Introduction
Prostate carcinoma is the second most common
malignancy in males and accounts for ~15% of all cancer cases in
males (1). Due to its high
prevalence and aggressive nature, prostate carcinoma is considered
as one of the leading causes of cancer-related deaths, particularly
in developed counties (2). The
majority of patients with prostate carcinoma are diagnosed at
advanced stages due to the low specificity of prostate specific
antigen (PSA) testing and the lack of identifiable symptoms at the
early stages (3,4). Therefore, the early and accurate
diagnosis of the disease may improve the survival time of patients.
However, this remains a challenge due to the complex mechanisms
underlying prostate carcinoma (5).
The identification of oncogenes and tumor
suppressors indicated that genetic factors play important roles in
the pathogenesis of prostate carcinoma (6). However, the limited number of oncogenes
and tumor suppressors involved in prostate carcinoma may not be
able to explain the complexity of the molecular pathways associated
with the disease. In recent years, long non-coding RNAs (lncRNAs)
have been identified as critical determinants in cancer biology due
to their functions in regulating cancer cell behaviors (7,8).
Therefore, studies on the involvement of lncRNAs in cancer biology
are required to improve the understanding of cancer development and
progression. Erbb4-IR is a recently identified lncRNA with
important functions in diabetic kidney injury (9). The deep sequencing data obtained in the
present study showed that Erbb4-IR was downregulated in prostate
carcinoma and was inversely associated with microRNA
(miR/miRNA)-21, a known oncogenic miRNA in prostate carcinoma
(10). Therefore, the present study
investigated the interactions between Erbb4-IR and miR-21 in
prostate carcinoma.
Materials and methods
Patient admission and follow-up
A total of 60 male patients (range, 42–67 years;
mean ± standard deviation, 55.2±6.7 years) with prostate carcinoma
were enrolled at The 940 Hospital of the Joint Logistics Support
Force of the Chinese People's Liberation Army between January 2010
and February 2013. The inclusion criteria were as follows: i)
Histopathologically-confirmed first diagnosis assessed by at least
3 experienced pathologists; and ii) no therapies received prior to
admission. The exclusion criteria were as follows: i)
Co-morbidities, including chronic diseases; and ii) treatment
received up to 3 months prior to admission. There were 12, 13, 20
and 15 cases at American Joint Committee on Cancer stage I, II, III
and IV (11), respectively. The
present study was approved by The 940 Hospital of the Joint
Logistics Support Force of the Chinese People's Liberation Army
Ethics Committee and written informed consent was obtained from all
patients prior to the study start. All patients were followed up
for 60 months after admission through outpatient visits or phone
calls. Patients lost to follow-up or who succumbed as a result of
other clinical conditions or accidents were excluded. A total of 54
patients completed the follow-up period.
Specimens and cell lines
Patients were subjected to prostate biopsies to
obtain prostate carcinoma and adjacent non-cancerous (within 2 cm
of the tumor margin) tissues prior to receiving treatment.
The 22Rv1 and DU145 prostate carcinoma cell lines
(American Type Culture Collection) were investigated in the present
study. The cells were cultured in Eagle's minimum essential medium
(Sigma-Aldrich; Merck KGaA) supplemented with 10% FBS
(Sigma-Aldrich; Merck KGaA) at 37°C in 5% CO2.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from tissue specimens and
22Rv1 and DU145 cells using TRIzol reagent (Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. RNAs
were precipitated using 80% ethanol to retain the miRNAs. Reverse
transcription was performed using AMV reverse transcriptase kit
(Sangon Biotech Co., Ltd.), under the following conditions: 25°C
for 10 min, 55°C for 20 min and 80°C for 10 min. qPCR was
subsequently performed using the QuantiTect SYBR Green PCR kit
(Qiagen, Inc.). Erbb4-IR expression was normalized to the
expression of endogenous 18S rRNA. miRNA reverse transcription was
performed using the TaqMan MicroRNA Reverse Transcription kit
(Thermo Fisher Scientific, Inc.). qPCR was performed using
MystiCq® microRNA® SYBR®Green qPCR
ReadyMix™ (Sigma-Aldrich; Merck KGaA). miR-21 expression was
normalized to the expression of endogenous U6. The following primer
sequences were used for PCR: Erbb4-IR forward,
5′-ACTCGCCACAGAAATCCAC-3′ and reverse, 5′-ACAACCCCAAACAAGCTGT-3′;
18S rRNA forward, 5′-CTACCACATCCAAGGAAGC-3′ and reverse,
5′-TTTTCGTCACTACCTCCCCG-3′; miR-21 forward,
5′-TAGCTTATCAGACTGATGTT-3′. The miR-21 reverse primer and the
forward and reverse U6 primers were from the MystiCq®
microRNA® SYBR® Green qPCR ReadyMix™ kit
(Sigma-Aldrich; Merck KGaA). The following thermocycling conditions
were used for PCR: Initial denaturation at 95°C for 1 min; 40
cycles of 95°C for 10 sec and 60°C for 45 sec. RT-qPCR was
performed in triplicate and expression levels were quantified using
the 2−ΔΔCq method (12).
Library preparation and deep
sequencing
The biopsies of 10 patients (range, 43–65 years;
mean ± standard deviation, 54.3±6.8 years) were subjected to deep
sequencing analysis. Following RNA extraction and reverse
transcription as aforementioned, the NEBNext® Ultra™
Directional RNA Library Prep kit for Illumina® (cat. no.
7420S; New England Biolabs) was used to prepare libraries and
High-seq (Illumina, Inc.) was used to sequence the libraries. Data
were analyzed using a Python-based pipeline (version 3.8,
http://www.python.org/downloads/release/python-380).
Transient transfection
Lipofectamine® 2000 reagent (Thermo
Fisher Scientific, Inc.) was used to perform all transient
transfections, according to the manufacturer's protocol. A miR-21
mimic and negative control miRNA were purchased from Sigma-Aldrich
(Merck KGaA). A pcDNA3.1 vector delivering Erbb4-IR was constructed
by Sangon Biotech Co., Ltd. Two control groups, including a
negative control (NC; empty vector or NC miRNA transfection) or
control (C; no transfection) were included. A total of 10 and 40 nM
of the vectors and miRNAs, respectively, were transfected into the
cells. Subsequent experiments were performed 24 h after
transfection.
Cell Counting Kit-8 (CCK-8) assay
22Rv1 and DU145 cells were harvested 24 h
post-transfection and 4×105 cells were suspended in 10
ml Eagle's minimum essential medium supplemented with 10% FBS to
create a cell suspension with a cell density of 4×104
cells/ml. The cell suspension was subsequently added to a 96-well
plate (100 µl/well). Cells were cultured at 37°C and 5%
CO2 and 10 µl CCK-8 solution (Sigma-Aldrich; Merck KGaA)
was added into each well every 24 h for a total of 96 h. The cells
were cultured for an additional 4 h, and 10 µl DMSO were added to
each well. The optical density values were measured at a wavelength
of 450 nm.
Cell apoptosis assay
22Rv1 and DU145 cells were harvested 24 h
post-transfection and 1×106 cells were washed with PBS
and mixed with 500 µl binding buffer, prior to incubation with FITC
labeled Annexin-V (5 µl) and PI solution (5 µl) for 10 min in the
dark. These steps were performed using the FITC Annexin V Apoptosis
Detection kit with PI (BioLegend, Inc.). Early apoptotic cells were
detected using a flow cytometer.
Statistical analysis
GraphPad Prism (version 6.0; GraphPad Software,
Inc.) was used for data analysis. Differences between prostate
carcinoma and adjacent non-cancerous tissues were analyzed using a
paired Student's t-test. Differences among multiple cell treatment
groups were analyzed using a one-way ANOVA followed by a Tukey post
hoc test. Linear regression was used for association analysis. The
54 patients who completed follow-up were divided into high (n=25)
and low (n=29) Erbb4-IR expression groups, based on Youden's index.
Based on follow-up data, survival curves were plotted and compared
using the Kaplan-Meier method and the χ2 test. All
experiments were performed in triplicate. P<0.05 was considered
to indicate a statistically significant difference.
Results
Erbb4-IR is downregulated in prostate
carcinoma
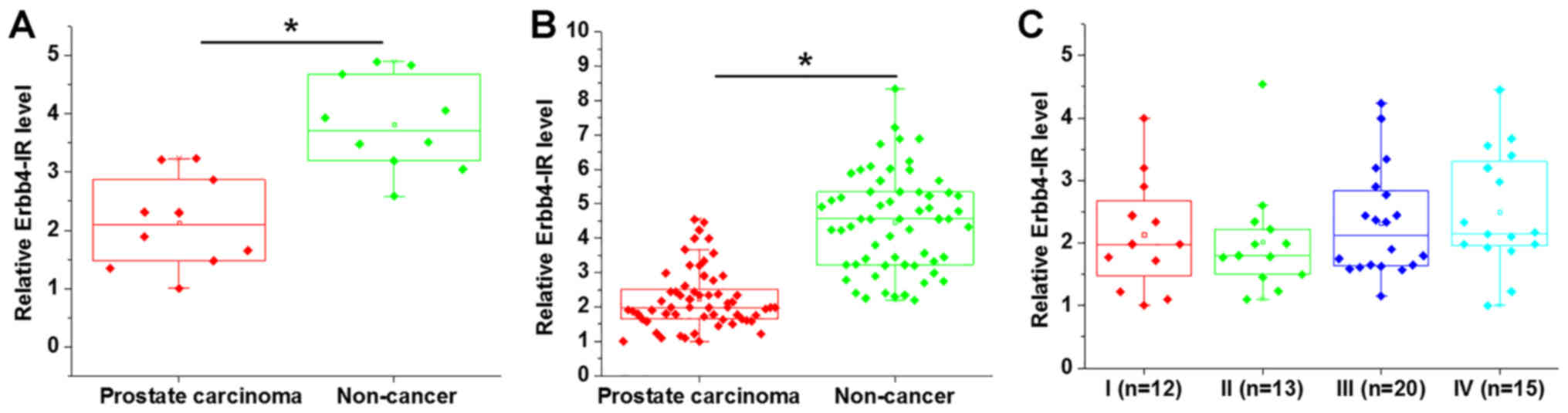
Deep sequencing analysis showed that the expression
levels of Erbb4-IR were significantly lower in prostate tissues
compared with adjacent non-cancerous tissues (Fig. 1A; P<0.05). Erbb4-IR expression in
prostate carcinoma and adjacent non-cancerous tissues was
subsequently detected by RT-qPCR. The data were analyzed using the
paired Student's t-test, and the results showed that Erbb4-IR was
significantly upregulated in prostate carcinoma tissues compared
with adjacent non-cancerous tissues (Fig. 1B; P<0.05). In addition, no
significant differences in the expression levels of Erbb4-IR were
observed in prostate carcinoma tissues obtained from patients with
different clinical stages (Fig. 1C;
P>0.05).
Low expression of Erbb4-IR in prostate
carcinoma tissues is closely associated with poor survival
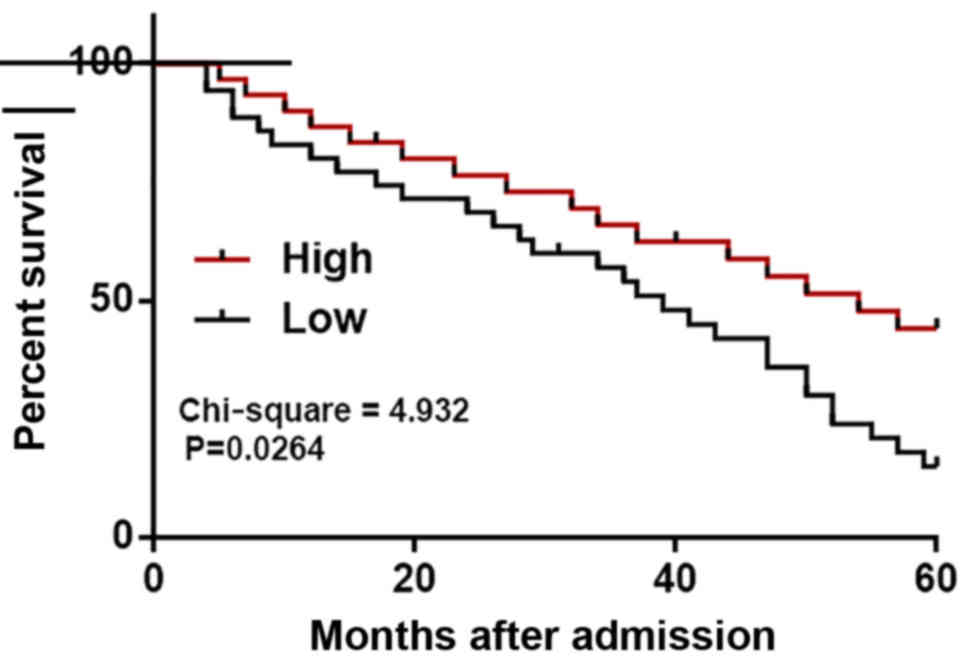
The 54 patients who completed the follow-up period
were divided into high (n=25) and low (n=29) Erbb4-IR expression
groups based on Youden's index. Based on follow-up data, the
survival curves revealed that patients with a low expression level
of Erbb4-IR had a significantly lower overall survival rate
compared with patients with a high expression level (Fig. 2).
miR-21 is upregulated in prostate
carcinoma tissues and is inversely associated with Erbb4-IR
expression
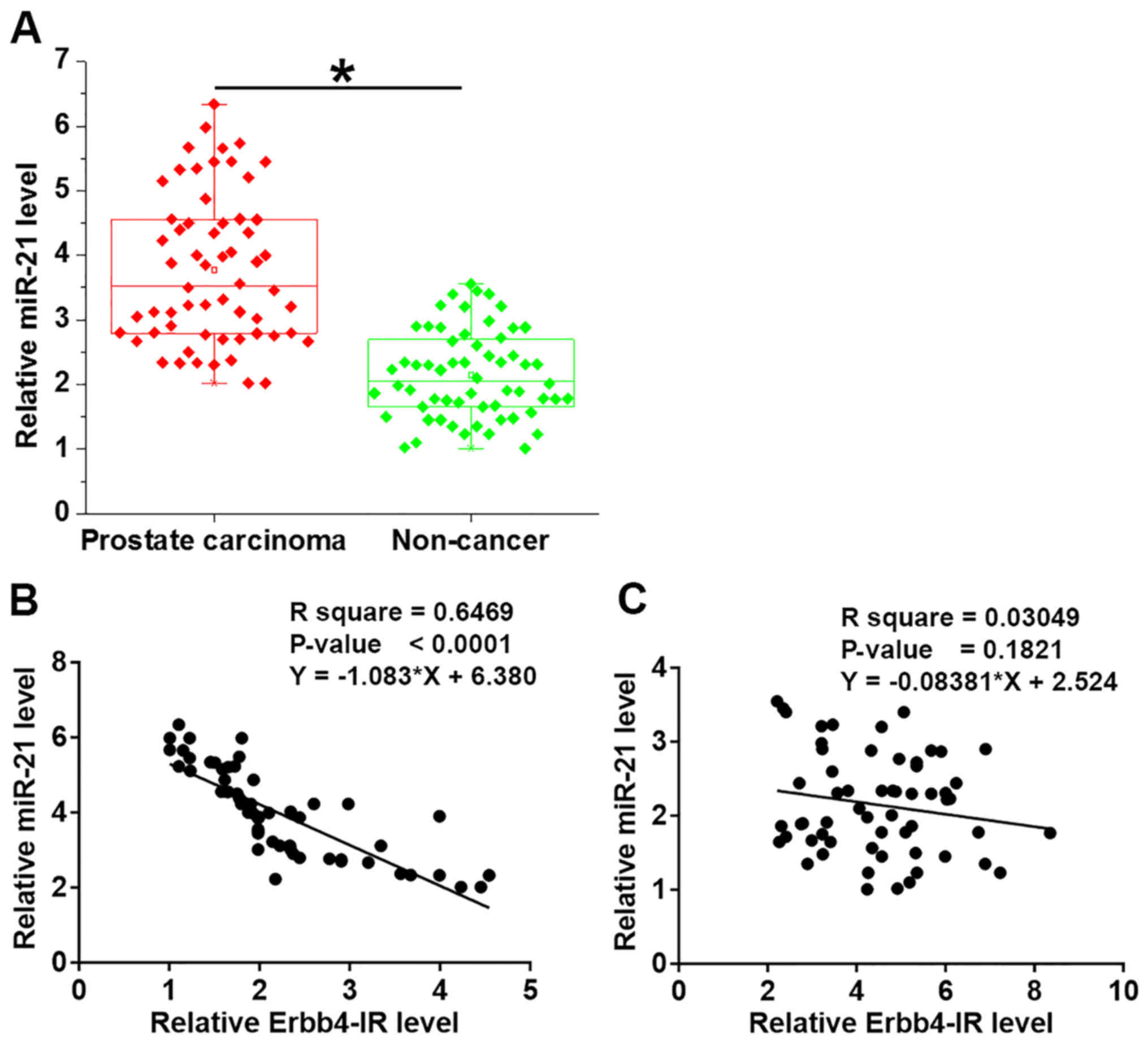
miR-21 expression in prostate carcinoma and adjacent
non-cancerous tissues was detected by RT-qPCR. The results revealed
that miR-21 was significantly downregulated in prostate carcinoma
tissues compared with the adjacent non-cancerous tissues (Fig. 3A; P<0.05). Linear regression
analysis was performed to explore the association between miR-21
and Erbb4-IR and revealed that miR-21 and Erbb4-IR were
significantly and inversely associated in prostate carcinoma
tissues (Fig. 3B), but not in
adjacent non-cancerous tissues (Fig.
3C).
Erbb4-IR overexpression downregulates
miR-21 expression
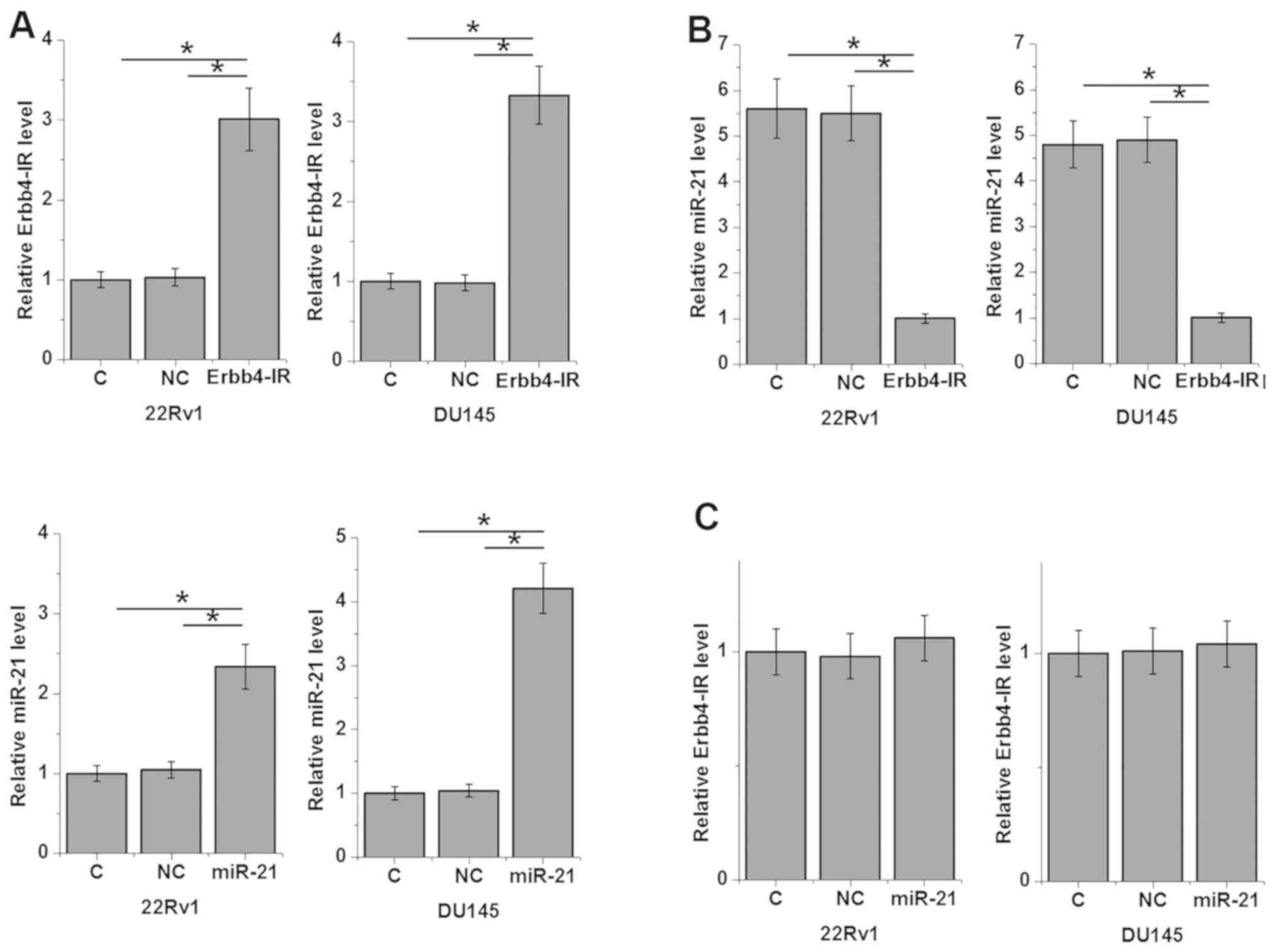
An Erbb4-IR-overexpression vector and miR-21 mimic
were transfected into 22Rv1 and DU145 cells. Compared with the two
control groups (C and NC), Erbb4-IR and miR-21 were significantly
upregulated in the overexpression group (Fig. 4A; P<0.05). Furthermore, Erbb4-IR
overexpression resulted in the downregulation of miR-21 (Fig. 4B; P<0.05), while miR-21
overexpression did not significantly affect Erbb4-IR expression
(Fig. 4C).
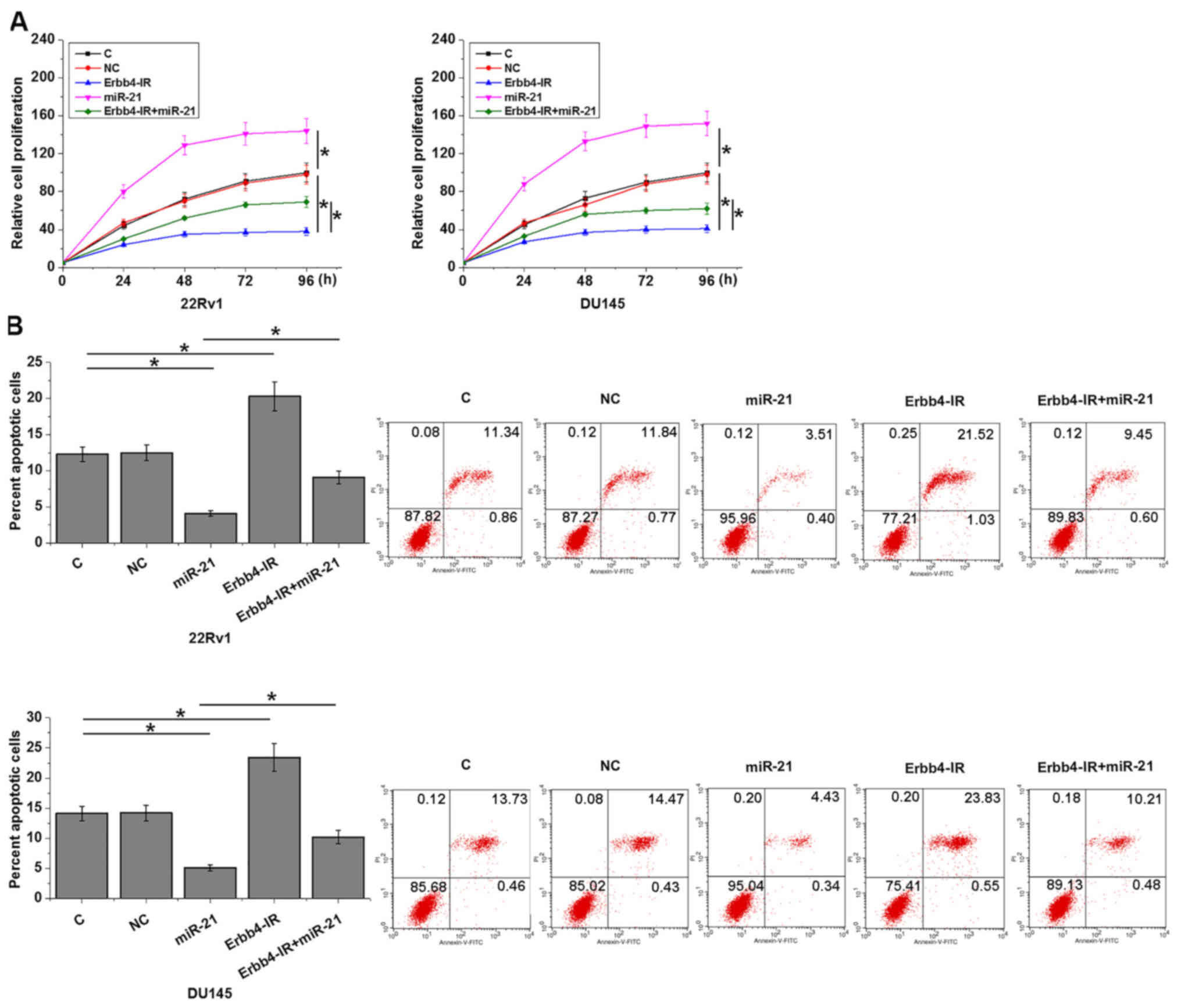
Erbb4-IR regulates cancer cell
proliferation and apoptosis through miR-21
Compared with the two control groups (C and NC),
Erbb4-IR overexpression promoted apoptosis (Fig. 5A) and inhibited the proliferation
(Fig. 5B) of prostate carcinoma
cells (P<0.05). miR-21 overexpression resulted in an opposite
effect and attenuated the effects of Erbb4-IR overexpression
(P<0.05).
Discussion
The function of lncRNA Erbb4-IR has only been
characterized in diabetic kidney injury (9), and its involvement in cancer biology is
unknown. To the best of our knowledge, the present study was the
first to show that Erbb4-IR was downregulated in prostate carcinoma
tissues. Furthermore, in vitro results revealed that the
overexpression of Erbb4-IR downregulated miR-21, a miRNA that has
been previously shown to promote prostate carcinoma (10).
Although the exact mechanism of Erbb4-IR in cancer
biology has not been elucidated, Erbb4-IR interacts with TGF-β
signaling (13), which has a
critical role in the development of most, if not all, types of
cancer (14). Therefore, Erbb4-IR
may be involved in carcinogenesis. The preliminary deep sequencing
data obtained in the present study revealed that Erbb4-IR was
downregulated in prostate carcinoma tissues compared with adjacent
non-cancerous tissues. Furthermore, low expression of Erbb4-IR in
prostate carcinoma tissues was significantly associated with poor
survival rate. In addition, the overexpression of Erbb4-IR promoted
apoptosis and inhibited the proliferation of prostate carcinoma
cells in vitro. Therefore, Erbb4-IR is likely to serve as a
tumor suppressor in prostate carcinoma, and the regulation of
Erbb4-IR expression may be a useful therapeutic strategy to
decrease cancer cell proliferation and increase apoptosis.
miR-21 is an oncogenic miRNA in several types of
cancer (13–15). Overexpression of miR-21 in cancer
cells not only regulates cell behavior, such as cell growth,
migration and invasion, but also regulates the sensitivity of
cancer cells to chemotherapy (15–17). The
present study revealed that the overexpression of miR-21 in
prostate carcinoma cells decreased apoptosis and increased
proliferation. A previous study revealed that miR-21 participates
in cancer biology primarily by downregulating the downstream tumor
suppression pathway associated with reversion inducing cysteine
rich protein with kazal motifs (18). However, the upstream regulators of
miR-21 remain largely unknown. The present study revealed that
Erbb4-IR was likely an upstream inhibitor of miR-21, however, the
underlying molecular mechanism was not investigated. It is known
that miR-21 may interact with TGF-β (19), which also interacts with Erbb4-IR
(11). Therefore, TGF-β may mediate
the interaction between Erbb4-IR and miR-21.
Future studies investigating additional more cell
lines are required to validate the results obtained in the present
study. In addition, in vivo studies will allow more in-depth
investigations.
In conclusion, the present study revealed that
Erbb4-IR was upregulated in prostate carcinoma and that
overexpression of Erbb4-IR may downregulate miR-21 to inhibit
prostate carcinoma.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JZ, QS and HQ conceived and designed the study. JZ,
QS, XL, HY, YW, LZ and SP analyzed and interpreted the data. JZ, XL
and HY drafted the manuscript. YW and HQ revised the manuscript.
All authors reviewed and approved the manuscript.
Ethics approval and consent to
participate
This study was approved by The 940 Hospital of the
Joint Logistics Support Force of the Chinese People's Liberation
Army Ethics Committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lippi G, Montagnana M, Guidi GC and
Plebani M: Prostate specific antigen-based screening for prostate
cancer in the third millennium: Useful or hype? Ann Med.
41:480–489. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Roddam AW, Duffy MJ, Hamdy FC, Ward AM,
Patnick J, Price CP, Rimmer J, Sturgeon C, White P and Allen NE;
NHS Prostate Cancer Risk Management Programme, : Use of
prostate-specific antigen (PSA) isoforms for the detection of
prostate cancer in men with a PSA level of 2–10 ng/ml: Systematic
review and meta-analysis. Eur Urol. 48:386–399. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shand RL and Gelmann EP: Molecular biology
of prostate-cancer pathogenesis. Curr Opin Urol. 16:123–131. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shen MM and Abate-Shen C: Molecular
genetics of prostate cancer: New prospects for old challenges.
Genes Dev. 24:1967–2000. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun SF, Tang PMK, Feng M, Xiao J, Huang
XR, Li P, Ma RCW and Lan HY: Novel lncRNA Erbb4-IR promotes
diabetic kidney injury in db/db mice by targeting miR-29b.
Diabetes. 67:731–744. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Folini M, Gandellini P, Longoni N, Profumo
V, Callari M, Pennati M, Colecchia M, Supino R, Veneroni S,
Salvioni R, et al: miR-21: An oncomir on strike in prostate cancer.
Mol Cancer. 9:122010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Egner JR: AJCC cancer staging manual.
JAMA. 304:1726–1727. 2010. View Article : Google Scholar
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Feng M, Tang PM, Huang XR, Sun SF, You YK,
Xiao J, Lv LL, Xu AP and Lan HY: TGF-β mediates renal fibrosis via
the Smad3-Erbb4-IR long noncoding RNA axis. Mol Ther. 26:148–161.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Colak S and Ten Dijke P: Targeting TGF-β
signaling in cancer. Trends Cancer. 3:56–71. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pfeffer SR, Yang CH and Pfeffer LM: The
role of miR-21 in cancer. Drug Dev Res. 76:270–277. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Si ML, Zhu S, Wu H, Lu Z, Wu F and Mo YY:
miR-21-mediated tumor growth. Oncogene. 26:2799–2803. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K
and Yang GH: MicroRNA-21 (miR-21) represses tumor suppressor PTEN
and promotes growth and invasion in non-small cell lung cancer
(NSCLC). Clin Chim Acta. 411:846–852. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Reis ST, Pontes-Junior J, Antunes AA,
Dall'Oglio MF, Dip N, Passerotti CC, Rossini GA, Morais DR,
Nesrallah AJ, Piantino C, et al: miR-21 may acts as an oncomir by
targeting RECK, a matrix metalloproteinase regulator, in prostate
cancer. BMC Urol. 12:142012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim YJ, Hwang SJ, Bae YC and Jung JS:
miR-21 regulates adipogenic differentiation through the modulation
of TGF-beta signaling in mesenchymal stem cells derived from human
adipose tissue. Stem Cells. 27:3093–3102. 2009.PubMed/NCBI
|