Introduction
In 2018, lung cancer (LC) was reported as the
leading cause of cancer-associated mortality worldwide (1), early screening of LC is regarded as a
significant means to improve patient survival. Over the past 20
years, various mechanisms of LC and their detection methods have
been comprehensively studied in liquid biopsy samples (2). A number of clinical molecular
diagnostic tests for LC have been developed based on these studies
(3–5). The objectives of these diagnostic
methods include early detection, screening for therapeutic targets,
profiling cancer panel, monitoring therapeutic effectiveness and
early recurrence detection. At the genetic level, the most relevant
genes that are assessed for LC include epidermal growth factor
receptor (EGFR), KRAS and anaplastic lymphoma kinase
(ALK) (6). At the protein
level, a number of antigen detection methods, including
carcinoembryonic antigen (CEA), cancer antigen 125 and cytokeratin
fragment 21-1, have also been studied to improve diagnostic
accuracy (7). At the cellular level,
the examination of circulating tumor cells (CTCs) has been shown to
provide prognostic information concerning tumor metastasis
(8). Moreover, exhaled volatile
organic compounds (VOCs) are considered to be optimal non-invasive
biomarkers since they are indicative of the mutations and
pathophysiological processes of LC (9).
Recent advances in the field of VOC biomarkers have
improved the sensitivity and specificity in the diagnosis of LC
(10). A VOC pattern or specific
VOCs can be detected and used as a potential guide for the
development of effective therapeutics (11). The use of mass spectrometers or
sensors for the precise detection of volatile biomarkers could
substantially aid the diagnostic process by supplementing the
standard screening methods in the early diagnosis and prognosis of
patients with LC.
In the current review, the related mechanisms of
molecular, cellular and volatile biomarkers for LC and techniques
used for their detection are discussed. In addition, the detection
characteristics of the different diagnostic methods are compared.
Important improvements are required in certain areas, including the
following: i) Exploring the relationship between liquid biopsies
and volatile biomarkers in standardized clinical trials of LC; ii)
customizing the anticancer strategies; and iii) developing more
sensitive and selective instruments for biomarker detection.
Emerging non-invasive detection methods for
LC
Methods of interest
Evaluation of the intratumor heterogeneity of
patients used to be based on surgical resection sampling and
multiple biopsies. However, recent advances in sampling are focused
on acquiring sufficient samples for various biomarkers using
minimally invasive or non-invasive methods, including liquid biopsy
for the detection of circulating tumor (ct)DNA. (12), circulating tumor cells (CTCs)
(13) and protein biomarkers
(14), and analysis of exhaled VOCs
(15). The specific characteristics
that were compared between these less invasive (nucleic acid,
protein and cell based) and non-invasive (VOCs based) detection
methods for the diagnosis of LC are listed in Table I.
 | Table I.Comparison of less invasive and
non-invasive detection methods for the diagnosis of lung
cancer. |
Table I.
Comparison of less invasive and
non-invasive detection methods for the diagnosis of lung
cancer.
| Sample | Analytical
methods | Sensitivity
(%) | Advantages | Disadvantages | (Refs.) |
|---|
| ctDNA | Multiplex-PCR;
NGS | 48.0–59.0 | Earlier detection
(~70 days prior to CT imaging) | Expensive; Limited
sensitivity; Require specialized skills and equipment; | (16,18,19,75) |
| Methylated DNA | Bisulfite
conversion + PCR, NGS; Restriction enzyme + PCR; Antibodies + PCR;
NGS | 70.0–87.8 | High sensitivity
and specificity | Require
standardization; Fragment (bisulfite conversion); Only recognize
special patterns (restriction enzyme); Low recovery rate
(antibodies) | (22,24,25,76) |
| microRNAs | NGS; RT-PCR;
Microarray | 80.0–91.5 | Stable; High
throughput | Require specialized
skills and equipment | (33,34,77,78) |
| Proteins | Microarray;
LC-MS/MS | 70.0–84.0 | Higher sensitivity;
High throughput; Rapid | Require validation;
Protein binding problems (Microarray); Difficult to quantitate
signals (MS) | (42,44,48,79) |
| CTCs | IF; FISH | 30.0–69.5 | High throughput;
High specificity; Co-localization | Limited
sensitivity; Require enrichment; Monitor only advanced types of
cancer; Heterogeneity effect | (51,80,81) |
| Exhaled VOCs | GC-MS;
Sensors/E-noses; PTR-MS; IMS; LPPI-MS | 81.0–96.5 | Simple, rapid;
Non-invasive; Non-expensive | Require
standardization | (59,61,63–65,
70,71,72,73) |
Nucleic acid-based detection
i) ctDNA markers. Liquid biopsy is thought to be one
of the potential options for the non-invasive diagnosis of LC.
ctDNA is secreted into the serum by necrotic or apoptotic cells,
which may provide effective means for tumor diagnoses (16). Additionally, the short half-life of
ctDNA (~2 h) renders it an ideal dynamic marker of tumors (17). Some genetic mutations, particularly
single nucleotide variants (SNVs) in ctDNA may be considered as
specific biomarkers for LC. A study assessing 100 early-stage
non-small cell lung cancer (NSCLC) specimens revealed that 48% of
patients had ≥2 detectable SNVs in ctDNA. Post-operative
next-generation sequencing (NGS) based on ctDNA analysis is able to
predict LC recurrence earlier than CT imaging, at ~70 days
(18). Furthermore, Cohen et
al (19) used CancerSEEK, a
PCR-based ctDNA method, for the detection of five cancer types,
namely ovarian, liver, stomach, pancreatic and esophageal cancer,
and reported that the accuracy of prediction varied with tumor
type. The performance of CancerSEEK in the analysis of 104 LC
samples was poor, with only 59% sensitivity; even when combined
with machine learning, the accuracy was lowest for LC.
Although ctDNA analysis for LC is promising, it
remains incomplete. Disadvantages of this method, which hamper its
widespread application, include the following: a) Poor detection
sensitivity; b) high cost; and c) limited clinical utility. More
specifically, the concentration of ctDNA in plasma is only 1%, and
as much as 10 ml plasma are required to obtain reliable results
when using the current ctDNA platforms. Regarding the cost of
liquid biopsy testing, this has been estimated at US$1,750 per
patient. Moreover, ctDNA analysis is technically complex and
requires specialized skills and equipment (18).
ii) Methylated DNA markers. Aberrant
hypermethylation of global DNA or specific CpG islands in promoter
regions has been considered a promising biomarker for different
cancer types, including ovarian, prostate, liver and cervical
cancer (20). Methylated DNA markers
in LC have been detected in various body fluid samples, including
blood, serum, pleural effusion and ascites (21). Improvements of this detection method
currently enable the determination of DNA methylation. Wielscher
et al (22) reported a panel
of four methylated genes, specifically homeobox D10, paired box 9,
protein tyrosine phosphatase receptor type 2 and stromal antigen 3,
as markers for LC detection. LC was efficiently differentiated from
other lung diseases and controls with a sensitivity of 87.8% and a
specificity of 90.2%. The commercial test kit Epi
proLung® (Epigenomics Inc.), used for the screening of
LC and based on the analysis of methylation in the short stature
homeobox 2 (SHOX2) gene, was approved by the Chinese Food
and Drug Administration (FDA) in July 2015 (23). To estimate the diagnostic efficacy of
SHOX2 DNA methylation, a meta-analysis was conducted in
2,296 subjects, including 1,129 patients with LC, which
demonstrated that this method had 70% sensitivity and 96%
specificity (24).
Three groups of methods are commonly employed to
distinguish methylated DNA from unmethylated DNA, including sodium
bisulfite conversion, restriction enzyme and specific antibodies
(25). DNA methylation may be
analyzed using various detection methods, including PCR,
microarrays, and NGS. However, each of these methods has certain
drawbacks; bisulfite conversion results in random DNA fragmentation
(26). In addition, restriction
enzyme-based methods can only detect specific patterns of CpG
sites. Antibody-based methods are limited by a low recovery rate
(27). Moreover, the establishment
of standardized protocols for methylation detection methods is
essential.
iii) MicroRNA (miRNA/miR) markers. miRNAs are small
non-coding RNAs that are capable of influencing cancer metabolism
by regulating tumor suppressor signaling pathways of glucose
metabolism or the expression of glycolytic enzymes (28). The miRNA expression profiles for LC
have been found to be present and stably expressed in bodily
fluids, including human serum/plasma and sputum (29). For instance, findings have
demonstrated a significantly decreased expression of miRNA-124-5p
and an increased expression of miRNA-124-2 and miRNA-124-3 in
patients with advanced-stage LC (30,31).
Moreover, miRNA expression patterns may serve as specific biomarker
signatures for the diagnosis and subtype differentiation of LC from
early stage to metastatic (32,33). In
a cohort of 180 subjects (92 patients; 88 healthy participants), a
plasma miRNA signature, consisting of miRs-126, −145, −210 and
−205-5p, was defined by NGS and shown to have 91.5% sensitivity and
96.2% specificity in the detection of LC (34). In another study, a panel of six
miRNAs, namely miR-17, miR-190b, miR-19a, miR-19b, miR-26b and
miR-375, was used for the diagnosis and further differentiation of
small cell lung cancer from NSCLC in 1,132 participants by
microarray analysis and was found to have 80% sensitivity and 80%
specificity (33).
In addition, numerous plasma miRNAs have been
identified using microarray platforms and are considered as plasma
biomarkers for LC (35,36). These miRNAs are usually further
verified by reverse transcription (RT)-quantitative PCR analysis.
Nevertheless, microarray-based methods are limited by relatively
low sensitivity and specificity. Therefore, they have yet to be
applied in clinical settings. By contrast, NGS is able to identify
LC-associated miRNAs with high throughput and high sensitivity and
specificity (37).
In combination with certain algorithms, miRNA
analyses may have powerful diagnostic and prognostic performance.
Sozzi et al (38) combined
miRNA analyses with low-dose computed tomography (LDCT) for the
detection of LC, which resulted in a 5-fold reduction of the
false-positive rate, from 19.4–3.7%.
Protein-based detection
Protein profiling is another diagnostic method based
on liquid biopsy (39). Numerous
studies on LC protein biomarkers have been conducted in blood,
urine, saliva and exhaled breath condensate (EBC) samples (40–42).
Common protein biomarkers for LC, including CEA and cytokeratin
fragment 21-1, are not sensitive enough to detect early tumors
(43). Therefore, newer protein
biomarkers with higher sensitivity have been developed.
Cytoskeleton-associated protein 4 (CKAP4) is expressed at
significantly higher levels in patients with LC than in the
controls. In a cohort comprising 271 patients with LC and 100
healthy controls, the sensitivity of reverse-phase protein array
for serum CKAP4 was 81.1%. Furthermore, CKAP4 has been reported to
reliably detect early-stage adenocarcinoma or squamous cell
carcinoma, with a sensitivity of 78.6% (44).
Compared with traditional serological markers,
exosomal proteins have certain advantages. Typically, there are
over 1×109 exosomes per ml of human blood. Furthermore,
approximately 80% salivary exosomal proteins are shared with serum
exosomes (45). Several proteins are
specifically expressed in exosomes, including Xbox-binding protein
1, glypican-1 and cGMP-dependent protein kinase 1, and have shown
higher sensitivity, specificity and stability compared with those
in serum for cancer screening (46,47). In
a cohort of 109 patients with NSCLC and 110 healthy controls, the
two groups were distinguished by extracellular vesicle array using
the New York esophageal squamous cell carcinoma-1 protein, which
was found to have 75.3% sensitivity (48). Although the mechanism of exosomal
protein-based diagnosis remains unclear, these biomarkers have
potential for use in the early clinical detection and diagnosis of
LC, provided that they are validated.
The evaluation of proteins found in EBC, saliva and
urinary samples of patients with LC is a non-invasive diagnostic
method. Lopez-Sanchez et al (42) collected EBC samples from 192
individuals (48 patients; 144 healthy controls) and analyzed them
by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The
expression levels of cytokeratins and other proteins were
significantly high in LC samples, yielding 70% sensitivity and 67%
specificity. Nolen et al (41) examined urinary samples of 234
individuals (83 patients; 151 healthy controls) by applying a panel
of three proteins, insulin-like growth factor binding protein 1,
secretory interleukin-1 receptor antagonist and cell adhesion
molecule 1, to differentiate NSCLC, with 84% sensitivity and 95%
specificity.
Although protein microarray analysis offers
comprehensive protein information with minimal sample requirement,
it could be optimized for the rapid screening of LC. However,
various issues regarding protein binding remain a challenge for
these protein detection arrays (49). Moreover, LC-MS/MS is an excellent
detection method for systematic analysis of protein profiling of
LC, though not a rapid and high-throughput platform in
translational medicine.
Cell-based detection
Tumors release certain malignant cells, termed CTCs,
into the vasculature. CTCs have been characterized based on their
migration and invasion abilities in the early stages of LC. High
CTC numbers correspond to an aggressive type of cancer, increased
metastasis and increased likelihood of relapse (50). Nevertheless, the number of CTCs in
patients may be as low as 1 CTC per ml of blood. Therefore, cell
enrichment is required, which typically depends on the antigens,
e.g., EpCAM and CD45, or inherent properties of CTCs, including
size, deformability or dielectric susceptibility (51). Following cell enrichment,
verification of CTCs is generally achieved by high-resolution
imaging combined with immunohistochemical enumeration.
The CellSearch® test (Menarini Silicon
Biosystems, Inc.), which is EpCAM-dependent, is the only
FDA-approved method for CTC detection that has been proven to
provide prognostic information in metastatic breast, colorectal and
prostate cancer. Regarding LC, however, this method has been found
to exhibit limited detection efficiency (52). In a cohort of 150 patients with LC,
the sensitivity of CellSearch® was only 30.4% (53). In another study, a novel, size-based
filtration platform was employed for the diagnosis of LC in 82
patients, which was shown to have improved sensitivity for CTC
detection (69.5%) (54).
CTC-based liquid biopsy has several advantages,
including high specificity, demonstrating the localization signal
as an integrated cell. However, the use of CTCs for LC screening is
currently impaired by its limited application in the clinical
setting. As negligible CTC counts are found in early-stage patients
with LC, CTC detection methods may only be efficient for patients
with advanced LC (54,55).
VOC-based detection
VOCs are a group of gaseous organic molecules with
relatively high vapor pressure or volatility. VOCs present in
bodily fluids reach the lungs via the bloodstream and are exhaled
through breath (15). Among them,
exogenous VOCs derived from cigarette smoke, pollution and
radiation target and damage DNA, proteins and polyunsaturated fatty
acids (PUFAs) in the body, thus boosting oxidative stress and
contributing to cancer development (56). By contrast, endogenous VOCs are
mainly derived from diverse metabolic pathways. Since endogenous
VOCs associated with certain metabolism may be altered by diseases,
different VOC profiles have been associated with various diseases,
including cancer (57).
A wide range of analytical techniques has been used
for the determination of volatile metabolites. Gas
chromatography-mass spectrometry (GC-MS) is considered the gold
standard for the detection of specific VOCs (58–60).
Phillips et al (61,62) applied the predictive model of 16 and
30 VOCs to differentiate patients with primary LC from the controls
in a cohort of 193 patients and 211 controls and it was found that
this model had approximately 85% sensitivity and approximately 80%
specificity. In another cohort consisting of 88 patients and 155
controls, three diagnostic models based on 23 VOCs were able to
easily identify patients, with 96.5% sensitivity and 97.5%
specificity (63).
Nonetheless, GC-MS is a method based on offline
analyses, which requires time-consuming preparations. On the
contrary, certain advanced analytical techniques, including proton
transfer reaction-mass spectrometry (PTR-MS), selected ion flow
tube-mass spectrometry (SIFT-MS) and low-pressure
photoionization-mass spectrometry (LPPI-MS) offer real-time
analysis and high sensitivity (64–66).
However, to the best of our knowledge, no systematic studies have
been performed for the discrimination of patients with LC by
PTR-MS, SIFT-MS or LPPI-MS.
Mass spectrometry-based techniques are expensive and
require complex instruments. By contrast, sensors/E-noses show
great potential for fast, easy and cost-effective diagnosis and
screening of LC (67). With the
advancement of sensing devices, electronics and signal processing,
the sizes of sensors/E-nose systems can be minimized along with
fast data processing to provide real-time results (67). When combined with pattern recognition
methods, sensors/E-noses are capable of distinguishing the exhaled
breath of patients with LC from the breath of healthy controls
without the need for dehumidification or pre-concentration of
biomarkers (68). Nevertheless, more
subjects are needed in order to increase the sensitivity and
negative predictive value (69).
Chang et al (11) examined
the breath samples of 37 patients with LC and 48 healthy controls
using a sensor system, which was found to have 79.0% sensitivity
and 72.0% specificity. Gasparri et al (70) reported the differentiation of 70
patients with LC from 76 healthy controls with 81% sensitivity and
91% specificity, using a gas sensor array composed of a matrix of
eight quartz microbalances. Shehada et al (71) collected breath samples from 149
volunteers with LC, specifically 40 patients with gastric cancer,
56 volunteers with non-cancerous lung diseases (asthma, chronic
obstructive pulmonary disease or both) and 129 healthy controls.
The self-developed silicon nanowire field effect transistor was
used to separate patients with LC from subjects in the control
group and were shown to have 87% sensitivity and 82%
specificity.
Similarly, ion mobility spectrometry (IMS) also
allows pattern recognition, rather than specific VOC
identification, and has the potential for miniaturization. Handa
et al (72) investigated the
breath samples of 50 patients with LC and 39 healthy controls by
IMS and revealed that this method had a sensitivity of 81.3% and
specificity of 89.7%. Nonetheless, sensors and IMS are limited in
terms of detection and, thus, are usually coupled with complex
algorithms (72–74). The advantages and disadvantages of
nucleic acid-based (75–78), protein-based (79) and cell-based (80,81)
detections were compared with VOC-based detection as displayed in
Table I.
Targeted therapies based on non-invasive
detection
Liquid biopsy-guided therapies
A liquid biopsy, which is a non-invasive sampling
procedure, is preferred for metastatic LC in order to obtain the
molecular characterization (Table
II). Several targeted methods, including PCR, NGS and
immuno-oncology, are employed to detect gene mutation targets
(EGFR, ALK, BRAF) from DNA/RNA or CTC samples (82). The application of liquid biopsy as a
guidance for targeted therapy would notably improve the overall
survival of patients with LC.
 | Table II.Comparison of the main targeted
therapies guided by liquid biopsy. |
Table II.
Comparison of the main targeted
therapies guided by liquid biopsy.
| Target | Frequency (%) | Drug used | Detection | (Refs.) |
|---|
| EGFR
mutation | 20–76 | Gefitinib,
erlotinib, afatinib, osimertinib | Plasma DNA;
CTCs | (84,85,91) |
| ALK
rearrangement | 4–6 | Crizotinib, Alecc,
Ceritinib, Lorlatinib. | Plasma DNA/RNA;
CTCs | (93,96,97) |
| BRAF
mutation | 1.6–1.8 | Vemurafenib,
dabrafenib sorafenib, trametinib | Plasma DNA;
CTCs | (102–105) |
i) EGFR mutation. Mutations in EGFR
are the only standardized therapeutic targets examined in clinical
practice. COBAS (Roche Diagnostics) and Therascreen® RGQ
PCR kit (Qiagen, Inc.) are two methods used for the detection of
EGFR mutations, which have been approved by the FDA in the
USA (83). EGFR mutations are
mostly detected in DNA samples extracted from plasma. By contrast,
CTC-based detection is limited by poor sensitivity and specificity
and the small quantity of CTCs in blood (84). EGFR mutations occur in 20–76%
patients with NSCLC and are more common among patients from the
Asia-Pacific region (85). Patients
with LC, who have EGFR mutations, are normally treated with
tyrosine kinase inhibitors (TKIs), including gefitinib, erlotinib
and afatinib (86). However,
sensitivity to TKI treatment varies with the type of EGFR
mutation (87,88). Monoclonal antibodies and TKIs are
usually applied to treat specific genetic alterations, including
EGFR and ALK, in both the first line and resistant
settings (89,90). Several new generation TKIs (e.g.,
osimertinib, olmutinib, nazartinib, avitinib and rociletinib) have
been developed for patients harboring EGFR resistance
mutations (5,91). In general, EGFR mutations that
confer sensitivity to TKI therapy seem to be greater than those
that confer resistance (92).
ii) ALK rearrangement. ALK
rearrangements act as an oncogenic driver in 4–6% of NSCLCs
(93). Several target methods are
used to evaluate the status of ALK by liquid biopsy,
including RT-PCR, NGS, and fluorescence in situ
hybridization. However, during the preprocessing steps, circulating
plasma RNAs are degraded rapidly, which results in lower
sensitivity (94,95). Conversely, CTC-based detection of
ALK status is more feasible and in accordance with tissue
biopsies (96). Previous studies
have evaluated several types of ALK inhibitors, including
crizotinib, alectinib, ceritinib and lorlatinib in patients with
LC/NSCLC (97–100). Most patients with NSCLC that have
ALK rearrangements respond well to ALK TKIs;
heterogeneous responses are also reported, though the reason
remains unknown (101).
iv) BRAF mutations. BRAF mutations can be
identified in 1.6–1.8% of patients with LC (102). These mutations act as an oncogenic
driver in NSCLC via the mitogen-activated protein kinase (MAPK)
pathway. Vemurafenib and dabrafenib are used to block MAPK
signaling in patients with LC (103). Furthermore, since these mutations
increase the kinase activity of BRAF towards
mitogen-activated protein kinase (MAPK) kinase (MEK), the
combination of MAPK pathway inhibition with BRAF and
MEK inhibitors may prove to be an effective therapeutic
strategy for LC. BRAF mutations can be detected in plasma
DNA or CTC samples. It has been reported that the combination of
dabrafenib and trametinib (a selective allosteric inhibitor of
MEK1/MEK2) (104) showed robust
antitumor activity and a manageable safety profile in patients with
NSCLC that harbored BRAF mutations (105).
Apart from the main oncogenic mutations that were
mentioned above, additional molecular targets for LC have been
identified, including ROS proto-oncogene 1, human epidermal growth
factor receptor 2, phosphatidylinositol-4,5-bisphosphate 3-kinase
catalytic subunit alpha, rearranged during transfection
proto-oncogene and hepatocyte growth factor receptor (106–108).
Although several monoclonal antibodies and tyrosine-kinase
inhibitor drugs have been developed to inhibit LC-related signaling
pathways, there is an urgent need for the development of detection
methods with improved sensitivity for the determination of specific
biomarkers and guidance of effective treatment options.
VOC production mechanisms and their
potential for targeted therapies
Different VOCs may contain metabolic information
concerning various types of human tissues and the storage
capacities of volatile organic substances significantly differ in
different types of human tissues. The time required to metabolize
these different VOCs from the human body also differs (109). Preliminary metabolic pathways of
several VOCs that have been studied include the following: i)
Hydrocarbons; ii) alcohols; iii) aldehydes; iv) branched aldehydes;
v) ketones; vi) esters; vii) nitriles and aromatics.
i) Hydrocarbon. In the human body, oxidative stress
is the main mechanism corresponding to the production of
hydrocarbons. Alkanes are mainly derived from the peroxidation of
PUFAs, which occurs in the cell and subcellular membranes. Lipid
peroxidation can cause tissue damage, which in turn may lead to
aging, inflammation, atherosclerosis, and cancer. The effects of
lipid peroxidation may be reduced and regulated by antioxidants
(57). Saturated alkanes including
ethane and pentane are considered the final products of lipid
peroxidation and have been widely recognized as volatile biomarkers
of this reaction in breath analysis (110). The production of C3-C11 saturated
alkanes is also associated with lipid peroxidation, though the
production of branched alkanes does not originate from this
mechanism (111). As hydrocarbons
have a lower solubility in blood, they can be excreted from the
body by exhaled breath in a matter of minutes (112).
ii) Alcohols. Alcohols are also considered products
of hydrocarbon metabolism and can be easily absorbed in the
digestive tract. Once absorbed, alcohols enter the bloodstream. As
short-chain alcohols are highly soluble in water, their absorption
into the bloodstream is rapid. The metabolism of alcohols varies
greatly due to the diversity of fat and water content among
individuals. Certain enzymes, including alcohol dehydrogenase and
cytochrome p450 family 2 subfamily E member 1 (CYP2E1), which is
mainly expressed in the liver, are involved in the metabolism of
alcohols (57).
iii) Aldehydes.Aldehydes may be produced through
normal physiological processes. Certain aldehydes serve important
roles in the physiological functions of the body, whereas others
are considered to be cytotoxic intermediates that perform specific
functions and serve roles in signal transmission, gene regulation
and cell proliferation (113,114).
In the human body, aldehydes may be derived from multiple processes
or sources, including the following: i) Alcohol metabolism; ii)
degradation of hydrogen peroxide-containing substances by CYP2E1, a
by-product of lipid peroxidation (115); iii) smoking, which produces a large
number of aldehydes; saturated and unsaturated aldehydes, including
formaldehyde, acetaldehyde, and acrolein, are produced from tobacco
burning (116); iv) by-products of
tobacco metabolism, which occurs in the cytochrome, which in turn
participates in detoxification (117); v) dietary intake (118).
iv) Ketones. Fatty acid oxidation in the body is
accelerated with the onset of cancer. As a result, a large number
of ketones are identified in cancer patients, the production of
which is closely related to the rapid decline of weight, a common
complication of cancer. The liver is a vital organ for ketone
metabolism (119). Since a
considerable amount of acetoacetate and b-hydroxybutyric acid are
synthesized in the liver, acetoacetate spontaneously undergoes
decarboxylation and acetone is consequently produced (120).
v) Esters. Large amounts of exogenous esters are
found in natural fats, oils, natural waxes and plant essential
oils. In the human body, esterases are capable of hydrolyzing
esters to alcohols and acids at body temperature, which is similar
to the process by which lipases hydrolyze fat (121).
vi) Nitriles and aromatic substances. Nitriles and
aromatic substances enter the human body via external pollutants,
including air pollution, drinking, smoking and radiation. Due to
their carcinogenicity, they have gained interest in the study of
cancer (11). They are highly active
and can cause damage to PUFAs, proteins and DNA. As these damages
accumulate in the body, normal repair functions cease to be
effective, leading to the development of a series of diseases,
including cancer.
Studying the VOC- and metabolite- profiles of LC
cells provides an alternative to invasive diagnosis. A recent study
revealed a possible association between VOCs and metabolites. It
was found that the combination of benzaldehyde, 2-ethylhexanol and
2, 4-decadien-1-ol could serve as potential VOC biomarkers for LC
(122). These VOCs are also
strongly negatively associated with the levels of certain amino
acids, glucose, cholesterol and several fatty acids. Another study
examined the VOC biomarkers that were associated with EGFR,
KRAS and ALK mutations in LC cell lines. Triethylamine,
benzaldehyde and decanal have the potential to become specific VOC
biomarkers for identifying and distinguishing these mutations in
patients with LC (123). Further
clinical studies are required to determine whether these cell-line
methods could be translated into clinical diagnostic tools.
The goal of personalized medicine is to treat
patients using their genetic profile. The gene-specific VOC
biomarkers are metabolized in different types of tissues,
transported to the lungs via blood circulation and exhaled from the
alveoli. These exhaled breath-detection methods are entirely
non-invasive, readily available and do not require any preparation
(57). Therefore, they have the
potential to provide indirect genetic and pathological information
prior to and during the treatment period.
In order to validate the efficacy of VOC profiles in
the exhaled breath as a means of diagnosing LC, advancements in the
following technologies are critically required: i) Standardized
clinical utility; and ii) advanced mass spectrometry with
ultra-high sensitivity. Recently, the excited state proton transfer
(ESPT) ionization technology was established by LPPI-MS,
discovering new mechanisms of chemi-ionization reactions and
offering new technological applications that have the potential to
greatly improve mass spectrometry sensitivity for detecting trace
gaseous organics (124,125). VUV excitation is applied to produce
a chemi-ionization reaction, which yields substantial
H3O+ ions, and the protonated analyte, an
equal amount of Cl−, may be produced with the aid of the
reorganization energy released from the formation of
CH2O and HCl. The sensitivity for VOC biomarkers is at
least 20 times higher than that for PTR-MS. Regarding oxygenated
VOCs, the signal intensities of oxygenated organics can be
amplified by more than two orders of magnitude.
The application of advanced techniques in volatile
fingerprinting, allows non-invasive, rapid and accurate detection,
thereby leading to early diagnosis and prognosis based on
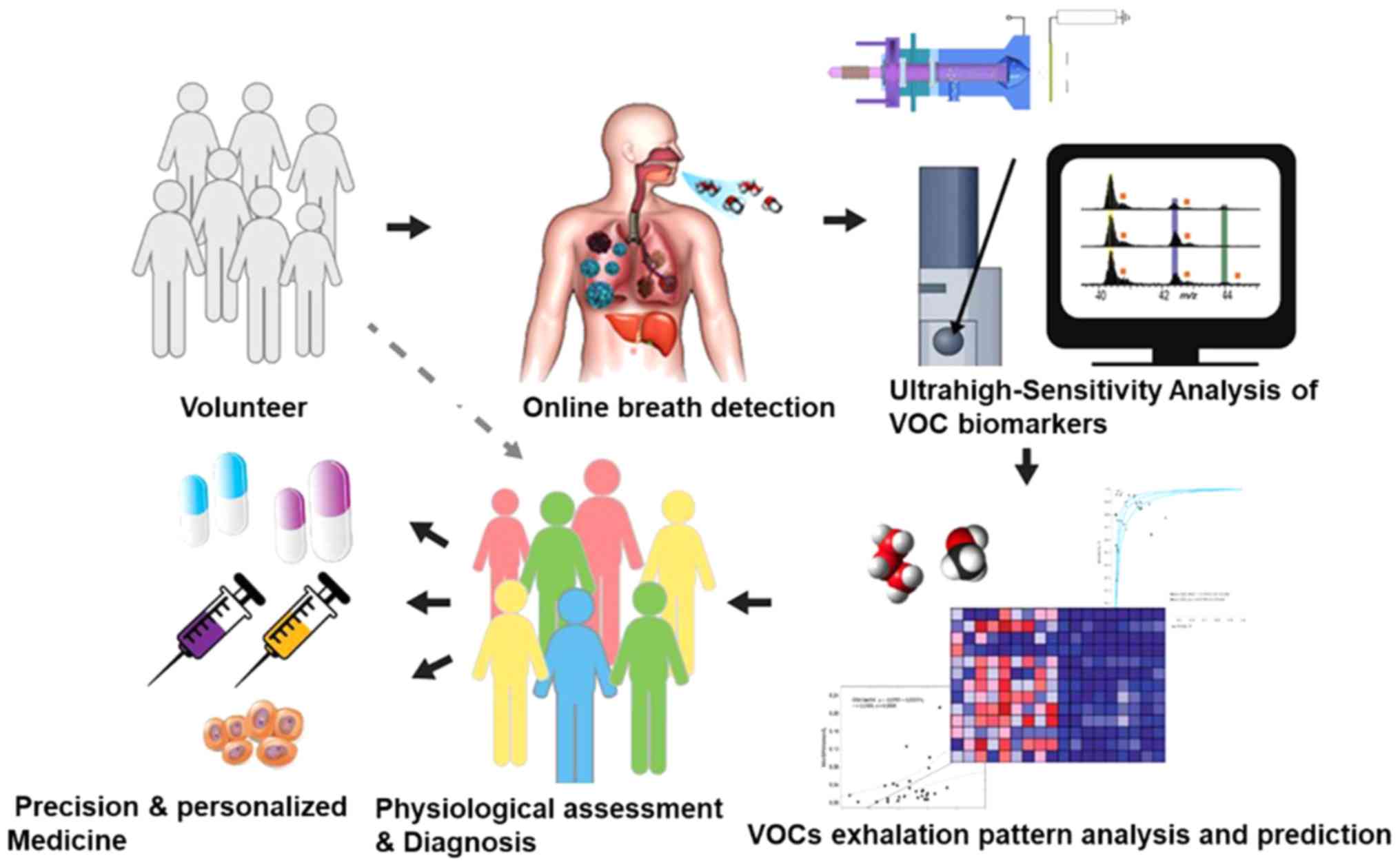
metabolomics. The process of VOC biomarker-based targeted therapies
for patients with LC is presented in Fig. 1. First, patients with LC and other
lung diseases and healthy controls are recruited. Secondly, exhaled
breath samples are collected from the volunteers and VOC biomarkers
are analyzed using ultra-high-sensitivity techniques, including
ESPT ionization. Subsequently, machine learning algorithms are
employed for the data mining of VOC profiles. In combination with
routine diagnostic methods, physiological assessment and diagnosis
are accomplished using VOC biomarkers. Improved precision of
diagnostic techniques and personalized medicine could lead to the
identification of additional VOC biomarkers and are anticipated to
open up connections between omics technologies (genomics,
proteomics and metabolomics), which may result in the faster and
more efficient discovery of other molecular markers and intervening
targets.
Challenges and future directions
Despite the vast potential of existing candidates
and methodologies, only a few non-invasive detection methods for
LC, including the Guardant360® panel (http://www.guardant360.com) and cobas®
EGFR Mutation Test v2 (Roche Molecular Diagnostics) are
currently being used in the clinical setting. In order to apply
potential non-invasive detection methods to clinical practice,
several milestones must be achieved. First, head-to-head
comparisons of different types of biomarkers in specific clinical
scenarios would be beneficial. Deep mining of data provided by
machine learning would also be useful. Secondly, the
instrumentation used for biomarker detection requires careful
reevaluation. Mass spectrometry, NGS, PCR, microarray and LDCT are
standard techniques that are widely used in biomarker detection.
However, in order to obtain accurate results in clinical practice,
the reliability of instruments and reproducibility of results
should be examined and optimized in clinical studies. Thirdly, the
interactions between liquid biopsy, imaging and VOC biomarkers
should also be identified in clinical studies. Moreover, the
combination of different biomarker tests could reduce
false-positive results and enable greater standardization of
diagnostic algorithms, thereby decreasing health care costs.
Although recent advances in the field of oncogenic
gene mutations have made it possible to realize molecular-targeted
therapy, monitoring the VOC signatures associated with
cancer-specific genetic mutations may be a faster and easier method
than conventional gene profiling. This technique would aid the
improvement of drug selection and detection of resistance, thereby
increasing the clinical benefits for patients with LC through
safer, more timely and effective interventions that could improve
their overall survival and quality of life.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural
Science Foundation of China (grant no. 81802981) and the Beijing
Municipal Science & Technology Commission (grant no.
Z181100003818008).
Availability of data and materials
Not applicable.
Authors' contributions
ZL contributed to the design and writing of the
manuscript, with support from ZZ and JH. BY and YC drafted the
initial manuscript and revised it critically for important
intellectual content. JS made substantial contributions to
conception and design, and gave final approval of the version to be
published. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Calvayrac O, Pradines A, Pons E, Mazieres
J and Guibert N: Molecular biomarkers for lung adenocarcinoma. Eur
Respir J. 49:16017342017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hyman DM, Puzanov I, Subbiah V, Faris JE,
Chau I, Blay JY, Wolf J, Raje NS, Diamond EL, Hollebecque A, et al:
Vemurafenib in multiple nonmelanoma cancers with BRAF V600
mutations. N Engl J Med. 373:726–736. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pao W, Miller V, Zakowski M, Doherty J,
Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, et al:
EGF receptor gene mutations are common in lung cancers from ‘never
smokers’ and are associated with sensitivity of tumors to gefitinib
and erlotinib. Proc Natl Acad Sci USA. 101:13306–13311. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Walter AO, Sjin RT, Haringsma HJ, Ohashi
K, Sun J, Lee K, Dubrovskiy A, Labenski M, Zhu Z, Wang Z, et al:
Discovery of a mutant-selective covalent inhibitor of EGFR that
overcomes T790M-mediated resistance in NSCLC. Cancer Discov.
3:1404–1415. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dugay F, Llamas-Gutierrez F, Gournay M,
Medane S, Mazet F, Chiforeanu DC, Becker E, Lamy R, Léna H,
Rioux-Leclercq N, et al: Clinicopathological characteristics of
ROS1- and RET-rearranged NSCLC in caucasian patients. Data from a
cohort of 713 non-squamous NSCLC lacking
KRAS/EGFR/HER2/BRAF/PIK3CA/ALK alterations. Oncotarget.
8:53336–53351. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Doseeva V, Colpitts T, Gao G, Woodcock J
and Knezevic V: Performance of a multiplexed dual analyte
immunoassay for the early detection of non-small cell lung cancer.
J Transl Med. 13:552015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Plaks V, Koopman CD and Werb Z: Cancer.
Circulating tumor cells. Science. 341:1186–1188. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rocco G, Pennazza G, Santonico M, Longo F,
Rocco R, Crucitti P and Antonelli Incalzi R: Breathprinting and
early diagnosis of lung cancer. J Thorac Oncol. 13:883–894. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
van der Schee MP, Paff T, Brinkman P, van
Aalderen WMC, Haarman EG and Sterk PJ: Breathomics in lung disease.
Chest. 147:224–231. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chang JE, Lee DS, Ban SW, Oh J, Jung MY,
Kim SH, Parka S, Persaude K and Jheon S: Analysis of volatile
organic compounds in exhaled breath for lung cancer diagnosis using
a sensor system. Sensors Actuators B Chem. 255:800–807. 2018.
View Article : Google Scholar
|
|
12
|
Chae YK and Oh MS: Detection of minimal
residual disease using ctDNA in lung cancer: Current evidence and
future directions. J Thorac Oncol. 14:16–24. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hou JM, Krebs M, Ward T, Sloane R, Priest
L, Hughes A, Clack G, Ranson M, Blackhall F and Dive C: Circulating
tumor cells as a window on metastasis biology in lung cancer. Am J
Pathol. 178:989–996. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ma S, Wang W, Xia B, Zhang S, Yuan H,
Jiang H, Meng W, Zheng X and Wang X: Multiplexed serum biomarkers
for the detection of lung cancer. EBioMedicine. 11:210–218. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Haick H, Broza YY, Mochalski P, Ruzsanyi V
and Amann A: Assessment, origin, and implementation of breath
volatile cancer markers. Chem Soc Rev. 43:1423–1449. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fiala C and Diamandis EP: Circulating
tumor DNA for personalized lung cancer monitoring. BMC Med.
15:1572017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Diehl F, Schmidt K, Choti MA, Romans K,
Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al:
Circulating mutant DNA to assess tumor dynamics. Nat Med.
14:985–990. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Abbosh C, Birkbak NJ, Wilson GA,
Jamal-Hanjani M, Constantin T, Salari R, Le Quesne J, Moore DA,
Veeriah S, Rosenthal R, et al: Corrigendum: Phylogenetic ctDNA
analysis depicts early-stage lung cancer evolution. Nature.
554:2642018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cohen JD, Li L, Wang Y, Thoburn C, Afsari
B, Danilova L, Douville C, Javed AA, Wong F, Mattox A, et al:
Detection and localization of surgically resectable cancers with a
multi-analyte blood test. Science. 359:926–930. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ehrlich M: DNA hypomethylation in cancer
cells. Epigenomics. 1:239–259. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ooki A, Maleki Z, Tsay JJ, Goparaju C,
Brait M, Turaga N, Nam HS, Rom WN, Pass HI, Sidransky D, et al: A
Panel of novel detection and prognostic methylated DNA markers in
primary non-small cell lung cancer and serum DNA. Clin Cancer Res.
23:7141–7152. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wielscher M, Vierlinger K, Kegler U,
Ziesche R, Gsur A and Weinhausel A: Diagnostic performance of
plasma DNA methylation profiles in lung cancer, pulmonary fibrosis
and COPD. EBioMedicine. 2:929–936. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ilse P, Biesterfeld S, Pomjanski N, Wrobel
C and Schramm M: Analysis of SHOX2 methylation as an aid to
cytology in lung cancer diagnosis. Cancer Genomics Proteomics.
11:251–258. 2014.PubMed/NCBI
|
|
24
|
Zhao QT, Guo T, Wang HE, Zhang XP, Zhang
H, Wang ZK, Yuan Z and Duan GC: Diagnostic value of SHOX2 DNA
methylation in lung cancer: A meta-analysis. Onco Targets Ther.
8:3433–3439. 2015.PubMed/NCBI
|
|
25
|
Lu Y, Li S, Zhu S, Gong Y, Shi J and Xu L:
Methylated DNA/RNA in body fluids as biomarkers for lung cancer.
Biol Proced Online. 19:22017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hernandez HG, Tse MY, Pang SC, Arboleda H
and Forero DA: Optimizing methodologies for PCR-based DNA
methylation analysis. Biotechniques. 55:181–197. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang ZH, Hu Y, Hua D, Wu YY, Song MX and
Cheng ZH: Quantitative analysis of multiple methylated genes in
plasma for the diagnosis and prognosis of hepatocellular carcinoma.
Exp Mol Pathol. 91:702–707. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen B, Li H, Zeng X, Yang P, Liu X, Zhao
X and Liang S: Roles of microRNA on cancer cell metabolism. J
Transl Med. 10:2282012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kim H, Yang JM, Jin Y, Jheon S, Kim K, Lee
CT, Chung JH and Paik JH: MicroRNA expression profiles and
clinicopathological implications in lung adenocarcinoma according
to EGFR, KRAS, and ALK status. Oncotarget. 8:8484–8498.
2017.PubMed/NCBI
|
|
31
|
Li D, Wei Y, Wang D, Gao H and Liu K:
MicroRNA-26b suppresses the metastasis of non-small cell lung
cancer by targeting MIEN1 via NF-KB/MMP-9/VEGF pathways. Biochem
Biophys Res Commun. 472:465–470. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dacic S, Kelly L, Shuai Y and Nikiforova
MN: MiRNA expression profiling of lung adenocarcinomas: Correlation
with mutational status. Mod Pathol. 23:1577–1582. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lu S, Kong H, Hou Y, Ge D, Huang W, Ou J,
Yang D, Zhang L, Wu G, Song Y, et al: Two plasma microRNA panels
for diagnosis and subtype discrimination of lung cancer. Lung
Cancer. 123:44–51. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Leng Q, Lin Y and Jiang F, Lee CJ, Zhan M,
Fang H, Wang Y and Jiang F: A plasma miRNA signature for lung
cancer early detection. Oncotarget. 8:111902–111911. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Arab A, Karimipoor M, Irani S, Kiani A,
Zeinali S, Tafsiri E and Sheikhy K: Potential circulating miRNA
signature for early detection of NSCLC. Cancer Genet.
216-217:150–158. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Halvorsen AR, Bjaanaes M, LeBlanc M, Holm
AM, Bolstad N, Rubio L, Peñalver JC, Cervera J, Mojarrieta JC,
López-Guerrero JA, et al: A unique set of 6 circulating microRNAs
for early detection of non-small cell lung cancer. Oncotarget.
7:37250–37259. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ma J, Mannoor K, Gao L, Tan A, Guarnera
MA, Zhan M, Shetty A, Stass SA, Xing L and Jiang F:
Characterization of microRNA transcriptome in lung cancer by
next-generation deep sequencing. Mol Oncol. 8:1208–1219. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sozzi G, Boeri M, Rossi M, Verri C,
Suatoni P, Bravi F, Roz L, Conte D, Grassi M, Sverzellati N, et al:
Clinical utility of a plasma-based miRNA signature classifier
within computed tomography lung cancer screening: A correlative
MILD trial study. J Clin Oncol. 32:768–773. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Molina-Vila MA: Liquid biopsy in lung
cancer: Present and future. Transl Lung Cancer Res. 5:452–454.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Han MK, Oh YH, Kang J, Kim YP, Seo S, Kim
J, Park K and Kim HS: Protein profiling in human sera for
identification of potential lung cancer biomarkers using antibody
microarray. Proteomics. 9:5544–5552. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nolen BM, Lomakin A, Marrangoni A,
Velikokhatnaya L, Prosser D and Lokshin AE: Urinary protein
biomarkers in the early detection of lung cancer. Cancer Prev Res
(Phila). 8:111–119. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lopez-Sanchez LM, Jurado-Gamez B,
Feu-Collado N, Valverde A, Canas A, Fernandez-Rueda JL, Aranda E
and Rodríguez-Ariza A: Exhaled breath condensate biomarkers for the
early diagnosis of lung cancer using proteomics. Am J Physiol Lung
Cell Mol Physiol. 313:L664–L676. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jung M, Kim SH, Lee YJ, Hong S, Kang YA,
Kim SK, Chang J, Rha SY, Kim JH, Kim DJ and Cho BC: Prognostic and
predictive value of CEA and CYFRA 21-1 levels in advanced non-small
cell lung cancer patients treated with gefitinib or erlotinib. Exp
Ther Med. 2:685–693. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yanagita K, Nagashio R, Jiang SX, Kuchitsu
Y, Hachimura K, Ichinoe M, Igawa S, Fukuda E, Goshima N, Satoh Y,
et al: Cytoskeleton-Associated protein 4 is a novel serodiagnostic
marker for lung cancer. Am J Pathol. 188:1328–1333. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sun Y, Liu S, Qiao Z, Shang Z, Xia Z, Niu
X, Qian L, Zhang Y, Fan L, Cao CX and Xiao H: Systematic comparison
of exosomal proteomes from human saliva and serum for the detection
of lung cancer. Anal Chim Acta. 982:84–95. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Melo SA, Luecke LB, Kahlert C, Fernandez
AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari
N, et al: Glypican-1 identifies cancer exosomes and detects early
pancreatic cancer. Nature. 523:177–182. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen IH, Xue L, Hsu CC, Paez JS, Pan L,
Andaluz H, Wendt MK, Iliuk AB, Zhu JK and Tao WA: Phosphoproteins
in extracellular vesicles as candidate markers for breast cancer.
Proc Natl Acad Sci USA. 114:3175–3180. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jakobsen KR, Paulsen BS, Baek R, Varming
K, Sorensen BS and Jorgensen MM: Exosomal proteins as potential
diagnostic markers in advanced non-small cell lung carcinoma. J
Extracell Vesicles. 4:266592015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kodadek T: Protein microarrays: Prospects
and problems. Chem Biol. 8:105–115. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Joosse SA, Gorges TM and Pantel K:
Biology, detection, and clinical implications of circulating tumor
cells. EMBO Mol Med. 7:1–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hamilton G and Rath B: Detection of
circulating tumor cells in non-small cell lung cancer. J Thorac
Dis. 8:1024–1028. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tanaka F, Yoneda K, Kondo N, Hashimoto M,
Takuwa T, Matsumoto S, Okumura Y, Rahman S, Tsubota N, Tsujimura T,
et al: Circulating tumor cell as a diagnostic marker in primary
lung cancer. Clin Cancer Res. 15:6980–6986. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sonn CH, Cho JH, Kim JW, Kang MS, Lee J
and Kim J: Detection of circulating tumor cells in patients with
non-small cell lung cancer using a size-based platform. Oncol Lett.
13:2717–2722. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Truini A, Alama A, Dal Bello MG, Coco S,
Vanni I, Rijavec E, Genova C, Barletta G, Biello F and Grossi F:
Clinical applications of circulating tumor cells in lung cancer
patients by cell search system. Front Oncol. 4:2422014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ambrosone CB: Oxidants and antioxidants in
breast cancer. Antioxid Redox Signal. 2:903–917. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hakim M, Broza YY, Barash O, Peled N,
Phillips M, Amann A and Haick H: Volatile organic compounds of lung
cancer and possible biochemical pathways. Chem Rev. 112:5949–5966.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Filipiak W, Sponring A, Filipiak A, Ager
C, Schubert J, Miekisch W, Amann A and Troppmair J: TD-GC-MS
analysis of volatile metabolites of human lung cancer and normal
cells in vitro. Cancer Epidemiol Biomarkers Prev. 19:182–195. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Buszewski B, Ligor T, Jezierski T,
Wenda-Piesik A, Walczak M and Rudnicka J: Identification of
volatile lung cancer markers by gas chromatography-mass
spectrometry: Comparison with discrimination by canines. Anal
Bioanal Chem. 404:141–146. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Rudnicka J, Walczak M, Kowalkowski T,
Jezierski T and Buszewski B: Determination of volatile organic
compounds as potential markers of lung cancer by gas
chromatography-mass spectrometry versus trained dogs. Sensors
Actuators B Chem. 202:615–621. 2014. View Article : Google Scholar
|
|
61
|
Phillips M, Altorki N, Austin JH, Cameron
RB, Cataneo RN, Kloss R, Maxfield RA, Munawar MI, Pass HI, Rashid
A, et al: Detection of lung cancer using weighted digital analysis
of breath biomarkers. Clin Chim Acta. 393:76–84. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Phillips M, Altorki N, Austin JH, Cameron
RB, Cataneo RN, Greenberg J, Kloss R, Maxfield RA, Munawar MI, Pass
HI, et al: Prediction of lung cancer using volatile biomarkers in
breath. Cancer Biomark. 3:95–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wang Y, Hu Y, Wang D, Yu K, Wang L, Zou Y,
Zhao C, Zhang X, Wang P and Ying K: The analysis of volatile
organic compounds biomarkers for lung cancer in exhaled breath,
tissues and cell lines. Cancer Biomark. 11:129–137. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Brunner C, Szymczak W, Hollriegl V, Mortl
S, Oelmez H, Bergner A, Huber RM, Hoeschen C and Oeh U:
Discrimination of cancerous and non-cancerous cell lines by
headspace-analysis with PTR-MS. Anal Bioanal Chem. 397:2315–2324.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Brůhová Michalčíková R, Dryahina K and
Španěl P: SIFT-MS quantification of several breath biomarkers of
inflammatory bowel disease, IBD: A detailed study of the ion
chemistry. Int J Mass Spectrom. 396:35–41. 2016. View Article : Google Scholar
|
|
66
|
Li Z, Xu C, Shu J, Yang B and Zou Y:
Doping-assisted low-pressure photoionization mass spectrometry for
the real-time detection of lung cancer-related volatile organic
compounds. Talanta. 165:98–106. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Behera B, Joshi R, Anil Vishnu GK,
Bhalerao S and Pandya HJ: Electronic-nose: A non-invasive
technology for breath analysis of diabetes and lung cancer
patients. J Breath Res. 13:0240012019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Peng G, Tisch U, Adams O, Hakim M, Shehada
N, Broza YY, Billan S, Abdah-Bortnyak R, Kuten A and Haick H:
Diagnosing lung cancer in exhaled breath using gold nanoparticles.
Nat Nanotechnol. 4:669–673. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Kort S, Brusse-Keizer M, Schouwink JH,
Gerritsen JW and Van dPJ: Detection of non-small cell lung cancer
by an electronic nose. Eur Respir J. 50 (Suppl 61):PA20322017.
|
|
70
|
Gasparri R, Santonico M, Valentini C,
Sedda G, Borri A, Petrella F, Maisonneuve P, Pennazza G, D'Amico A,
Di Natale C, et al: Volatile signature for the early diagnosis of
lung cancer. J Breath Res. 10:0160072016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Shehada N, Cancilla JC, Torrecilla JS,
Pariente ES, Bronstrup G, Christiansen S, Johnson DW, Leja M,
Davies MP, Liran O, et al: Silicon nanowire sensors enable
diagnosis of patients via exhaled breath. ACS Nano. 10:7047–7057.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Handa H, Usuba A, Maddula S, Baumbach JI,
Mineshita M and Miyazawa T: Exhaled breath analysis for lung cancer
detection using ion mobility spectrometry. PLoS One. 9:e1145552014.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhong X, Li D, Du W, Yan M, Wang Y, Huo D
and Hou C: Rapid recognition of volatile organic compounds with
colorimetric sensor arrays for lung cancer screening. Anal Bioanal
Chem. 410:3671–3681. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Queralto N, Berliner AN, Goldsmith B,
Martino R, Rhodes P and Lim SH: Detecting cancer by breath volatile
organic compound analysis: A review of array-based sensors. J
Breath Res. 8:0271122014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Aravanis AM, Lee M and Klausner RD:
Next-Generation sequencing of circulating tumor DNA for early
cancer detection. Cell. 168:571–574. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wang J, Han X and Sun Y: DNA methylation
signatures in circulating cell-free DNA as biomarkers for the early
detection of cancer. Sci China Life Sci. 60:356–362. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Iqbal MA, Arora S, Prakasam G, Calin GA
and Syed MA: MicroRNA in lung cancer: Role, mechanisms, pathways
and therapeutic relevance. Mol Aspects Med. 70:3–20. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Keller A, Leidinger P, Gislefoss R, Haugen
A, Langseth H, Staehler P, Lenhof HP and Meese E: Stable serum
miRNA profiles as potential tool for non-invasive lung cancer
diagnosis. RNA Biol. 8:506–516. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Li A, Zhang T, Zheng M, Liu Y and Chen Z:
Exosomal proteins as potential markers of tumor diagnosis. J
Hematol Oncol. 10:1752017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Harouaka R, Kang Z, Zheng SY and Cao L:
Circulating tumor cells: Advances in isolation and analysis, and
challenges for clinical applications. Pharmacol Ther. 141:209–221.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Cabel L, Proudhon C, Gortais H, Loirat D,
Coussy F, Pierga JY and Bidard FC: Circulating tumor cells:
Clinical validity and utility. Int J Clin Oncol. 22:421–430. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Hofman P: Liquid biopsy and therapeutic
targets: Present and future issues in thoracic oncology. Cancers
(Basel). 9:E1542017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Tsui DW and Berger MF: Profiling non-small
cell lung cancer: From tumor to blood. Clin Cancer Res. 22:790–792.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Sundaresan TK, Sequist LV, Heymach JV,
Riely GJ, Janne PA, Koch WH, Sullivan JP, Fox DB, Maher R,
Muzikansky A, et al: Detection of T790M, the acquired resistance
EGFR mutation, by tumor biopsy versus noninvasive blood-based
analyses. Clin Cancer Res. 22:1103–1110. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Midha A, Dearden S and McCormack R: EGFR
mutation incidence in non-small-cell lung cancer of adenocarcinoma
histology: A systematic review and global map by ethnicity
(mutMapII). Am J Cancer Res. 5:2892–2911. 2015.PubMed/NCBI
|
|
86
|
Liang W, Wu X, Fang W, Zhao Y, Yang Y, Hu
Z, Xue C, Zhang J, Zhang J, Ma Y, et al: Network meta-analysis of
erlotinib, gefitinib, afatinib and icotinib in patients with
advanced non-small-cell lung cancer harboring EGFR mutations. PLoS
One. 9:e852452014. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Brandao EP, Pantarotto MG and Cruz M: A
novel EGFR mutation in exon 18 with high sensitivity to EGFR TKI
treatment with reduced dose. J Thorac Oncol. 7:e322012. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Kobayashi Y, Togashi Y, Yatabe Y, Mizuuchi
H, Jangchul P, Kondo C, Shimoji M, Sato K, Suda K, Tomizawa K, et
al: EGFR exon 18 mutations in lung cancer: Molecular predictors of
augmented sensitivity to afatinib or neratinib as compared with
first- or third-generation TKIs. Clin Cancer Res. 21:5305–5313.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Wang J, Wang B, Chu H and Yao Y: Intrinsic
resistance to EGFR tyrosine kinase inhibitors in advanced
non-small-cell lung cancer with activating EGFR mutations. Onco
Targets Ther. 9:3711–3726. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Lindeman NI, Cagle PT, Beasley MB, Chitale
DA, Dacic S, Giaccone G, Jenkins RB, Kwiatkowski DJ, Saldivar JS,
Squire J, et al: Molecular testing guideline for selection of lung
cancer patients for EGFR and ALK tyrosine kinase inhibitors:
Guideline from the college of American pathologists, international
association for the study of lung cancer, and association for
molecular pathology. J Thorac Oncol. 8:823–859. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Tan CS, Kumarakulasinghe NB, Huang YQ, Ang
YLE, Choo JR, Goh BC and Soo RA: Third generation EGFR TKIs:
Current data and future directions. Mol Cancer. 17:292018.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Karachaliou N, Molina-Vila MA and Rosell
R: The impact of rare EGFR mutations on the treatment response of
patients with non-small cell lung cancer. Expert Rev Respir Med.
9:241–244. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Noh KW, Lee MS, Lee SE, Song JY, Shin HT,
Kim YJ, Oh DY, Jung K, Sung M, Kim M, et al: Molecular breakdown: A
comprehensive view of anaplastic lymphoma kinase (ALK)-rearranged
non-small cell lung cancer. J Pathol. 243:307–319. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Vendrell JA, Taviaux S, Beganton B,
Godreuil S, Audran P, Grand D, Clermont E, Serre I, Szablewski V,
Coopman P, et al: Detection of known and novel ALK fusion
transcripts in lung cancer patients using next-generation
sequencing approaches. Sci Rep. 7:125102017. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Hofman P: ALK status assessment with
liquid biopsies of lung cancer patients. Cancers (Basel).
9:E1062017. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Pailler E, Oulhen M, Borget I, Remon J,
Ross K, Auger N, Billiot F, Ngo Camus M, Commo F, Lindsay CR, et
al: Circulating tumor cells with aberrant ALK copy number predict
progression-free survival during crizotinib treatment in
ALK-rearranged non-small cell lung cancer patients. Cancer Res.
77:2222–2230. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Passaro A, Lazzari C, Karachaliou N,
Spitaleri G, Pochesci A, Catania C, Rosell R and de Marinis F:
Personalized treatment in advanced ALK-positive non-small cell lung
cancer: From bench to clinical practice. Onco Targets Ther.
9:6361–6376. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Peters S, Camidge DR, Shaw AT, Gadgeel S,
Ahn JS, Kim DW, Ou SI, Pérol M, Dziadziuszko R, Rosell R, et al:
Alectinib versus crizotinib in untreated ALK-Positive
non-small-cell lung cancer. N Engl J Med. 377:829–838. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Shen L and Ji HF: Ceritinib in
ALK-rearranged non-small-cell lung cancer. N Engl J Med.
370:25372014. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Waqar SN and Morgensztern D: Lorlatinib: A
new-generation drug for ALK-positive NSCLC. Lancet Oncol.
19:1555–1557. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Kim RN, Choi YL, Lee MS, Lira ME, Mao M,
Mann D, Stahl J, Licon A, Choi SJ, Van Vrancken M, et al:
SEC31A-ALK fusion gene in lung adenocarcinoma. Cancer Res Treat.
48:398–402. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Barlesi F, Mazieres J, Merlio JP,
Debieuvre D, Mosser J, Lena H, Ouafik L, Besse B, Rouquette I,
Westeel V, et al: Routine molecular profiling of patients with
advanced non-small-cell lung cancer: Results of a 1-year nationwide
programme of the French cooperative thoracic intergroup (IFCT).
Lancet. 387:1415–1426. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Sanchez-Torres JM, Viteri S, Molina MA and
Rosell R: BRAF mutant non-small cell lung cancer and treatment with
BRAF inhibitors. Transl Lung Cancer Res. 2:244–250. 2013.PubMed/NCBI
|
|
104
|
Planchard D, Groen HJM, Kim TM, Rigas JR,
Souquet PJ, Baik CS, Bariesi F, Mazières J, Quoix EA, Curtis CM, et
al: Interim results of a phase II study of the BRAF inhibitor
(BRAFi) dabrafenib (D) in combination with the MEK inhibitor
trametinib (T) in patients (pts) with BRAF V600E mutated (mut)
metastatic non-small cell lung cancer (NSCLC). J Clin Oncol. 33 (15
Suppl):S80062015. View Article : Google Scholar
|
|
105
|
Planchard D, Besse B, Groen HJM, Souquet
PJ, Quoix E, Baik CS, Barlesi F, Kim TM, Mazieres J, Novello S, et
al: Dabrafenib plus trametinib in patients with previously treated
BRAF(V600E)-mutant metastatic non-small cell lung cancer: An
open-label, multicentre phase 2 trial. Lancet Oncol. 17:984–993.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Chuang JC, Stehr H, Liang Y, Das M, Huang
J, Diehn M, Wakelee HA and Neal JW: ERBB2-Mutated metastatic
non-small cell lung cancer: Response and resistance to targeted
therapies. J Thorac Oncol. 12:833–842. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Salgia R: MET in lung cancer: Biomarker
selection based on scientific rationale. Mol Cancer Ther.
16:555–565. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Gainor JF and Shaw AT: Novel targets in
non-small cell lung cancer: ROS1 and RET fusions. Oncologist.
18:865–875. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Mochalski P, King J, Haas M, Unterkofler
K, Amann A and Mayer G: Blood and breath profiles of volatile
organic compounds in patients with end-stage renal disease. BMC
Nephrol. 15:432014. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Terelius Y and Ingelman-Sundberg M:
Metabolism of n-pentane by ethanol-inducible cytochrome P-450 in
liver microsomes and reconstituted membranes. Eur J Biochem.
161:303–308. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Kohlmuller D and Kochen W: Is n-pentane
really an index of lipid peroxidation in humans and animals? A
methodological reevaluation. Anal Biochem. 210:268–276. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Risby TH and Sehnert SS: Clinical
application of breath biomarkers of oxidative stress status. Free
Radic Biol Med. 27:1182–1192. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Marchitti SA, Brocker C, Stagos D and
Vasiliou V: Non-P450 aldehyde oxidizing enzymes: The aldehyde
dehydrogenase superfamily. Expert Opin Drug Metab Toxicol.
4:697–720. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Rahman I, van Schadewijk AA, Crowther AJ,
Hiemstra PS, Stolk J, MacNee W and De Boer WI: 4-Hydroxy-2-nonenal,
a specific lipid peroxidation product, is elevated in lungs of
patients with chronic obstructive pulmonary disease. Am J Respir
Crit Care Med. 166:490–495. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Vaz AD and Coon MJ: Hydrocarbon formation
in the reductive cleavage of hydroperoxides by cytochrome P-450.
Proc Natl Acad Sci USA. 84:1172–1176. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Branton PJ, McAdam KG, Winter DB, Liu C,
Duke MG and Proctor CJ: Reduction of aldehydes and hydrogen cyanide
yields in mainstream cigarette smoke using an amine functionalised
ion exchange resin. Chem Cent J. 5:152011. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Kang JO, Slater G, Aufses AH Jr and Cohen
G: Production of ethane by rats treated with the colon carcinogen,
1, 2-dimethylhydrazine. Biochem Pharmacol. 37:2967–2971. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Burdock GA: Fenaroli's handbook of flavor
ingredients. CRC Press; 2016, View Article : Google Scholar
|
|
119
|
Smith D, Wang T and Spanel P: On-line,
simultaneous quantification of ethanol, some metabolites and water
vapour in breath following the ingestion of alcohol. Physiol Meas.
23:477–489. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Xu ZQ, Broza YY, Ionsecu R, Tisch U, Ding
L, Liu H, Song Q, Pan YY, Xiong FX, Gu KS, et al: A
nanomaterial-based breath test for distinguishing gastric cancer
from benign gastric conditions. Br J Cancer. 108:941–950. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Eckel RH: Lipoprotein lipase A
multifunctional enzyme relevant to common metabolic diseases. New
Engl J Med. 320:1060–1068. 1989.PubMed/NCBI
|
|
122
|
Jia Z, Zhang H, Ong CN, Patra A, Lu Y, Lim
CT and Venkatesan T: Detection of lung cancer: Concomitant volatile
organic compounds and metabolomic profiling of six cancer cell
lines of different histological origins. ACS Omega. 3:5131–5140.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Peled N, Barash O, Tisch U, Ionescu R,
Broza YY, Ilouze M, Mattei J, Bunn PA Jr, Hirsch FR and Haick H:
Volatile fingerprints of cancer specific genetic mutations.
Nanomedicine. 9:758–766. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Yang B, Zhang H, Shu J, Ma P, Zhang P,
Huang J, Li Z and Xu C: Vacuum-ultraviolet-excited and
CH2Cl2/H2O-amplified
ionization-coupled mass spectrometry for oxygenated organics
analysis. Anal Chem. 90:1301–1308. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Huang J, Yang B, Shu J, Zhang Z, Li Z and
Jiang K: Kinetic understanding of the ultrahigh ionization
efficiencies (up to 28%) of excited-state
CH2Cl2-induced associative ionization: A case
study with nitro compounds. Anal Chem. 91:5605–5612. 2019.
View Article : Google Scholar : PubMed/NCBI
|