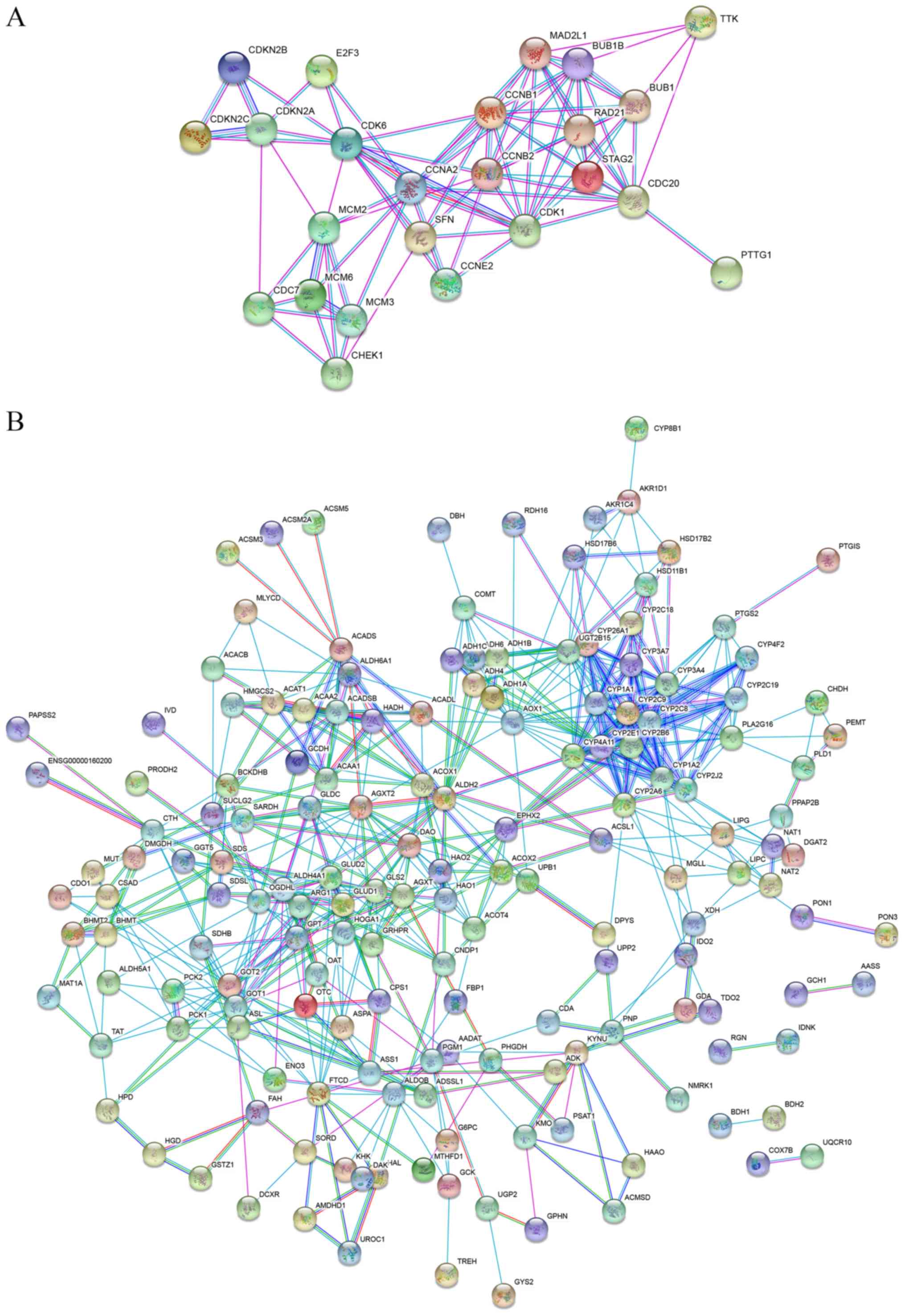
|
1
|
Wu CC, Wu DW, Lin YY, Lin PL and Lee H:
Hepatitis B virus X protein represses LKB1 expression to promote
tumor progression and poor postoperative outcome in hepatocellular
carcinoma. Surgery. 163:1040–1046. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Duan L, Wu R, Zhang X, Wang D, You Y,
Zhang Y, Zhou L and Chen W: HBx-induced S100A9 in NF-κB dependent
manner promotes growth and metastasis of hepatocellular carcinoma
cells. Cell Death Dis. 9:6292018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tsunematsu S, Suda G, Yamasaki K, Kimura
M, Izumi T, Umemura M, Ito J, Sato F, Nakai M, Sho T, et al:
Hepatitis B virus X protein impairs alpha-interferon signaling via
up-regulation of suppressor of cytokine signaling 3 and protein
phosphatase 2A. J Med Virol. 89:267–275. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kapoor NR, Chadha R, Kumar S, Choedon T,
Reddy VS and Kumar V: The HBx gene of hepatitis B virus can
influence hepatic microenvironment via exosomes by transferring its
mRNA and protein. Virus Res. 240:166–174. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Casciano JC and Bouchard MJ: Hepatitis B
virus X protein modulates cytosolic Ca(2+) signaling in primary
human hepatocytes. Virus Res. 246:23–27. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang XY, Li D, Chen ZX, Huang YH, Gao WY,
Zheng BY and Wang XZ: Hepatitis B Virus X protein elevates
Parkin-mediated mitophagy through Lon Peptidase in starvation. Exp
Cell Res. 368:75–83. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hensel KO, Cantner F, Bangert F, Wirth S
and Postberg J: Episomal HBV persistence within transcribed host
nuclear chromatin compartments involves HBx. Epigenetics Chromatin.
11:342018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li C, Lin C, Cong X and Jiang Y: PDK1-WNK1
signaling is affected by HBx and involved in the viability and
metastasis of hepatic cells. Oncol Lett. 15:5940–5946.
2018.PubMed/NCBI
|
|
9
|
Jin Y, Wu D, Yang W, Weng M, Li Y, Wang X,
Zhang X, Jin X and Wang T: Hepatitis B virus × protein induces
epithelial-mesenchymal transition of hepatocellular carcinoma cells
by regulating long non-coding RNA. Virol J. 14:2382017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu F, Song H, Xiao Q, Li N, Zhang H, Cheng
G and Tan G: Type III interferon-induced CBFbeta inhibits HBV
replication by hijacking HBx. Cell Mol Immunol. 17:357–366. 2019.
View Article : Google Scholar
|
|
11
|
Liu Y, Yao W, Si L, Hou J, Wang J, Xu Z,
Li W, Chen J, Li R, Li P, et al: Chinese herbal extract Su-duxing
had potent inhibitory effects on both wild-type and
entecavir-resistant hepatitis B virus (HBV) in vitro and
effectively suppressed HBV replication in mouse model. Antiviral
Res. 155:39–47. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ko C, Chakraborty A, Chou WM, Hasreiter J,
Wettengel JM, Stadler D, Bester R, Asen T, Zhang K, Wisskirchen K,
et al: Hepatitis B virus genome recycling and de novo secondary
infection events maintain stable cccDNA levels. J Hepatol.
69:1231–1241. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Han Q, Wang X, Liao X, Han C, Yu T, Yang
C, Li G, Han B, Huang K, Zhu G, et al: Diagnostic and prognostic
value of WNT family gene expression in hepatitis B virusrelated
hepatocellular carcinoma. Oncol Rep. 42:895–910. 2019.PubMed/NCBI
|
|
14
|
Tian Y and Ou JH: Genetic and epigenetic
alterations in hepatitis B virus-associated hepatocellular
carcinoma. Virol Sin. 30:85–91. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xie X, Xu X, Sun C and Yu Z: Hepatitis B
virus X protein promotes proliferation of hepatocellular carcinoma
cells by upregulating miR-181b by targeting ING5. Biol Chem.
399:611–619. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hamamoto H, Maemura K, Matsuo K, Taniguchi
K, Tanaka Y, Futaki S, Takeshita A, Asai A, Hayashi M, Hirose Y, et
al: Delta-like 3 is silenced by HBx via histone acetylation in
HBV-associated HCCs. Sci Rep. 8:48422018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang Q, Wang Q, Zhang Q, Lin SY, Zhu Y,
Yang X and Guo AY: Gene expression, regulation of DEN and HBx
induced HCC mice models and comparisons of tumor, para-tumor and
normal tissues. BMC Cancer. 17:8622017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
He X, Zhang C, Shi C and Lu Q:
Meta-analysis of mRNA expression profiles to identify
differentially expressed genes in lung adenocarcinoma tissue from
smokers and non-smokers. Oncol Rep. 39:929–938. 2018.PubMed/NCBI
|
|
19
|
Yildiz G, Arslan-Ergul A, Bagislar S, Konu
O, Yuzugullu H, Gursoy-Yuzugullu O, Ozturk N, Ozen C, Ozdag H,
Erdal E, et al: Genome-wide transcriptional reorganization
associated with senescence-to-immortality switch during human
hepatocellular carcinogenesis. PLoS One. 8:e640162013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Melis M, Diaz G, Kleiner DE, Zamboni F,
Kabat J, Lai J, Mogavero G, Tice A, Engle RE, Becker S, et al:
Viral expression and molecular profiling in liver tissue versus
microdissected hepatocytes in hepatitis B virus-associated
hepatocellular carcinoma. J Transl Med. 12:2302014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schulze K, Imbeaud S, Letouze E,
Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C,
Shinde J, Soysouvanh F, et al: Exome sequencing of hepatocellular
carcinomas identifies new mutational signatures and potential
therapeutic targets. Nat Genet. 47:505–511. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang M, Gong Q, Zhang J, Chen L, Zhang Z,
Lu L, Yu D, Han Y, Zhang D, Chen P, et al: Characterization of gene
expression profiles in HBV-related liver fibrosis patients and
identification of ITGBL1 as a key regulator of fibrogenesis. Sci
Rep. 7:434462017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang H, Huo X, Yang XR, He J, Cheng L,
Wang N, Deng X, Jin H, Wang N, Wang C, et al: STAT3-mediated
upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer
metastasis by regulating SOX4. Mol Cancer. 16:1362017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Grinchuk OV, Yenamandra SP, Iyer R, Singh
M, Lee HK, Lim KH, Chow PK and Kuznetsov VA: Tumor-adjacent tissue
co-expression profile analysis reveals pro-oncogenic ribosomal gene
signature for prognosis of resectable hepatocellular carcinoma. Mol
Oncol. 12:89–113. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Phipson B, Lee S, Majewski IJ, Alexander
WS and Smyth GK: Robust hyperparameter estimation protects against
hypervariable genes and improves power to detect differential
expression. Ann Appl Stat. 10:946–963. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
He B, Yin J, Gong S, Gu J, Xiao J, Shi W,
Ding W and He Y: Bioinformatics analysis of key genes and pathways
for hepatocellular carcinoma transformed from cirrhosis. Medicine
(Baltimore). 96:e69382017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xia Y, Cheng X, Li Y, Valdez K, Chen W and
Liang TJ: Hepatitis B Virus Deregulates the Cell Cycle To Promote
Viral Replication and a Premalignant Phenotype. J Virol. 92:2018.
View Article : Google Scholar
|
|
29
|
Chen XL, Zhou L, Yang J, Shen FK, Zhao SP
and Wang YL: Hepatocellular carcinoma-associated protein markers
investigated by MALDI-TOF MS. Mol Med Rep. 3:589–596. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lewis AG, Flanagan J, Marsh A, Pupo GM,
Mann G, Spurdle AB, Lindeman GJ, Visvader JE, Brown MA and
Chenevix-Trench G; Kathleen Cuningham Foundation Consortium for
Research into Familial Breast Cancer, : Mutation analysis of
FANCD2, BRIP1/BACH1, LMO4 and SFN in familial breast cancer. Breast
Cancer Res. 7:R1005–1016. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lee IN, Chen CH, Sheu JC, Lee HS, Huang
GT, Yu CY, Lu FJ and Chow LP: Identification of human
hepatocellular carcinoma-related biomarkers by two-dimensional
difference gel electrophoresis and mass spectrometry. J Proteome
Res. 4:2062–2069. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Reis H, Putter C, Megger DA, Bracht T,
Weber F, Hoffmann AC, Bertram S, Wohlschlager J, Hagemann S,
Eisenacher M, et al: A structured proteomic approach identifies
14-3-3 sigma as a novel and reliable protein biomarker in panel
based differential diagnostics of liver tumors. Biochim Biophys
Acta. 1854:641–650. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Saeki A, Tamura S, Ito N, Kiso S, Matsuda
Y, Yabuuchi I, Kawata S and Matsuzawa Y: Frequent impairment of the
spindle assembly checkpoint in hepatocellular carcinoma. Cancer.
94:2047–2054. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Seike M, Gemma A, Hosoya Y, Hosomi Y,
Okano T, Kurimoto F, Uematsu K, Takenaka K, Yoshimura A, Shibuya M,
et al: The promoter region of the human BUBR1 gene and its
expression analysis in lung cancer. Lung Cancer. 38:229–234. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhuang L, Yang Z and Meng Z: Upregulation
of BUB1B, CCNB1, CDC7, CDC20, and MCM3 in tumor tissues predicted
worse overall survival and disease-free survival in hepatocellular
carcinoma patients. Biomed Res Int. 2018:78973462018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu AW, Cai J, Zhao XL, Xu AM, Fu HQ, Nian
H and Zhang SH: The clinicopathological significance of BUBR1
overexpression in hepatocellular carcinoma. J Clin Pathol.
62:1003–1008. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sun B, Lin G, Ji D, Li S, Chi G and Jin X:
Dysfunction of sister chromatids separation promotes progression of
hepatocellular carcinoma according to analysis of gene expression
profiling. Front Physiol. 9:10192018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Janssen A, van der Burg M, Szuhai K, Kops
GJ and Medema RH: Chromosome segregation errors as a cause of DNA
damage and structural chromosome aberrations. Science.
333:1895–1898. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Miao R, Luo H, Zhou H, Li G, Bu D, Yang X,
Zhao X, Zhang H, Liu S, Zhong Y, et al: Identification of
prognostic biomarkers in hepatitis B virus-related hepatocellular
carcinoma and stratification by integrative multi-omics analysis. J
Hepatol. 61:840–849. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pineda-Solis K and McAlister V: Wading
through the noise of ‘multi-omics’ to identify prognostic
biomarkers in hepatocellular carcinoma. Hepatobiliary Surg Nutr.
4:293–294. 2015.PubMed/NCBI
|
|
41
|
Baffy G: Decoding multifocal
hepatocellular carcinoma: An opportune pursuit. Hepatobiliary Surg
Nutr. 4:206–210. 2015.PubMed/NCBI
|
|
42
|
Feo F and Pascale RM: Multifocal
hepatocellular carcinoma: Intrahepatic metastasis or multicentric
carcinogenesis? Ann Transl Med. 3:42015.PubMed/NCBI
|
|
43
|
Wang G, Chen H, Huang M, Wang N, Zhang J,
Zhang Y, Bai G, Fong WF, Yang M and Yao X: Methyl protodioscin
induces G2/M cell cycle arrest and apoptosis in HepG2 liver cancer
cells. Cancer Lett. 241:102–109. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Nimeus-Malmstrom E, Koliadi A, Ahlin C,
Holmqvist M, Holmberg L, Amini RM, Jirstrom K, Warnberg F,
Blomqvist C, Ferno M and Fjällskog ML: Cyclin B1 is a prognostic
proliferation marker with a high reproducibility in a
population-based lymph node negative breast cancer cohort. Int J
Cancer. 127:961–967. 2010.PubMed/NCBI
|
|
45
|
Soria JC, Jang SJ, Khuri FR, Hassan K, Liu
D, Hong WK and Mao L: Overexpression of cyclin B1 in early-stage
non-small cell lung cancer and its clinical implication. Cancer
Res. 60:4000–4004. 2000.PubMed/NCBI
|
|
46
|
Begnami MD, Fregnani JH, Nonogaki S and
Soares FA: Evaluation of cell cycle protein expression in gastric
cancer: cyclin B1 expression and its prognostic implication. Hum
Pathol. 41:1120–1127. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Weng L, Du J, Zhou Q, Cheng B, Li J, Zhang
D and Ling C: Identification of cyclin B1 and Sec62 as biomarkers
for recurrence in patients with HBV-related hepatocellular
carcinoma after surgical resection. Mol Cancer. 11:392012.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fang Y, Yu H, Liang X, Xu J and Cai X:
Chk1-induced CCNB1 overexpression promotes cell proliferation and
tumor growth in human colorectal cancer. Cancer Biol Ther.
15:1268–1279. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cheng P, Li Y, Yang L, Wen Y, Shi W, Mao
Y, Chen P, Lv H, Tang Q and Wei Y: Hepatitis B virus X protein
(HBx) induces G2/M arrest and apoptosis through sustained
activation of cyclin B1-CDK1 kinase. Oncol Rep. 22:1101–1107.
2009.PubMed/NCBI
|
|
50
|
Chen QF, Xia JG, Li W, Shen LJ, Huang T
and Wu P: Examining the key genes and pathways in hepatocellular
carcinoma development from hepatitis B viruspositive cirrhosis. Mol
Med Rep. 18:4940–4950. 2018.PubMed/NCBI
|
|
51
|
Li H, Zhao X, Li C, Sheng C and Bai Z:
Integrated analysis of lncRNA-associated ceRNA network reveals
potential biomarkers for the prognosis of hepatitis B virus-related
hepatocellular carcinoma. Cancer Manag Res. 11:877–897. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chae S, Ji JH, Kwon SH, Lee HS, Lim JM,
Kang D, Lee CW and Cho H: HBxAPalpha/Rsf-1-mediated HBx-hBubR1
interactions regulate the mitotic spindle checkpoint and chromosome
instability. Carcinogenesis. 34:1680–1688. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Slagle BL and Bouchard MJ: Role of HBx in
hepatitis B virus persistence and its therapeutic implications.
Curr Opin Virol. 30:32–38. 2018. View Article : Google Scholar : PubMed/NCBI
|