK2
(23). Certain circRNAs, such as
circ-SNF 2 histone linker PHD RING helicase (SHPRH), which has an
internal ribosome entry site (IRES)-driven small open reading frame
(sORF) for the translation of SHPRH-146aa, is a tumour suppressor
in glioma (52). Moreover, it was
revealed that IRESs and the N6-methyladenosine modification may
drive sORFs for the translation of circRNAs (Fig. 2D) (53).
In summary, the aforementioned reasons may explain
why exosomes have more abundant levels of specific circRNAs
compared with producer cells, and why the expression of circRNAs is
higher compared with corresponding linear RNAs.
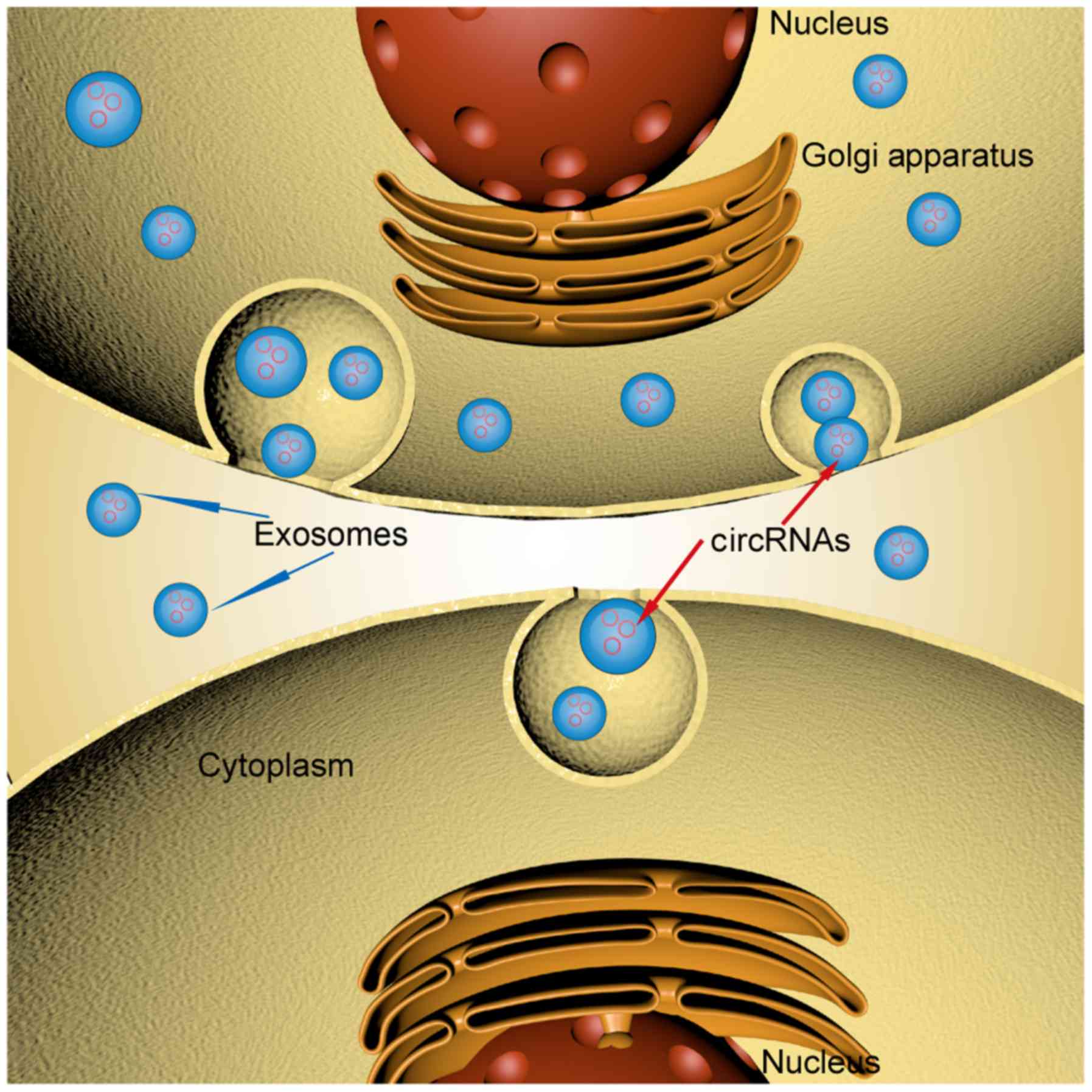
Exosomes contain numerous different varieties of
bioactive cargoes derived from cells, and these cargoes are
heterogeneous, reflecting the type and state of their cells of
origin (65). However, exosomes with
different origins may contain common cargoes, and cargoes in
exosomes derived from the same cell may differ. Hence, exosomes
contain specific cellular RNA subsets that are distinct or
tissue-specific. The detection of exosomal RNAs by
transcript-specific reverse transcription-quantitative PCR or
high-throughput analyses revealed a difference between circRNA
species in exosomes and the cytoplasm (6), which also reveals that circRNAs are
actively, not passively, incorporated into exosomes and that this
sorting process is selective (Fig.
3) (66). Despite having a
substantial understanding of cargo molecule transport from cells to
exosomes, knowledge of the basic mechanisms underlying cargo
selection remains limited.
A number of defined RBPs can be used to recognize
and sort RNAs with specific binding motifs into exosomes (67–69).
These specific motifs are shared by RNAs in EVs, which may
facilitate their targeting to EVs. For instance, RBP hnRNPA2B1 can
bind and transfer miRNAs into exosomes via conserved sequences
known as exosomes motifs, which are enriched in exosomes but not in
cells (70). Moreover, RBP hnRNPA2B1
was reported to specifically regulate the inclusion of lncARSR into
exosomes, and their specific binding depends on the sequence at the
5′ end (58). However, the mechanism
by which RBPs interact with the endosomal system is yet to be
elucidated. Moreover, because the RNA-binding complex ESCRT-II
serves a canonical role in MVB biogenesis (1), it may assist in selecting RNAs for
incorporation into EVs. A study on circRNA sorting into exosomes
indicated that this process was, at least partially, regulated by
associated miRNA levels in the producer cells (6). Moreover, exosomal circRNAs are
considered to retain biological activity and can abrogate the
growth inhibition induced by miR-7 in recipient cells (6).
In summary, the complex mechanisms by which RNAs are
selectively and specifically sorted into exosomes require further
clarification. Several types of sorting mechanisms may be
simultaneously active. Therefore, it is hypothesized that exosomal
ncRNA sorting will become a future research hotspot.
The early diagnosis of cancer has long been a focus
of research worldwide. In recent years, an increasing number of
novel and accessible methods for cancer diagnosis have continued to
emerge (71). Notably, exosomes can
be detected in nearly all types of human bodily fluids, such as
blood (72), breast milk (73), bile, saliva (74), tears, urine (75), semen (76), ascites, synovial fluid, cerebrospinal
fluid (77), amniotic fluid
(78), bronchoalveolar lavage fluid
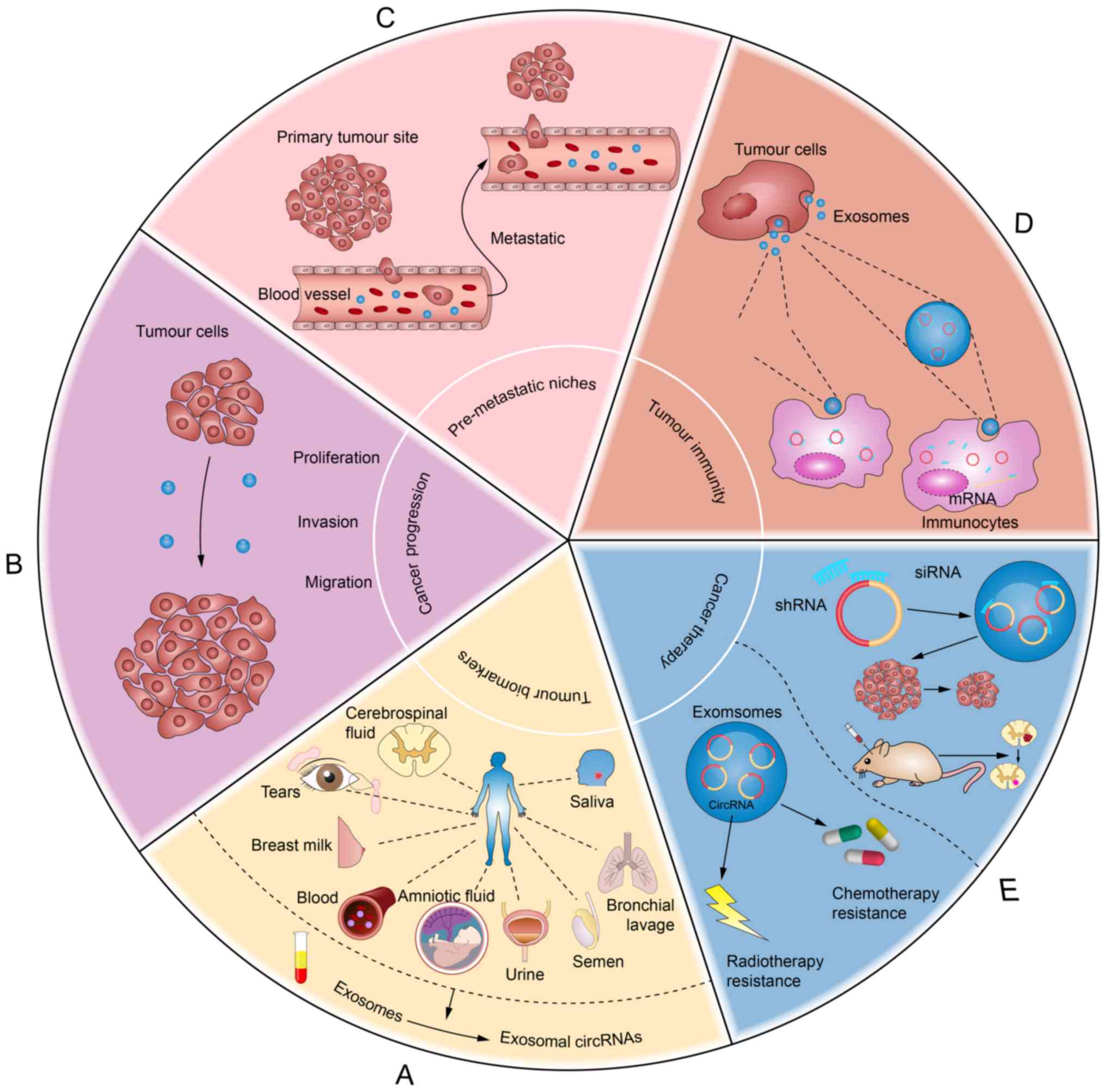
(79) and faeces (Fig. 4A). Compared with biomarkers in tumour
tissues, which can only be obtained by biopsy, exosomal biomarkers
can be detected using samples that are easier and less invasive to
obtain. This is an advantage of using exosomes as a biomarker. As
an exosomal cargo, circRNAs themselves have several notable
characteristics, including high abundance and diversity, stability,
conservation and localization and expression specificity (18). In recent years, numerous studies have
explored the advantages of exosomal circRNAs. These features
indicate that circRNAs may also be suitable as potential diagnostic
biomarkers for cancer (19,23) or other diseases such as coronary
artery disease (80), Alzheimer's
disease (81) and systemic lupus
erythematosus (82). For instance,
high expression of blood exosomal circ-PDE8A was revealed to be
associated with lymphatic invasion and tumour stage, as well as
poor survival in patients with PDAC. Therefore, this exosomal
circRNA may be used as a marker for determining diagnosis or
progression in patients with PDAC (32). In another study, it was observed that
exosomes from pancreatic cancer overexpressed circ-IRAS (34). Circ-IARS could enter HUVECs via
exosomes and promote tumor invasion and metastasis (34). Exosomal circRNA_100284 was reported
to induce cell cycle acceleration and promote cell proliferation
via miR-217, which targets enhancer of zeste homologue 2 (EZH2) in
cancers (83). Exosomal
circRNA_100284 may also serve as a biomarker for arsenite exposure
(83). Recently, researchers
demonstrated that human plasma is rich in mRNA, circRNA and lncRNA
via EV long RNA sequencing (exLR-Seq), and these RNAs may be useful
as biomarkers for tumour diagnosis and prognosis. The
aforementioned study reported that 8 exLRs may be used as
diagnostic biomarkers, having utility beyond the current diagnostic
approaches (such as α-feto protein) for hepatocellular carcinoma
(HCC), with high sensitivity and specificity (84). These findings have crucial functional
and clinical implications.
In summary, exosomal circRNAs are clinically
valuable as a new generation of biomarkers for the early diagnosis
and prognosis of various diseases, particularly cancer, as well as
the evaluation of therapeutic effects (12).
Several studies have reported that circRNAs can
function in cancer by either promoting or inhibiting tumour
progression and regulating the biological behaviour of malignant
tumours, including cell proliferation, migration, invasion and
metastasis (26,85,86).
However, circRNAs in exosomes are still rarely studied. Notably,
exosomal circRNAs appear to play a pleiotropic role in cancer due
to their involvement in intercellular communication (22,87). On
the one hand, exosomes containing circRNAs promote the progression
of cancer, the generation of premetastatic niches and the
occurrence of metastasis (33,34). On
the other hand, exosomes may potentially function in tumour immune
regulation and cancer therapy (35,36).
circRNAs in exosomes derived from patients with
multiple tumours are distinct from those of healthy individuals
(88), indicating that they may have
great clinical significances and research value. A number of
studies have investigated the mechanism by which exosomal circRNAs
function in cancer development. A study revealed that the exosomal
circ-PDE8A is released by tumours facilitates invasive development
in a miR-338-MET transcriptional regulator MACC1-MET proto-oncogene
receptor tyrosine kinase pathway-dependent manner in pancreatic
cancer (PC), revealing that circ-PDE8A enhances tumour invasiveness
via the communication mediated by exosomes (32). Another study reported that exosomal
circRNA_100284 accelerated the cell division cycle and promoted
proliferation, via sponging miR-217. Overexpression of
circRNA_100284 enhanced the invasion and migration of, and
increased the formation of tumour colonies, by stimulating the
downstream signalling pathway and increasing the expression of EZH2
and cyclin-D1 in human hepatic cells (83). Furthermore, this circRNA also affects
the malignant transformation of cells (83). In another study, gastric cancer (GC)
cell-derived exosomes transferred ciRS-133 into preadipocytes,
which facilitated preadipocyte differentiation into brown-like
cells via the activation of PR/SET domain 16 and the inhibition of
miR-133 (8). Additionally, this type
of exosome can participate in white adipose tissue browning and
affect cancer cachexia (8), a
syndrome characterized by weight loss due to muscle and/or fat
loss. Cancer cachexia results in functional impairment, decreased
physical ability and it is associated with a poor prognosis
(89). The same mechanisms have been
reported in other studies (90,91). It
can be concluded from the aforementioned studies that exosomes can
directly transfer circRNAs from cancer cells to surrounding cells,
and then further mediate the biological functions of recipient
cells. Meanwhile, circRNAs serve as competing endogenous RNAs or
miRNA sponges to promote the progression of multiple cancer
types.
In summary, exosomal circRNAs influence the
promotion of malignant transformation, proliferation, invasiveness
and migration in cancer, and are associated with cancer cachexia.
Notably, acting as miRNA sponges is the most common mechanism.
Further investigation is needed to explore other exosomal circRNAs
that serve a role in promoting cancer progression (Fig. 4B).
Exosomes are vital in mediating cell-to-cell
communication and delivering cargo from donor cells to recipient
cells, regardless of whether the recipient cells are located in a
remote or nearby tissue, known as horizontal transfer (92). In cancer, exosomes are involved in
the communication between tumour cells and the surrounding
microenvironment (93). The
interaction between malignant and surrounding tumour or normal
cells can greatly affect tumour progression (94). Studies have reported that exosomal
circRNAs have pivotal functions in premetastatic niche formation
and metastasis (Fig. 4C) (12). For example, exosomes derived from
cells with high metastatic potential and abundant circPTGR1 can
potentially downregulate the interaction between miR-449a and MET
in recipient cells. Thus, affecting cells with low metastatic
potential and disrupting tumour microenvironment homeostasis, as
well as promoting HCC progression (30). In addition, a previous study revealed
that exosomal communication between GC cells aids in the transfer
of circNRIP1. Exosomal circNRIP1 may sponge miR-149-5p to
facilitate GC metastasis via the AKT1/mTOR signalling pathway
(95). Pathological EMT is a key
factor in the process of tumour progression and metastasis.
Recently, a study observed that the level of circPRMT5 was
increased in serum and urine exosomes from patients with UCB, and
this was associated with cancer metastasis (96). CircPRMT5 may promote the EMT of UCB
cells and result in an aggressive phenotype via sponging
miR-30c.
Thus, exosomal circRNAs have a significant influence
in promoting the generation of premetastatic niches and the
occurrence of tumour metastasis.
Increasing studies have revealed that exosomes are
closely associated with tumour immunity, resulting in either tumour
suppression or tumour promotion (97–99).
Whether they originate from the tumour or from immune cells
determines the role of exosomes in the tumour immune response
(100).
Exosomes from tumour cells have unique
immunoregulatory effects on the immune system, and they can be used
as a medium for regulating tumour cell-regulatory T cell (Treg)
communication (55). For example,
nasopharyngeal carcinoma (NPC)-exosome may induce expansion of the
Treg population and enhance the suppressive functions of Tregs. The
interactions of NPC-exosome and Tregs may be associated with the
tumour microenvironment, allowing immune suppression as well as
evading host immune surveillance (101). EVs derived from colorectal cancer
(CRC) cells may induce phenotypic alteration of the T cells to
Treg-like cells by activating transforming growth factor-β/SMAD
signalling and inactivating stress-activated protein kinase (SAPK)
signalling 9 (102). Additionally,
these Treg-like cells may promote tumour cell proliferation
(102).
Accumulating evidence has revealed that exosomes and
ncRNAs (miRNA and lncRNA) are critical to Treg cell homeostasis
(103). For example, tumour
exosomal miRNAs may induce immune tolerance and then exert an
adverse immune effect (104).
However, studies on exosomal circRNA regulation of Treg cells are
relatively new, and further research is needed.
Tumour cell-derived exosomes may carry abnormally
expressed circRNAs and shuttle them to surrounding cells (including
immunocytes). circRNAs may become tumour antigens or bind miRNAs or
proteins to regulate immunocompetence when reaching the target
immunocytes. Furthermore, there are important interactions between
circRNAs and miRNAs, as described above: circRNAs in exosomes may
initially bind tumour-specific miRNAs or mRNAs via the
circRNA-miRNA-mRNA axis, assisting in the shuttling of miRNAs
between carcinoma cells and immunocytes, as well as enhancing their
stability as exosomes transit the intercellular space (36). When exosomes fuse with the target
immunocytes, they release their cargo, which perform notable
functions in tumour immunity (Fig.
4D) (36). For example,
miRNA-155 can be upregulated to potentiate immunotherapies of
tumor-specific CD8+ T cells for cancer (105). Tumor-derived miR-214 targets tensin
homolog (PTEN) resulting in an increase in Treg cell number and
enhanced immune suppression (55).
It is predicted that exosomal circRNAs are a promising target for
tumour immunotherapy.
These findings indicate that malignant carcinoma
cells and immunocytes exist continually in a dynamic equilibrium,
in which exosomes serve a crucial role in maintaining homeostasis
between these two cell types.
The primary prospects of exosome treatment include
participating in intercellular information exchange and targeted
drug delivery. As exosomes are nanoscale biological vesicles, their
cargo is often protected so they have an innate advantage as a
useful therapeutic vector (106).
In addition, exosomes, as mediators of communication between cells,
are highly safe, bioavailable and exhibit low systematic
immunogenicity and toxicity compared with conventional targeting
vectors (107). Exosomes have
exhibited a more notable positive therapeutic effect on cancer
compared with direct delivery of chemotherapy (107). Exosomes may also be used to solve a
number of the issues surrounding currently used drug therapy
methods, which involve programmable RNA, which has low uptake
efficiency, is highly cytotoxic and is thus unsuitable for clinical
practice (108). Researchers have
proposed and verified the effectiveness of a new strategy for the
generation of large amounts of RBC-derived EVs for delivering RNA
drugs (109). The use of gold
nanoparticle targeting approaches to generate specific types of
exosomes offers an alternative for the selective, targeted
elimination of cancer cells (110).
In summary, exosomes are vesicles that can be artificially
engineered into useful treatment vectors as a therapeutic agent
against cancer (13,109,111,112).
Regarding pharmaceutical cargo, exosomes can be
loaded with chemotherapeutic drugs (113) for targeted therapy and
immunoadjuvant-mediated immunotherapy (106), as well as ncRNAs and small
interfering RNAs (siRNAs) (114).
The transfer of siRNA can promote the exchange of genetic
information and make a substantial difference in cellular
biological behaviour. For example, nanovesicles that mimic exosomes
can achieve targeted delivery of RNA interference chemotherapeutic
drugs to malignant tumours, and evidence suggests that exosomes
carrying siRNA may target c-Myc (115).
Exosomal delivery of circRNA would have a number of
therapeutic advantages. Primarily, the control of natural circRNA
expression in specific tissues and cells is likely to reduce
adverse effects compared with synthetic molecular drugs. Moreover,
one common phenomenon and major function of circRNA is their role
as a miRNA sponge. Therefore, research on endogenous circRNA sponge
structures may assist in the design and development of potent
artificial sponges to regulate the function of miRNAs in disease.
Additionally, the off-target effects of circRNAs may be low
compared with siRNAs and miRNAs due to their short length. Indeed,
off-target effects are a significant problem that limits the
translation of small molecule RNAs into clinical practice (116). However, off-target effects do not
negatively affect circRNA therapy, as circRNAs have a stable and
specific structure (18). In
addition, circRNA is not easily digested by ribonucleases and has a
natural structure that makes it more stable than linear RNA. Due to
the construction of engineered exogenous circRNAs, mRNA cyclization
can effectively solve the issues facing linear mRNA, such as its
shorter half-life, enabling it to permanently and efficiently
express proteins in eukaryotic cells (117). In summary, it is likely that
exosomal circRNA therapy will be a future gene therapy used for the
treatment of various diseases.
Exosomes can also encapsulate both siRNAs and short
hairpin RNAs (shRNAs) that are specifically designed to target and
reduce the levels of specific circRNAs in tumour cells that have
detrimental effects on patient outcomes (35). These siRNAs or shRNAs sponge miRNAs
and promote the expression of antioncogenes that indirectly inhibit
damage caused by circRNAs (95,118).
For example, three shRNAs that span the back-splicing site of the
circRNA c transferrin receptor (cTFRC) were designed in a previous
study to inhibit its expression. The results indicated that the
downregulation of cTFRC suppresses bladder cancer cell invasion
(118). Moreover, similar studies
of circNRIP1 have been reported, and circNRIP1 silencing via
circNRIP1 siRNA transfection notably inhibited the proliferation,
invasion and migration of GC cells (95). This phenomenon was also observed in
prostate cancer (119), CRC
(120), PC (32), UCB (96), HCC (30,121)
and glioblastoma (122,123).
Therapeutic resistance is a major problem in the
treatment of cancer. Notably, exosomal circRNAs were demonstrated
to influence radioresistance in a study by Zhao et al
(28), who used RNA-Seq to analyse
the circRNAs present in EVs from U251 and radioresistant U251
cells. CircATP8B4 was demonstrated to promote glioma
radioresistance by serving as a miR-766 sponge, and circATP8B4 in
EVs may be a potential biomarker for glioma radioresistance. It was
reported that exosome-transmitted miRNA may promote
chemosensitivity in multiple cancer types such as colorectal cancer
and head and neck cancer (124,125),
and recent studies have revealed that circRNAs mediate chemotherapy
resistance in various types of cancer such as ovarian cancer and
thyroid carcinoma (126–128). Hence, there is a novel hypothesis
that exosomal circRNAs are influence chemotherapy resistance, but
this still requires further experimental confirmation (Fig. 4E).
In summary, exosomes carrying various circRNAs may
assist in promoting cancer progression. However, exosomes loaded
with circRNA and engineered siRNAs that target specific circRNAs
may be valuable in the development of drugs to inhibit tumour
progression, and for accurate and effective therapy.
Exosomes encapsulate and transfer a myriad of
functional molecular cargoes and serve as crucial mediators of
intercellular communication. As an important cargo of exosomes,
circRNAs have been demonstrated to serve crucial regulatory
functions in various aspects of cancer, such as tumorigenesis,
proliferation, migration, invasion, metastasis, apoptosis and
resistance to chemotherapy and radiation, as well as influencing
overall cancer prognosis. However, exosomal circRNA can function to
either promote or inhibit cancer. Exosomes harbouring circRNAs can
promote the progression of cancer, the generation of premetastatic
niches and the occurrence of metastasis. They can also function in
tumour immune regulation and cancer therapy. Due to their unique
features and high specificity, the combination of exosomes and
circRNAs lends favour to their potential clinical application as
cancer diagnostic and prognostic markers.
Notably, although numerous studies have assessed the
roles of circRNAs as well as exosomal ncRNAs in cancer, relatively
few studies have investigated the functions of exosomal circRNAs.
In the near future, scientists worldwide will devote increased
attention to exosomal circRNAs research to explore their functions
and roles in various diseases, especially in cancer. In addition,
cells clear circRNAs via EVs or exosomes, a process that may be
used to alleviate circRNA accumulation. Thus, elucidating the
mechanisms underlying circRNA clearance and release via exosomes
still requires further investigation.
The ultimate goal of medical scientific research is
to contribute to improved clinical outcomes for patients. Although
studies have already demonstrated the potential functions of
circRNAs in cancer diagnosis and therapy, their use as targets in
clinical applications, appears to be a promising area of future
research, particularly due to the low risk of adverse effects. The
use of exosomes as natural drug vectors for the delivery of
pharmaceutical cargo and their use in the development of targeted
therapeutics should be further studied to enhance delivery
efficiency. It is hypothesized that the continued efforts of
researchers worldwide will result in therapies involving the
delivery of specific circRNAs via exosomes in the near future.
Not applicable.
The present study was financially supported by
grants from the government-funded Provincial Clinical Medicine
Talent Programs in 2017 (grant no. 3010601198073), the Science and
Technology Capacity Improvement Projects of Hebei University of
Chinese Medicine in 2019 (grant no. KTZ2019019), the Outstanding
Student Scientific Research Ability Improvement Projects of Hebei
University of Chinese Medicine in 2019 (grant no. YXZ2019002) and
the Graduate Innovative Ability Training Projects of Hebei Province
in 2020 (grant nos XCXZZBS2020002 and hbu2020bs003).
Not applicable.
XG and XL were the major contributors in the writing
and revision of the manuscript. YZ made substantial contributions
to the conception or design of the work. QL revised this article
critically for important intellectual content. YG analysed the
data. CF gave the final approval of the version to be published. HW
gave approval for the final version of the manuscript. All authors
read and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
van Niel G, D'Angelo G and Raposo G:
Shedding light on the cell biology of extracellular vesicles. Nat
Rev Mol Cell Biol. 19:213–228. 2018. View Article : Google Scholar
|
|
2
|
Thery C, Zitvogel L and Amigorena S:
Exosomes: Composition, biogenesis and function. Nat Rev Immunol.
2:569–579. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Trajkovic K, Hsu C, Chiantia S, Rajendran
L, Wenzel D, Wieland F, Schwille P, Brugger B and Simons M:
Ceramide triggers budding of exosome vesicles into multivesicular
endosomes. Science. 319:1244–1247. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang K, Fang C, Yi K, Liu X, Qi H, Tan Y,
Zhou J, Li Y, Liu M, Zhang Y, et al: The role of PTRF/Cavin1 as a
biomarker in both glioma and serum exosomes. Theranostics.
8:1540–1557. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wei Z, Batagov AO, Schinelli S, Wang J,
Wang Y, El Fatimy R, Rabinovsky R, Balaj L, Chen CC, Hochberg F, et
al: Coding and noncoding landscape of extracellular RNA released by
human glioma stem cells. Nat Commun. 8:11452017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J,
Zhou K, Liu X, Ren X, Wang F, et al: Cancer-derived exosomal
miR-25-3p promotes pre-metastatic niche formation by inducing
vascular permeability and angiogenesis. Nat Commun. 9:53952018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang H, Zhu L, Bai M, Liu Y, Zhan Y, Deng
T, Yang H, Sun W, Wang X, Zhu K, et al: Exosomal circRNA derived
from gastric tumor promotes white adipose browning by targeting the
miR-133/PRDM16 pathway. Int J Cancer. 144:2501–2515. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li L, Li C, Wang S, Wang Z, Jiang J, Wang
W, Li X, Chen J, Liu K, Li C and Zhu G: Exosomes derived from
hypoxic oral squamous cell carcinoma cells deliver miR-21 to
normoxic cells to elicit a prometastatic phenotype. Cancer Res.
76:1770–1780. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Silachev DN, Goryunov KV, Shpilyuk MA,
Beznoschenko OS, Morozova NY, Kraevaya EE, Popkov VA, Pevzner IB,
Zorova LD, Evtushenko EA, et al: Effect of MSCs and MSC-derived
extracellular vesicles on human blood coagulation. Cells.
8:E2582019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shabbir A, Cox A, Rodriguez-Menocal L,
Salgado M and Van Badiavas E: Mesenchymal stem cell exosomes induce
proliferation and migration of normal and chronic wound
fibroblasts, and enhance angiogenesis in vitro. Stem Cells Dev.
24:1635–1647. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen G, Huang AC, Zhang W, Zhang G, Wu M,
Xu W, Yu Z, Yang J, Wang B, Sun H, et al: Exosomal PD-L1
contributes to immunosuppression and is associated with anti-PD-1
response. Nature. 560:382–386. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kamerkar S, LeBleu VS, Sugimoto H, Yang S,
Ruivo CF, Melo SA, Lee JJ and Kalluri R: Exosomes facilitate
therapeutic targeting of oncogenic KRAS in pancreatic cancer.
Nature. 546:498–503. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tian T, Zhang HX, He CP, Fan S, Zhu YL, Qi
C, Huang NP, Xiao ZD, Lu ZH, Tannous BA and Gao J: Surface
functionalized exosomes as targeted drug delivery vehicles for
cerebral ischemia therapy. Biomaterials. 150:137–149. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pan Y, Hui X, Hoo RLC, Ye D, Chan CYC,
Feng T, Wang Y, Lam KSL and Xu A: Adipocyte-secreted exosomal
microRNA-34a inhibits M2 macrophage polarization to promote
obesity-induced adipose inflammation. J Clin Invest. 129:834–849.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen LL and Yang L: Regulation of circRNA
biogenesis. RNA Biol. 12:381–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cocquerelle C, Mascrez B, Hetuin D and
Bailleul B: Mis-splicing yields circular RNA molecules. FASEB J.
7:155–160. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Han B, Chao J and Yao H: Circular RNA and
its mechanisms in disease: From the bench to the clinic. Pharmacol
Ther. 187:31–44. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao
L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al: The
landscape of circular RNA in cancer. Cell. 176:869–881.e13. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rybak-Wolf A, Stottmeister C, Glazar P,
Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss
R, et al: Circular RNAs in the mammalian brain are highly abundant,
conserved, and dynamically expressed. Mol Cell. 58:870–885. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cheng X, Zhang L, Zhang K, Zhang G, Hu Y,
Sun X, Zhao C, Li H, Li YM and Zhao J: Circular RNA VMA21 protects
against intervertebral disc degeneration through targeting miR-200c
and X linked inhibitor-of-apoptosis protein. Ann Rheum Dis.
77:770–779. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P
and Yang BB: Foxo3 circular RNA retards cell cycle progression via
forming ternary complexes with p21 and CDK2. Nucleic Acids Res.
44:2846–2858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guarnerio J, Zhang Y, Cheloni G, Panella
R, Mae Katon J, Simpson M, Matsumoto A, Papa A, Loretelli C, Petri
A, et al: Intragenic antagonistic roles of protein and circRNA in
tumorigenesis. Cell Res. 29:628–640. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang PF, Wei CY, Huang XY, Peng R, Yang
X, Lu JC, Zhang C, Gao C, Cai JB, Gao PT, et al: Circular RNA
circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress
hepatocellular carcinoma progression. Mol Cancer. 18:1052019.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dong W, Bi J, Liu H, Yan D, He Q, Zhou Q,
Wang Q, Xie R, Su Y, Yang M, et al: Circular RNA ACVR2A suppresses
bladder cancer cells proliferation and metastasis through
miR-626/EYA4 axis. Mol Cancer. 18:952019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang X, Li Z, Zhang Q, Wang W, Li B, Wang
L, Xu Z, Zeng A, Zhang X, Zhang X, et al: Circular RNA AKT3
upregulates PIK3R1 to enhance cisplatin resistance in gastric
cancer via miR-198 suppression. Mol Cancer. 18:712019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao M, Xu J, Zhong S, Liu Y, Xiao H, Geng
L and Liu H: Expression profiles and potential functions of
circular RNAs in extracellular vesicles isolated from
radioresistant glioma cells. Oncol Rep. 41:1893–1900.
2019.PubMed/NCBI
|
|
29
|
Liang WC, Wong CW, Liang PP, Shi M, Cao Y,
Rao ST, Tsui SK, Waye MM, Zhang Q, Fu WM and Zhang JF: Translation
of the circular RNA circβ-catenin promotes liver cancer cell growth
through activation of the Wnt pathway. Genome Biol. 20:842019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo
J, Zhang Y, Li H, Zhang Q, Yang Y and Chen G: Three isoforms of
exosomal circPTGR1 promote hepatocellular carcinoma metastasis via
the miR449a-MET pathway. EBioMedicine. 40:432–445. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie
JW, Wang JB, Lin JX, Chen QY, Cao LL, et al: Circular RNA
circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to
facilitate gastric cancer invasion and metastasis. Cancer Lett.
471:38–48. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Z, Yanfang W, Li J, Jiang P, Peng T,
Chen K, Zhao X, Zhang Y, Zhen P, Zhu J and Li X: Tumor-released
exosomal circular RNA PDE8A promotes invasive growth via the
miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett.
432:237–250. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu
K, Fan Q, Li J, Ning T, Tian F, et al: Exosome circRNA secreted
from adipocytes promotes the growth of hepatocellular carcinoma by
targeting deubiquitination-related USP7. Oncogene. 38:2844–2859.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li J, Li Z, Jiang P, Peng M, Zhang X, Chen
K, Liu H, Bi H, Liu X and Li X: Circular RNA IARS (circ-IARS)
secreted by pancreatic cancer cells and located within exosomes
regulates endothelial monolayer permeability to promote tumor
metastasis. J Exp Clin Cancer Res. 37:1772018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bai H, Lei K, Huang F, Jiang Z and Zhou X:
Exo-circRNAs: A new paradigm for anticancer therapy. Mol Cancer.
18:562019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu Z, Li P, Fan L and Wu M: The potential
role of circRNA in tumor immunity regulation and immunotherapy.
Front Immunol. 9:92018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ivanov A, Memczak S, Wyler E, Torti F,
Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M,
Dieterich C and Rajewsky N: Analysis of intron sequences reveals
hallmarks of circular RNA biogenesis in animals. Cell Rep.
10:170–177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Aktas T, Avsar Ilik I, Maticzka D,
Bhardwaj V, Pessoa Rodrigues C, Mittler G, Manke T, Backofen R and
Akhtar A: DHX9 suppresses RNA processing defects originating from
the Alu invasion of the human genome. Nature. 544:115–119. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Barrett SP, Wang PL and Salzman J:
Circular RNA biogenesis can proceed through an exon-containing
lariat precursor. Elife. 4:e075402015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Noto JJ, Schmidt CA and Matera AG:
Engineering and expressing circular RNAs via tRNA splicing. RNA
Biol. 14:978–984. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lu Y, Deng X, Xiao G, Zheng X, Ma L and
Huang W: Circ_0001730 promotes proliferation and invasion via the
miR-326/Wnt7B axis in glioma cells. Epigenomics. 11:1335–1352.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Abdelmohsen K, Panda AC, Munk R,
Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM,
Martindale JL and Gorospe M: Identification of HuR target circular
RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA
Biol. 14:361–369. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kong P, Yu Y, Wang L, Dou YQ, Zhang XH,
Cui Y, Wang HY, Yong YT, Liu YB, Hu HJ, et al: Circ-Sirt1 controls
NF-κB activation via sequence-specific interaction and enhancement
of SIRT1 expression by binding to miR-132/212 in vascular smooth
muscle cells. Nucleic Acids Res. 47:3580–3593. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang M, Huang N, Yang X, Luo J, Yan S,
Xiao F, Chen W, Gao X, Zhao K, Zhou H, et al: A novel protein
encoded by the circular form of the SHPRH gene suppresses glioma
tumorigenesis. Oncogene. 37:1805–1814. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N6-methyladenosine. Cell Res.
27:626–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Valadi H, Ekstrom K, Bossios A, Sjostrand
M, Lee JJ and Lotvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yin Y, Cai X, Chen X, Liang H, Zhang Y, Li
J, Wang Z, Chen X, Zhang W, Yokoyama S, et al: Tumor-secreted
miR-214 induces regulatory T cells: A major link between immune
evasion and tumor growth. Cell Res. 24:1164–1180. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yin J, Zeng A, Zhang Z, Shi Z, Yan W and
You Y: Exosomal transfer of miR-1238 contributes to
temozolomide-resistance in glioblastoma. EBioMedicine. 42:238–251.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Shao N, Xue L, Wang R, Luo K, Zhi F and
Lan Q: MiR-454-3p is an exosomal biomarker and functions as a tumor
suppressor in glioma. Mol Cancer Ther. 18:459–469. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y,
Chen W, Liu F, Sun W, Li XF, et al: Exosome-transmitted lncARSR
promotes sunitinib resistance in renal cancer by acting as a
competing endogenous RNA. Cancer Cell. 29:653–668. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Dou Y, Cha DJ, Franklin JL, Higginbotham
JN, Jeppesen DK, Weaver AM, Prasad N, Levy S, Coffey RJ, Patton JG
and Zhang B: Circular RNAs are down-regulated in KRAS mutant colon
cancer cells and can be transferred to exosomes. Sci Rep.
6:379822016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Liang D, Tatomer DC, Luo Z, Wu H, Yang L,
Chen LL, Cherry S and Wilusz JE: The output of protein-coding genes
shifts to circular RNAs when the pre-mRNA processing machinery is
limiting. Mol Cell. 68:940–954.e3. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhang Y, Xue W, Li X, Zhang J, Chen S,
Zhang JL, Yang L and Chen LL: The biogenesis of nascent circular
RNAs. Cell Rep. 15:611–624. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lasda E and Parker R: Circular RNAs
co-precipitate with extracellular vesicles: A possible mechanism
for circRNA clearance. PLoS One. 11:e01484072016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Hong BS, Cho JH, Kim H, Choi EJ, Rho S,
Kim J, Kim JH, Choi DS, Kim YK, Hwang D and Gho YS: Colorectal
cancer cell-derived microvesicles are enriched in cell
cycle-related mRNAs that promote proliferation of endothelial
cells. BMC Genomics. 10:5562009. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
He C, Zheng S, Luo Y and Wang B: Exosome
theranostics: Biology and translational medicine. Theranostics.
8:237–255. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Raposo G and Stoorvogel W: Extracellular
vesicles: Exosomes, microvesicles, and friends. J Cell Biol.
200:373–383. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Perez-Boza J, Lion M and Struman I:
Exploring the RNA landscape of endothelial exosomes. RNA.
24:423–435. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Santangelo L, Giurato G, Cicchini C,
Montaldo C, Mancone C, Tarallo R, Battistelli C, Alonzi T, Weisz A
and Tripodi M: The RNA-binding protein SYNCRIP is a component of
the hepatocyte exosomal machinery controlling microRNA sorting.
Cell Rep. 17:799–808. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Shurtleff MJ, Temoche-Diaz MM, Karfilis
KV, Ri S and Schekman R: Y-box protein 1 is required to sort
microRNAs into exosomes in cells and in a cell-free reaction.
Elife. 5:e192762016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Villarroya-Beltri C, Gutierrez-Vazquez C,
Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N,
Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M and
Sanchez-Madrid F: Sumoylated hnRNPA2B1 controls the sorting of
miRNAs into exosomes through binding to specific motifs. Nat
Commun. 4:29802013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Scilla KA and Rolfo C: The role of
circulating tumor DNA in lung cancer: Mutational analysis,
diagnosis, and surveillance now and into the future. Curr Treat
Options Oncol. 20:612019. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Caby MP, Lankar D, Vincendeau-Scherrer C,
Raposo G and Bonnerot C: Exosomal-like vesicles are present in
human blood plasma. Int Immunol. 17:879–887. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Qin W, Tsukasaki Y, Dasgupta S,
Mukhopadhyay N, Ikebe M and Sauter ER: Exosomes in human breast
milk promote EMT. Clin Cancer Res. 22:4517–4524. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Katsiougiannis S, Chia D, Kim Y, Singh RP
and Wong DT: Saliva exosomes from pancreatic tumor-bearing mice
modulate NK cell phenotype and antitumor cytotoxicity. FASEB J.
31:998–1010. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
McKiernan J, Donovan MJ, O'Neill V,
Bentink S, Noerholm M, Belzer S, Skog J, Kattan MW, Partin A,
Andriole G, et al: A novel urine exosome gene expression assay to
predict high-grade prostate cancer at initial biopsy. JAMA Oncol.
2:882–889. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Barcelo M, Castells M, Bassas L, Vigues F
and Larriba S: Semen miRNAs contained in exosomes as non-invasive
biomarkers for prostate cancer diagnosis. Sci Rep. 9:137722019.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Kong FL, Wang XP, Li YN and Wang HX: The
role of exosomes derived from cerebrospinal fluid of spinal cord
injury in neuron proliferation in vitro. Artif Cells Nanomed
Biotechnol. 46:200–205. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Dixon CL, Sheller-Miller S, Saade GR,
Fortunato SJ, Lai A, Palma C, Guanzon D, Salomon C and Menon R:
Amniotic fluid exosome proteomic profile exhibits unique pathways
of term and preterm labor. Endocrinology. 159:2229–2240. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Levanen B, Bhakta NR, Torregrosa Paredes
P, Barbeau R, Hiltbrunner S, Pollack JL, Skold CM, Svartengren M,
Grunewald J, Gabrielsson S, et al: Altered microRNA profiles in
bronchoalveolar lavage fluid exosomes in asthmatic patients. J
Allergy Clin Immunol. 131:894–903. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Wang L, Shen C, Wang Y, Zou T, Zhu H, Lu
X, Li L, Yang B, Chen J, Chen S, et al: Identification of circular
RNA Hsa_circ_0001879 and Hsa_circ_0004104 as novel biomarkers for
coronary artery disease. Atherosclerosis. 286:88–96. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhang Y, Yu F, Bao S and Sun J: Systematic
characterization of circular RNA-associated ceRNA network
identified novel circRNA biomarkers in Alzheimer's disease. Front
Bioeng Biotechnol. 7:2222019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Li H, Li K, Lai W, Li X, Wang H, Yang J,
Chu S, Wang H, Kang C and Qiu Y: Comprehensive circular RNA
profiles in plasma reveals that circular RNAs can be used as novel
biomarkers for systemic lupus erythematosus. Clin Chim Acta.
480:17–25. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Dai X, Chen C, Yang Q, Xue J, Chen X, Sun
B, Luo F, Liu X, Xiao T, Xu H, et al: Exosomal circRNA_100284 from
arsenite-transformed cells, via microRNA-217 regulation of EZH2, is
involved in the malignant transformation of human hepatic cells by
accelerating the cell cycle and promoting cell proliferation. Cell
Death Dis. 9:4542018. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Li Y, Zhao J, Yu S, Wang Z, He X, Su Y,
Guo T, Sheng H, Chen J, Zheng Q, et al: Extracellular vesicles long
RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human
blood as potential biomarkers for cancer diagnosis. Clin Chem.
65:798–808. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Chen J, Chen T, Zhu Y, Li Y, Zhang Y, Wang
Y, Li X, Xie X, Wang J, Huang M, et al: CircPTN sponges
miR-145-5p/miR-330-5p to promote proliferation and stemness in
glioma. J Exp Clin Cancer Res. 38:3982019. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Meng Q, Li S, Liu Y, Zhang S, Jin J, Zhang
Y, Guo C, Liu B and Sun Y: Circular RNA circSCAF11 accelerates the
glioma tumorigenesis through the miR-421/SP1/VEGFA Axis. Mol Ther
Nucleic Acids. 17:669–677. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Ma P, Pan Y, Li W, Sun C, Liu J, Xu T and
Shu Y: Extracellular vesicles-mediated noncoding RNAs transfer in
cancer. J Hematol Oncol. 10:572017. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wang LP, Peng XY, Lv XQ, Liu L, Li XL, He
X, Lv F, Pan Y, Wang L, Liu KF and Zhang XM: High throughput
circRNAs sequencing profile of follicle fluid exosomes of
polycystic ovary syndrome patients. J Cell Physiol. Feb
18–2019.(Epub ahead of print).
|
|
89
|
Dunne RF, Loh KP, Williams GR, Jatoi A,
Mustian KM and Mohile SG: Cachexia and sarcopenia in older adults
with cancer: A comprehensive review. Cancers (Basel). 11:E18612019.
View Article : Google Scholar
|
|
90
|
Su Y, Lv X, Yin W, Zhou L, Hu Y, Zhou A
and Qi F: circRNA Cdr1as functions as a competitive endogenous RNA
to promote hepatocellular carcinoma progression. Aging (Albany NY).
11:8182–8203. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Guan X, Zong ZH, Liu Y, Chen S, Wang LL
and Zhao Y: CircPUM1 promotes tumorigenesis and progression of
ovarian cancer by sponging miR-615-5p and miR-6753-5p. Mol Ther
Nucleic Acids. 18:882–892. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Kim KM, Abdelmohsen K, Mustapic M,
Kapogiannis D and Gorospe M: RNA in extracellular vesicles. Wiley
Interdiscip Rev RNA. 8:2017. View Article : Google Scholar
|
|
93
|
Hu C, Chen M, Jiang R, Guo Y, Wu M and
Zhang X: Exosome-related tumor microenvironment. J Cancer.
9:3084–3092. 2018. View Article : Google Scholar
|
|
94
|
Bebelman MP, Smit MJ, Pegtel DM and Baglio
SR: Biogenesis and function of extracellular vesicles in cancer.
Pharmacol Ther. 188:1–11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zhang X, Wang S, Wang H, Cao J, Huang X,
Chen Z, Xu P, Sun G, Xu J, Lv J and Xu Z: Circular RNA circNRIP1
acts as a microRNA-149-5p sponge to promote gastric cancer
progression via the AKT1/mTOR pathway. Mol Cancer. 18:202019.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Chen X, Chen RX, Wei WS, Li YH, Feng ZH,
Tan L, Chen JW, Yuan GJ, Chen SL, Guo SJ, et al: PRMT5 circular RNA
promotes metastasis of urothelial carcinoma of the bladder through
sponging miR-30c to induce epithelial-mesenchymal transition. Clin
Cancer Res. 24:6319–6330. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
LeBleu VS and Kalluri R: Exosomes exercise
inhibition of anti-tumor immunity during chemotherapy. Immunity.
50:547–549. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Lema DA and Burlingham WJ: Role of
exosomes in tumour and transplant immune regulation. Scand J
Immunol. 90:e128072019. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Wolfers J, Lozier A, Raposo G, Regnault A,
Thery C, Masurier C, Flament C, Pouzieux S, Faure F, Tursz T, et
al: Tumor-derived exosomes are a source of shared tumor rejection
antigens for CTL cross-priming. Nat Med. 7:297–303. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Xie Y, Dang W, Zhang S, Yue W, Yang L,
Zhai X, Yan Q and Lu J: The role of exosomal noncoding RNAs in
cancer. Mol Cancer. 18:372019. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Mrizak D, Martin N, Barjon C,
Jimenez-Pailhes AS, Mustapha R, Niki T, Guigay J, Pancre V, de
Launoit Y, Busson P, et al: Effect of nasopharyngeal
carcinoma-derived exosomes on human regulatory T cells. J Natl
Cancer Inst. 107:3632015. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Yamada N, Kuranaga Y, Kumazaki M,
Shinohara H, Taniguchi K and Akao Y: Colorectal cancer cell-derived
extracellular vesicles induce phenotypic alteration of T cells into
tumor-growth supporting cells with transforming growth
factor-β1-mediated suppression. Oncotarget. 7:27033–27043. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Li P, Liu C, Yu Z and Wu M: New insights
into regulatory T cells: Exosome- and non-coding RNA-mediated
regulation of homeostasis and resident treg cells. Front Immunol.
7:5742016. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Que RS, Lin C, Ding GP, Wu ZR and Cao LP:
Increasing the immune activity of exosomes: The effect of
miRNA-depleted exosome proteins on activating dendritic
cell/cytokine-induced killer cells against pancreatic cancer. J
Zhejiang Univ Sci B. 17:352–360. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Dudda JC, Salaun B, Ji Y, Palmer DC,
Monnot GC, Merck E, Boudousquie C, Utzschneider DT, Escobar TM,
Perret R, et al: MicroRNA-155 is required for effector CD8+ T cell
responses to virus infection and cancer. Immunity. 38:742–753.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Morishita M, Takahashi Y, Matsumoto A,
Nishikawa M and Takakura Y: Exosome-based tumor antigens-adjuvant
co-delivery utilizing genetically engineered tumor cell-derived
exosomes with immunostimulatory CpG DNA. Biomaterials. 111:55–65.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Kim MS, Haney MJ, Zhao Y, Yuan D, Deygen
I, Klyachko NL, Kabanov AV and Batrakova EV: Engineering
macrophage-derived exosomes for targeted paclitaxel delivery to
pulmonary metastases: In vitro and in vivo evaluations.
Nanomedicine. 14:195–204. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Aqil F, Munagala R, Jeyabalan J, Agrawal
AK, Kyakulaga AH, Wilcher SA and Gupta RC: Milk exosomes-natural
nanoparticles for siRNA delivery. Cancer Lett. 449:186–195. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Usman WM, Pham TC, Kwok YY, Vu LT, Ma V,
Peng B, Chan YS, Wei L, Chin SM, Azad A, et al: Efficient RNA drug
delivery using red blood cell extracellular vesicles. Nat Commun.
9:23592018. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Roma-Rodrigues C, Raposo LR, Cabral R,
Paradinha F, Baptista PV and Fernandes AR: Tumor microenvironment
modulation via gold nanoparticles targeting malicious exosomes:
Implications for cancer diagnostics and therapy. Int J Mol Sci.
18:E1622017. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Kim SM, Yang Y, Oh SJ, Hong Y, Seo M and
Jang M: Cancer-derived exosomes as a delivery platform of
CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J
Control Release. 266:8–16. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Qi H, Liu C, Long L, Ren Y, Zhang S, Chang
X, Qian X, Jia H, Zhao J, Sun J, et al: Blood exosomes endowed with
magnetic and targeting properties for cancer therapy. ACS Nano.
10:3323–3333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Agrawal AK, Aqil F, Jeyabalan J, Spencer
WA, Beck J, Gachuki BW, Alhakeem SS, Oben K, Munagala R, Bondada S
and Gupta RC: Milk-derived exosomes for oral delivery of
paclitaxel. Nanomedicine. 13:1627–1636. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
O'Loughlin AJ, Mager I, de Jong OG, Varela
MA, Schiffelers RM, El Andaloussi S, Wood MJA and Vader P:
Functional delivery of lipid-conjugated siRNA by extracellular
vesicles. Mol Ther. 25:1580–1587. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Lunavat TR, Jang SC, Nilsson L, Park HT,
Repiska G, Lasser C, Nilsson JA, Gho YS and Lotvall J: RNAi
delivery by exosome-mimetic nanovesicles - Implications for
targeting c-Myc in cancer. Biomaterials. 102:231–238. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Jackson AL, Burchard J, Leake D, Reynolds
A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et
al: Position-specific chemical modification of siRNAs reduces
‘off-target’ transcript silencing. RNA. 12:1197–1205. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Wesselhoeft RA, Kowalski PS and Anderson
DG: Engineering circular RNA for potent and stable translation in
eukaryotic cells. Nat Commun. 9:26292018. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Su H, Tao T, Yang Z, Kang X, Zhang X, Kang
D, Wu S and Li C: Circular RNA cTFRC acts as the sponge of
microRNA-107 to promote bladder carcinoma progression. Mol Cancer.
18:272019. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Chen S, Huang V, Xu X, Livingstone J,
Soares F, Jeon J, Zeng Y, Hua JT, Petricca J, Guo H, et al:
Widespread and functional RNA circularization in localized prostate
cancer. Cell. 176:831–843.e22. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T,
Sun H, Pan Y, He B and Wang S: CircHIPK3 promotes colorectal cancer
growth and metastasis by sponging miR-7. Cell Death Dis. 9:4172018.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Bai N, Peng E, Qiu X, Lyu N, Zhang Z, Tao
Y, Li X and Wang Z: CircFBLIM1 act as a ceRNA to promote
hepatocellular cancer progression by sponging miR-346. J Exp Clin
Cancer Res. 37:1722018. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Wang R, Zhang S, Chen X, Li N, Li J, Jia
R, Pan Y and Liang H: CircNT5E acts as a sponge of miR-422a to
promote glioblastoma tumorigenesis. Cancer Res. 78:4812–4825. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Wang R, Zhang S, Chen X, Li N, Li J, Jia
R, Pan Y and Liang H: EIF4A3-induced circular RNA MMP9 (circMMP9)
acts as a sponge of miR-124 and promotes glioblastoma multiforme
cell tumorigenesis. Mol Cancer. 17:1662018. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Liu T, Zhang X, Du L, Wang Y, Liu X, Tian
H, Wang L, Li P, Zhao Y, Duan W, et al: Exosome-transmitted
miR-128-3p increase chemosensitivity of oxaliplatin-resistant
colorectal cancer. Mol Cancer. 18:432019. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Qin X, Guo H, Wang X, Zhu X, Yan M, Wang
X, Xu Q, Shi J, Lu E, Chen W and Zhang J: Exosomal miR-196a derived
from cancer-associated fibroblasts confers cisplatin resistance in
head and neck cancer through targeting CDKN1B and ING5. Genome
Biol. 20:122019. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Geng X, Jia Y, Zhang Y, Shi L, Li Q, Zang
A and Wang H: Circular RNA: Biogenesis, degradation, functions and
potential roles in mediating resistance to anticarcinogens.
Epigenomics. 12:267–283. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Zhao Z, Ji M, Wang Q, He N and Li Y:
Circular RNA Cdr1as Upregulates SCAI to suppress cisplatin
resistance in ovarian cancer via miR-1270 suppression. Mol Ther
Nucleic Acids. 18:24–33. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Liu F, Zhang J, Qin L, Yang Z, Xiong J,
Zhang Y, Li R, Li S, Wang H, Yu B, et al: Circular RNA EIF6
(Hsa_circ_0060060) sponges miR-144-3p to promote the
cisplatin-resistance of human thyroid carcinoma cells by autophagy
regulation. Aging (Albany NY). 10:3806–3820. 2018. View Article : Google Scholar : PubMed/NCBI
|