Introduction
In 2018, lung cancer was reported as having the
highest morbidity and mortality rates of all types of malignancy
worldwide, posing a notable risk to human health (1). Although the methods of clinical
diagnosis and treatment of lung cancer are improving, the prognosis
for patients with lung cancer remains poor. The mortality rate of
lung cancer is increasing and the 5-year survival rate is estimated
to approximately 18% (2). Therefore,
novel targets for drug treatment and prognosis of lung cancer are
needed.
Myosin heavy chain 11 (MYH11), which is encoded by
the MYH11 gene, is a smooth muscle myosin belonging to the
myosin heavy chain family (3). MYH11
is a contractile protein that slides past actin filaments to induce
muscle contraction via adenosine triphosphate hydrolysis (4,5).
Previous findings have shown that in aortic tissue, destruction of
MYH11 can lead to vascular smooth muscle cell loss, disorganization
and hyperplasia, which is one of the mechanisms leading to thoracic
aortic aneurysms and dissections (6,7). In
previous years, studies have indicated that mutations of the
MYH11 gene are also associated with various types of cancer,
including colorectal (8,9), bladder (10), laryngeal (11) and head and neck cancer (12). This may be due to the role of
MYH11 role in cell migration, interaction with cell adhesion
proteins and tumor suppression (13,14).
However, the association between the expression levels of
MYH11 and lung cancer has rarely been investigated. To the
best of our knowledge, only Ma et al (15) investigated the key role of
MYH11 in non-small-cell lung cancer. In the present study,
the Oncomine, Kaplan-Meier plotter, cBioportal, Coexpedia and
Metascape databases were used in order to explore the biological
significance of MYH11 in lung cancer and provide a
scientific basis of the underlying molecular mechanism of
MYH11 function in the pathogenesis of lung cancer.
Materials and methods
Oncomine analysis
Oncomine is a large database of cancer microarray
experiments and a web-based data-mining platform that is used to
screen for differences in gene expression levels between different
tumor tissues (16,17). The query terms were set as follows:
i) gene: MYH11; and ii) analysis type: Lung cancer vs.
normal analysis. P<0.05 was considered to indicate a
statistically significant difference, fold change was set to 2 and
gene rank to 5%. An unpaired t-test was used for the comparison of
two groups.
Kaplan-Meier plotter database
Kaplan-Meier plotter online database (kmplot.com) was used to assess the effects of
MYH11 expression levels on survival and prognosis (18). The database was used to query
relevant data of lung cancer and the mean expression levels of the
gene probes. The correlation between MYH11 expression levels
and the survival rate of patients with lung cancer was
investigated. P<0.05 was considered to indicate a statistically
significant difference. Overall survival, first progression and
post progression survival curves were generated using the
Kaplan-Meier method and evaluated using the log-rank test.
cBioportal analysis
In order to find mutations and copy number
alterations (CNAs) of MYH11, genomic data were retrieved
from The Cancer Genome Atlas (TCGA) lung cancer datasets using
cBioportal (cbioportal.org) (19). The location and frequency of
MYH11 alterations (amplifications, deep deletions and
missense mutations) and copy number variance data were
assessed.
Coexpedia database
The co-expression genes of MYH11 in Gene
Expression Omnibus (GEO) database were analyzed using Coexpedia
(coexpedia.org/) (20). Coexpedia is a database of
context-associated co-expression networks constructed from
individual series of microarray samples for humans. The total score
for each gene was the sum of edge-weights (log-likelihood score)
and all connected genes in the network.
Metascape analysis
Metascape (metascape.org)
is a powerful web-based tool for gene annotation and gene list
enrichment analysis, including biological process (BP), molecular
function (MF), cellular component (CC) and a Kyoto Encyclopedia of
Genes and Genomes (KEGG) pathway analysis (21). In the present study, Metascape was
used to conduct a pathway and process enrichment analysis of
MYH11 and co-expression genes significantly associated with
MYH11 alterations. In addition, protein-protein interaction
(PPI) networks were also analyzed.
Results
Expression levels of MYH11 in
different types of cancer
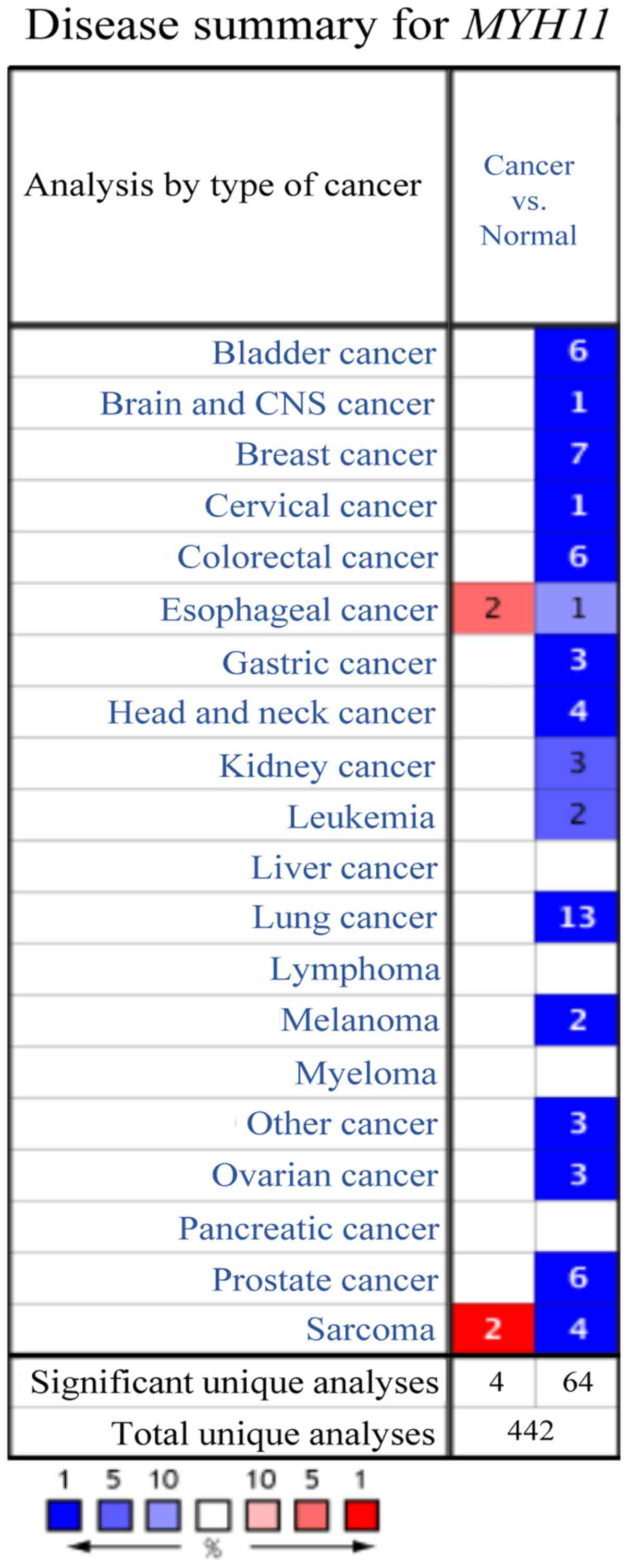
In the Oncomine database, a total of 442 studies
were available to compare MHY11 gene expression levels
between different types of cancer and normal tissues. A total of 68
studies were identified as statistically significant in the
Oncomine database. In 64 and 4 studies, MYH11 expression
levels were significantly down- and upregulated in cancer tissues
compared with normal tissues, respectively. In addition, in 13
studies, decreased MYH11 gene expression levels were noted
in lung cancer compared with normal lung tissues (Fig. 1).
Expression of MYH11 in lung
cancer
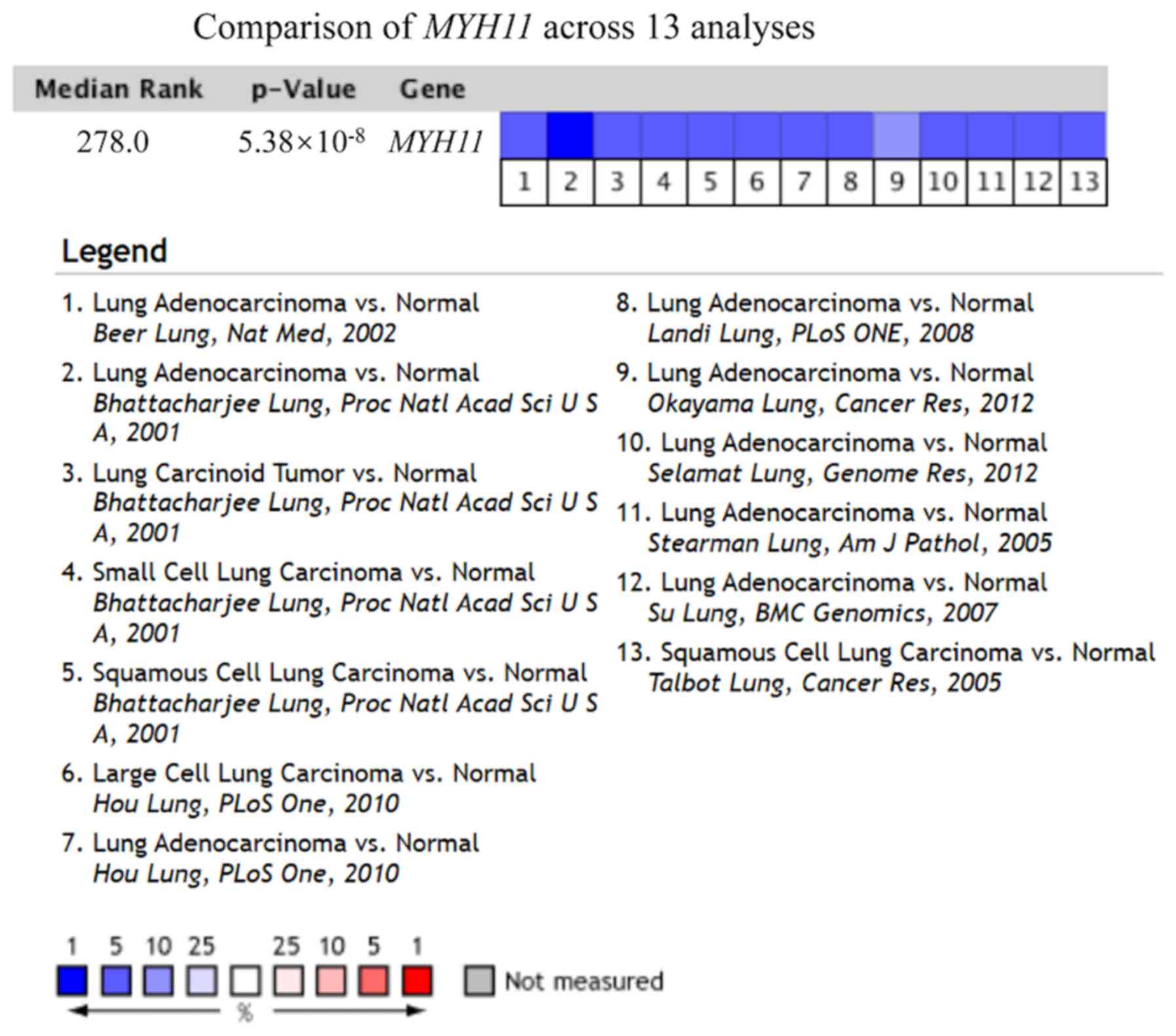
In the Oncomine database, MYH11 gene
expression levels in all studies investigating lung cancer were
obtained. From November 2001 to the present, thirteen studies met
the screening threshold criteria, with a total sample size of 1,122
cases (22–30). As shown in Fig. 2 and Table
I, the output results indicated that MYH11 gene
expression levels were significantly downregulated in lung cancer
tissue compared with normal lung tissue in all 13 studies (median
rank=278.0; P=5.38×10−8; Fig.
2).
 | Table I.Oncomine analysis of 8 datasets
analyzing MYH11 gene expression levels in different lung
cancer subtypes. The data were compared with the expression levels
of MYH11 in normal lung tissues. |
Table I.
Oncomine analysis of 8 datasets
analyzing MYH11 gene expression levels in different lung
cancer subtypes. The data were compared with the expression levels
of MYH11 in normal lung tissues.
| Author, year | Type of lung
cancer | Fold change | P-value | t-test | (Refs.) |
|---|
| Bhattacharjee et
al, 2001 | Lung
adenocarcinoma | −20.207 |
6.39×10−9 | −8.181 | (22) |
| Bhattacharjee et
al, 2001 | Squamous cell lung
carcinoma | −21.362 |
5.16×10−8 | −7.066 | (22) |
| Bhattacharjee et
al, 2001 | Small cell lung
carcinoma | −12.541 |
1.09×10−5 | −5.795 | (22) |
| Bhattacharjee et
al, 2001 | Lung carcinoid
tumor | −25.905 |
3.97×10−9 | −8.204 | (22) |
| Beer et al,
2002 | Lung
adenocarcinoma | −24.567 |
3.17×10−12 | −10.557 | (23) |
| Landi et al,
2008 | Lung
adenocarcinoma | −2.979 |
6.23×10−25 | −13.475 | (24) |
| Stearman et
al, 2005 | Lung
adenocarcinoma | −16.652 |
5.14×10−8 | −7.805 | (25) |
| Talbot et
al, 2005 | Squamous cell lung
carcinoma | −2.149 |
5.38×10−8 | −6.285 | (26) |
| Hou et al,
2010 | Large cell lung
carcinoma | −5.286 |
3.28×10−14 | −12.556 | (27) |
| Hou et al,
2010 | Lung
adenocarcinoma | −3.289 |
1.7×10−15 | −9.7 | (27) |
| Su et al,
2007 | Lung
adenocarcinoma | −7.195 |
2.72×10−8 | −6.672 | (28) |
| Selamat et
al, 2012 | Lung
adenocarcinoma | −2.007 |
1.62×10−17 | −10.215 | (29) |
| Okayama et
al, 2012 | Lung
adenocarcinoma | −2.717 |
5.2×10−10 | −8.48 | (30) |
Expression levels of MYH11 in lung
cancer subtypes
In the Oncomine database, MYH11 gene
expression levels results were derived from 13 studies
investigating lung cancer and the relevant data were compared and
analyzed using box-plot analysis (Fig.
3). The aforementioned studies included comparisons between
different types of lung cancer (squamous cell lung carcinoma, lung
adenocarcinoma, small cell lung carcinoma and large cell lung
carcinoma) and normal lung tissue (Table
I). The results suggested that the MYH11 gene expression
levels in different lung cancer subtypes were significantly
decreased compared with those noted in normal lung tissue
(P<0.05; Table I). Compared with
normal lung tissue, the specific output results for the lung
adenocarcinoma samples in Bhattacharjee et al (22), Beer et al (23), Landi et al (24), Stearman et al (25), Hou et al (27), Su et al (28), Selamat et al (29) and Okayama et al (30) were 6.39×10−9,
3.17×10−12, 6.23×10−25, 5.14×10−8,
1.7×10−15, 2.72×10-8, 1.62×10−17 and
5.2×10−10, respectively. Compared with normal lung
tissues, the p-values were as follows for the squamous cell lung
carcinoma samples in Bhattacharjee et al
(5.16×10−8) and Talbot et al (26) (5.38×10−8), the small cell
lung carcinoma samples in Bhattacharjee et al (22) (1.09×10−5), the lung
carcinoid tumor samples in Bhattacharjee et al (22) (3.97×10−9) and the large
cell lung carcinoma samples in Hou et al (27) (3.28×10−14).
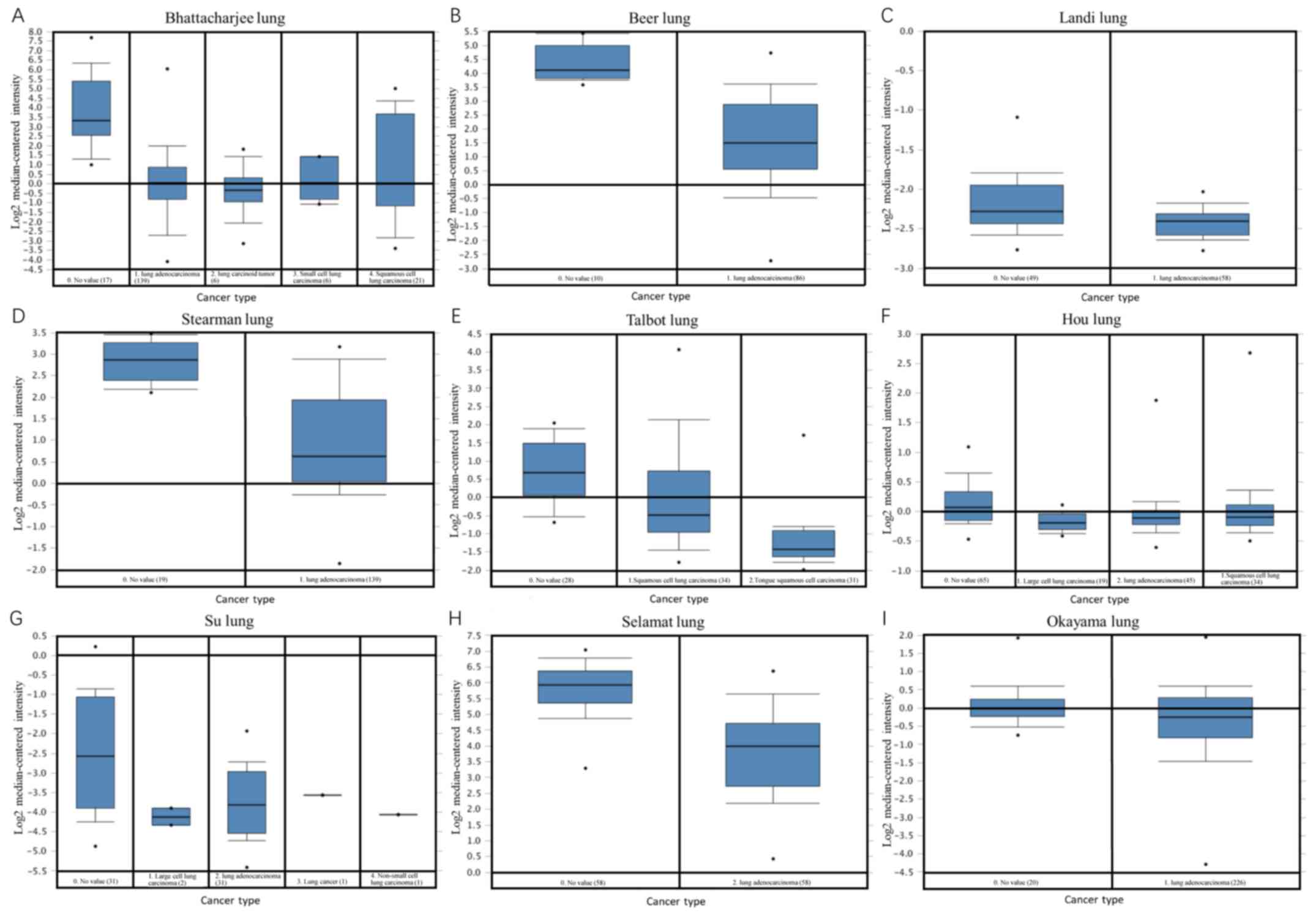 | Figure 3.Oncomine analysis. A total of eight
datasets, including (A) Bhattacharjee lung, (B) Beer lung, (C)
Landi lung, (D) Stearman lung, (E) Talbot lung, (F) Hou lung, (G)
Su lung, (H) Selamat lung and (I) Okayama lung datasets were
extracted from the Oncomine database for analyzing MYH11
gene expression levels in different lung cancer subtypes compared
with normal lung tissues. The expression profiles were
median-centered, log2-transformed and uploaded onto the Oncomine
database. This analysis revealed that MYH11 gene expression
levels were significantly downregulated in several lung cancer
subtypes. MYH11, myosin heavy chain 11 gene. |
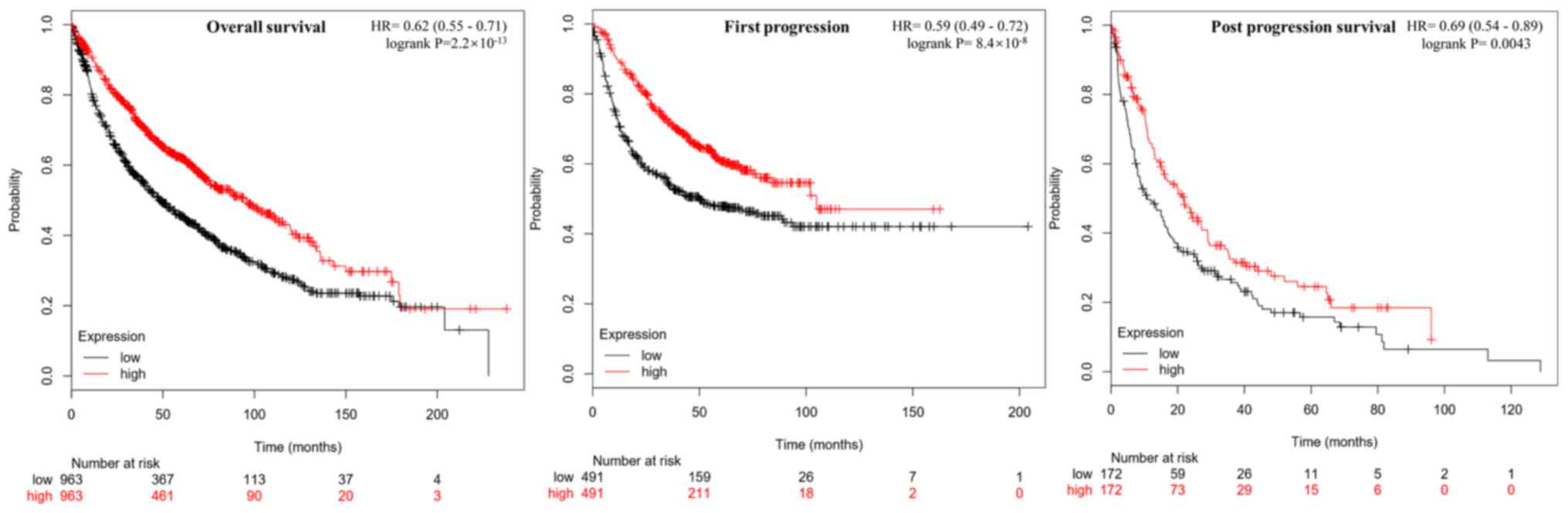
Correlation between MYH11 expression
levels and prognosis of lung cancer
The correlation between MYH11 gene expression
levels and lung cancer survival rate were analyzed using the
Kaplan-Meier plotter analysis tool (Fig.
4). MYH11 expression levels were positively correlated
with overall survival (P=2.2×10−13), first progression
(P=8.4×10−8) and post progression survival (P=0.0043) in
patients with lung cancer. These results suggested that
MYH11 exhibited tumor suppressive roles in lung cancer.
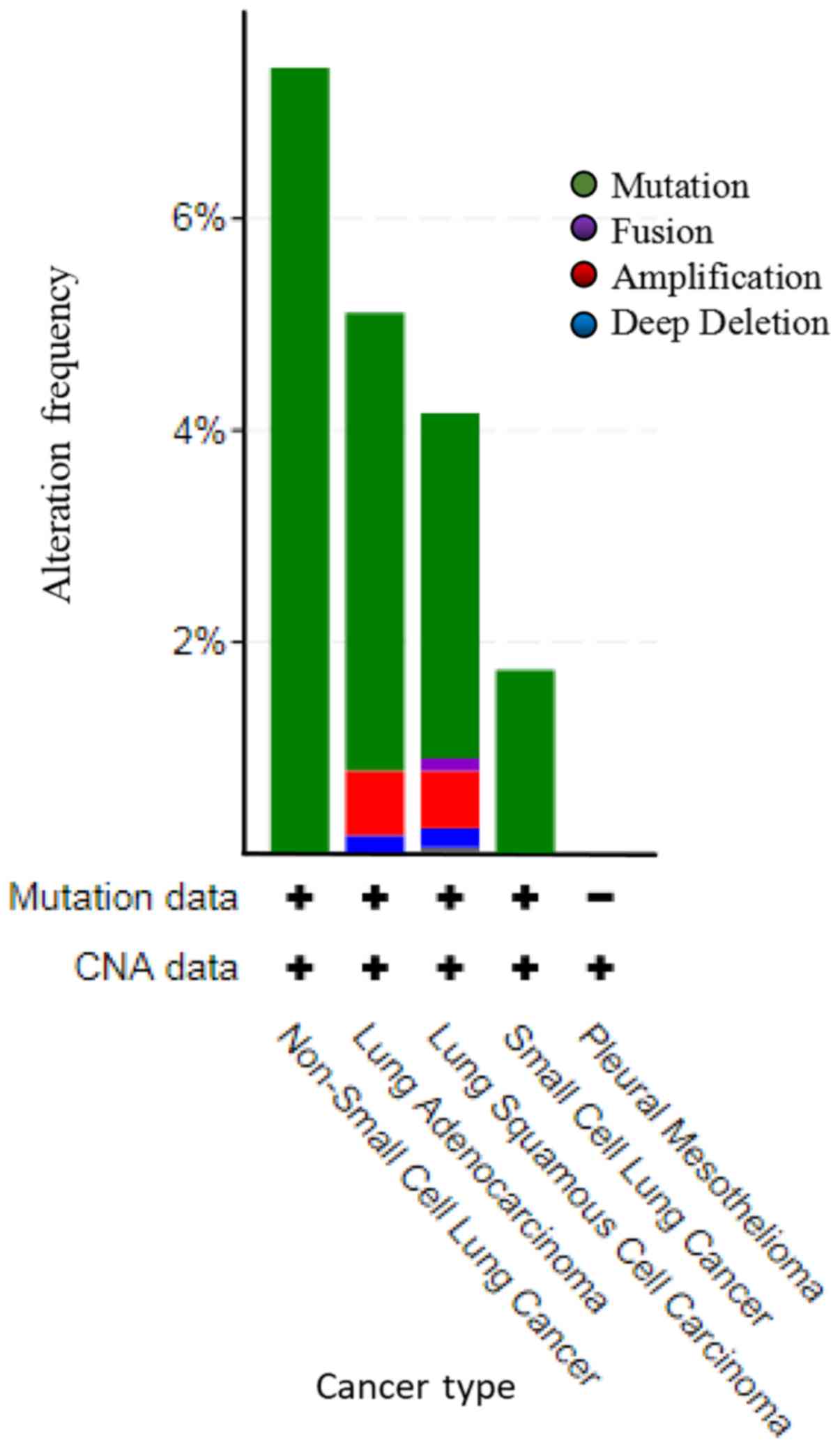
Mutations and CNAs of MYH11 in lung
cancer
TCGA datasets of all lung cancer samples were
selected to investigate mutations and CNAs in the MYH11
gene. A total of 4,582 cases in 21 studies were included for this
analysis (Fig. 5). Among the lung
cancer cases with gene alteration of MYH11, mutation was the
most common alteration type. A total of 30 cases of non-small cell
lung cancer were amplified, accounting for 7.43%. In lung
adenocarcinoma, 99 cases were amplified, 14 cases were deep
deletion, and 4 cases were fusion, which accounted for 4.32, 0.61
and 0.17%, respectively. In lung squamous cell carcinoma, 54 cases
were amplified, 9 cases were deep deletion, 3 cases were fusion and
2 cases were multiple alterations, which accounted for 3.26, 0.54,
0.18 and 0.12%, respectively. A total of 4 cases of small cell lung
cancer were amplified, accounting for 1.74% (Fig. 5 and Table
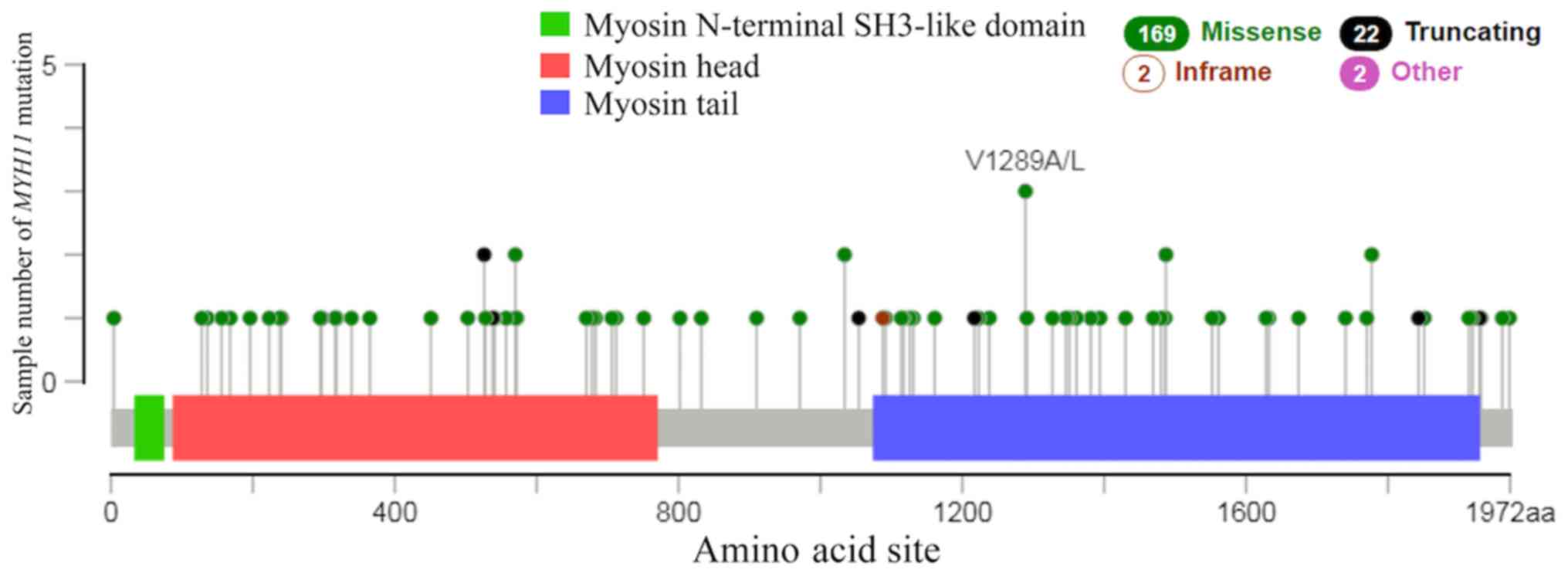
II). There were 169 cases of missense, 22 cases of truncating
and 2 cases of in-frame mutations in lung cancer (Fig. 6). The results of the present study
demonstrated that the most common type of mutation in lung cancer
was missense mutation. A previous study reported that MYH11
missense mutations in breast cancer are associated with tumor
development (31).
 | Table II.Mutations and copy number alterations
of MYH11 in different types of lung cancer. The alteration
frequency of MYH11 in different lung cancer studies. |
Table II.
Mutations and copy number alterations
of MYH11 in different types of lung cancer. The alteration
frequency of MYH11 in different lung cancer studies.
|
| Lung cancer
subtype |
|---|
|
|
|
|---|
|
| Non-small
cell, |
Adenocarcinoma, | Squamous cell
carcinoma, | Small cell, |
|---|
| Mutation type | n (%) | n (%) | n (%) | n (%) |
| Amplification | 30 (7.43) | 99 (4.32) | 54 (3.26) | 4 (1.74) |
| Deep deletion | – | 14 (0.61) | 9
(0.54) | – |
| Fusion | – | 4
(0.17) | 3
(0.18) | – |
| Multiple
alterations | – | – | 2
(0.12) | – |
Enrichment analysis of MYH11
The visualization of co-expressed genes was ranked
according to the online Coexpedia instructions. The total score for
each gene was a sum of edge-weights (log likelihood score) and all
connected genes in the network. Then, the functions of MYH11
and the 30 genes with the highest scores were predicted by
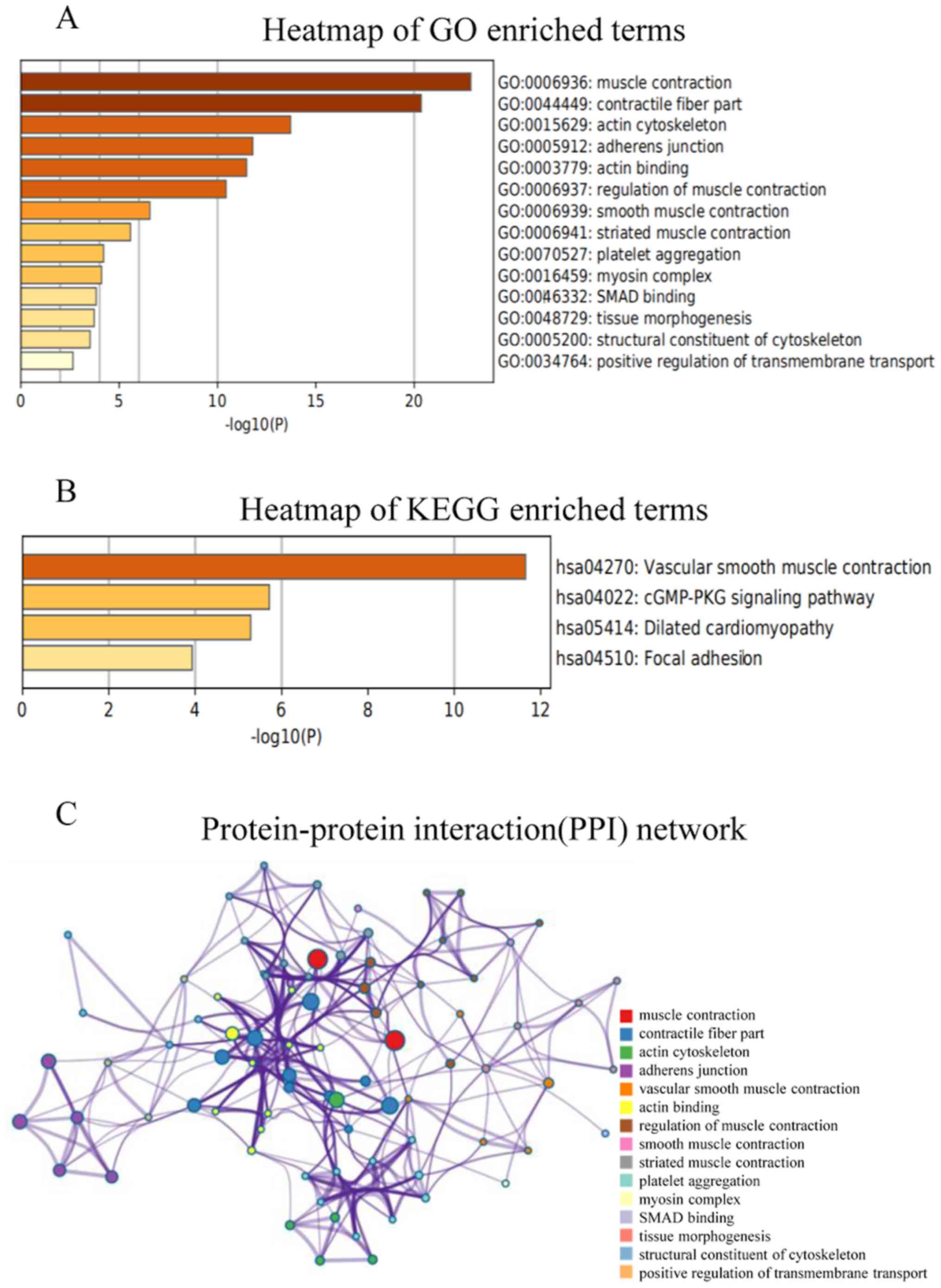
analyzing GO and KEGG data using Metascape (Fig. 7A and B). For GO term enrichment
analysis, the target genes were mainly enriched in ‘muscle
contraction’, ‘contractile fiber part’, ‘actin cytoskeleton’,
‘adherens junction’, ‘actin binding’, ‘regulation of muscle
contraction’, ‘smooth muscle contraction’, ‘striated muscle
contraction’, ‘platelet aggregation’, ‘myosin complex’, ‘SMAD
binding’, ‘tissue morphogenesis’, ‘structural constituent of
cytoskeleton’ and ‘positive regulation of transmembrane transport’
(Fig. 7A). KEGG analysis revealed
that the target genes were mostly enriched in ‘vascular smooth
muscle contraction’, ‘cGMP-PKG signaling pathway’, ‘dilated
cardiomyopathy’ and ‘focal adhesion’ (Fig. 7B). Subsequently, the PPI networks and
their connections were identified using Metascape. The results
demonstrated that the main functions associated with MYH11 protein
were ‘muscle contraction’, ‘contractile fiber part’, ‘actin
cytoskeleton’, ‘adherens junction’, ‘vascular smooth muscle
contraction’, ‘actin binding’, ‘regulation of muscle contraction’,
‘smooth muscle contraction’, ‘striated muscle contraction’,
‘platelet aggregation’, ‘myosin complex’, ‘SMAD binding’, ‘tissue
morphogenesis’, ‘structural constituent of cytoskeleton’ and
‘positive regulation of transmembrane transport’ (Fig. 7C).
Discussion
The MYH11 gene, which encodes a smooth muscle
myosin protein, is located at chromosome 16p13.13-13.12 (32). It has been shown that myosin proteins
are involved in muscle movement (33). Issouf et al (34) reported that MYH11 is a gene
that functions in the molecular mechanisms underlying the
contraction of airway smooth muscle in asthma. However, previous
studies suggested that MYH11 was also involved in cell
adhesion, migration and tumor suppression (13,14).
Previous studies demonstrated that mutations in the MYH11
gene could promote cancer formation. For example, the
CBFB/MYH11 fusion gene has been implicated in the onset of
acute myeloid leukemia (35,36). In addition, MYH11 gene
expression levels have been associated with the prognostic outcome
of the bladder urothelial carcinoma, notably in patients with
advanced tumors (37). Seitz et
al (38) demonstrated that
MYH11 gene expression levels were downregulated in breast
tumor and metastasis, using microarray and correlation analyses.
Disregulated MYH11 gene expression levels have also been
associated with the occurrence of intestinal tumors by affecting
the cellular energy balance (39).
In the present study, MYH11 gene expression
levels were significantly decreased in lung cancer tissues compared
with that noted in normal lung tissues. These results indicated
that the decreased MYH11 gene expression levels were
associated with the onset of lung cancer. In addition, decreased
MYH11 expression levels were associated with a less
favorable prognosis of patients with lung cancer. These results are
in accordance with previous studies from Wang et al
(9) in colorectal cancer and Ma
et al (15) in non-small-cell
lung cancer, indicating that MYH11 gene expression levels
could be used as a novel biomarker in predicting the prognosis of
lung cancer.
Among the lung cancer cases with gene alteration of
MYH11, mutation was the most common of all alteration types.
Gene-set enrichment analysis showed that the target genes were
mainly enriched in ‘muscle contraction’, ‘contractile fiber part’,
‘actin cytoskeleton’ and ‘adherens junction’. It has been proposed
that myosin-dependent contractile activity in non-muscle cells,
similar to that observed in muscle, may also be involved in cell
migration (14). Consistent with
these findings, the results of the present study suggest that
MYH11 may increase cell migration and adhesion in a mutated
manner, in order to increase tumor invasion. In conclusion, the
results of the present study demonstrate the potential of
MYH11 as a novel target for the treatment of lung cancer.
Despite the use of a large sample size, which increases the
accuracy of the results, the present study failed to identify the
underlying molecular mechanism of MYH11 in the development
of lung cancer, thus further verification is required.
Acknowledgements
Not applicable.
Funding
The present study was supported by The Foundation of
the Priority Academic Program Development of Jiangsu Higher
Education Institutions (grant no. ZYX03KF022), The Fifth ‘333’
Project of Jiangsu Province (grant no. BRA2016522), Subject of
Jiangsu Province Hospital of Traditional Chinese Medicine (grant
no. Y2017CX55) and The Studio of National famous TCM specialist
Liu-ShenLin confirmed by State Administration of Traditional
Chinese Medicine.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
MN, XP, HT and XZ designed the present study and
drafted the initial manuscript. MX, SL, WS and JW interpreted the
data. All authors contributed to analyzing the data, and have read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Matsuoka R, Yoshida MC, Furutani Y,
Imamura S, Kanda N, Yanagisawa M, Masaki T and Takao A: Human
smooth muscle myosin heavy chain gene mapped to chromosomal region
16q12. Am J Med Genet. 46:61–67. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huxley AF and Niedergerke R: Structural
changes in muscle during contraction: Interference microscopy of
living muscle fibres. Nature. 173:971–973. 1954. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huxley H and Hanson J: Changes in the
cross-striations of muscle during contraction and stretch and their
structural interpretation. Nature. 173:973–976. 1954. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhu L, Vranckx R, Khau Van Kien P, Lalande
A, Boisset N, Mathieu F, Wegman M, Glancy L, Gasc JM, Brunotte F,
et al: Mutations in myosin heavy chain 11 cause a syndrome
associating thoracic aortic aneurysm/aortic dissection and patent
ductus arteriosus. Nat Genet. 38:343–349. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pannu H, Tran-Fadulu V, Papke CL, Scherer
S, Liu Y, Presley C, Guo D, Estrera AL, Safi HJ, Brasier AR, et al:
MYH11 mutations result in a distinct vascular pathology driven by
insulin-like growth factor 1 and angiotensin II. Hum Mol Genet.
16:2453–2462. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xi WD, Liu YJ, Sun XB, Shan J, Yi L and
Zhang TT: Bioinformatics analysis of RNA-seq data revealed critical
genes in colon adenocarcinoma. Eur Rev Med Pharmacol Sci.
21:3012–3020. 2017.PubMed/NCBI
|
|
9
|
Wang RJ, Wu P, Cai GX, Wang ZM, Xu Y, Peng
JJ, Sheng WQ, Lu HF and Cai SJ: Down-regulated MYH11 expression
correlates with poor prognosis in stage II and III colorectal
cancer. Asian Pac J Cancer Prev. 15:7223–7228. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu J, Zhou L, Song Z, Xiong M, Zhang Y,
Yang Y, Chen K and Chen Z: The identification of new biomarkers for
bladder cancer: A study based on TCGA and GEO datasets. J Cell
Physiol. Feb 18–2019.(Epub ahead of print). View Article : Google Scholar
|
|
11
|
Su J, Zhang Y, Su H, Zhang C and Li W: A
recurrence model for laryngeal cancer based on SVM and gene
function clustering. Acta Otolaryngol. 137:557–562. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Islam T, Rahman R, Gov E, Turanli B,
Gulfidan G, Haque A, Arga KY and Haque Mollah N: Drug targeting and
biomarkers in head and neck cancers: Insights from systems biology
analyses. OMICS. 22:422–436. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Krendel M and Mooseker MS: Myosins: Tails
(and Heads) of functional diversity. Physiology (Bethesda).
20:239–251. 2005.PubMed/NCBI
|
|
14
|
Pollard TD and Weihing RR: Actin and
myosin and cell movemen. CRC Crit Rev Biochem. 2:1–65. 2008.
View Article : Google Scholar
|
|
15
|
Ma Q, Xu Y, Liao H, Cai Y, Xu L, Xiao D,
Liu C, Pu W, Zhong X and Guo X: Identification and validation of
key genes associated with non-small-cell lung cancer. J Cell
Physiol. 234:22742–22752. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Findeisen P, Röckel M, Nees M, Röder C,
Kienle P, Von Knebel Doeberitz M, Kalthoff H and Neumaier M:
Systematic identification and validation of candidate genes for
detection of circulating tumor cells in peripheral blood specimens
of colorectal cancer patients. Int J Oncol. 33:1001–1010.
2008.PubMed/NCBI
|
|
17
|
Rhodes DR, Kalyana-Sundaram S, Mahavisno
V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ,
Kincead-Beal C, et al: Oncomine 3. 0: Genes, pathways, and networks
in a collection of 18, 000 cancer gene expression profiles.
Neoplasia. 9:166–180. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nagy A, Lánczky A, Menyhárt O and Győrffy
B: Validation of miRNA prognostic power in hepatocellular carcinoma
using expression data of independent datasets. Sci Rep. 8:92272018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang S, Kim CY, Hwang S, Kim E, Kim H,
Shim H and Lee I: COEXPEDIA: Exploring biomedical hypotheses via
co-expressions associated with medical subject headings (MeSH).
Nucleic Acids Res. 45:D389–D936. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bhattacharjee A, Richards WG, Staunton J,
Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et
al: Classification of human lung carcinomas by mRNA expression
profiling reveals distinct adenocarcinoma subclasses. Proc Natl
Acad Sci USA. 98:13790–13795. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Beer DG, Kardia SL, Huang CC, Giordano TJ,
Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al:
Gene-expression profiles predict survival of patients with lung
adenocarcinoma. Nat Med. 8:816–824. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Landi MT, Dracheva T, Rotunno M, Figueroa
JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, et
al: Gene expression signature of cigarette smoking and its role in
lung adenocarcinoma development and survival. PLoS One.
3:e16512008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Stearman RS, Dwyer-Nield L, Zerbe L,
Blaine SA, Chan Z, Bunn PA Jr, Johnson GL, Hirsch FR, Merrick DT,
Franklin WA, et al: Analysis of orthologous gene expression between
human pulmonary adenocarcinoma and a carcinogen-induced murine
model. Am J Pathol. 167:1763–1775. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Talbot SG, Estilo C, Maghami E, Sarkaria
IS, Pham DK, O-charoenrat P, Socci ND, Ngai I, Carlson D, Ghossein
R, et al: Gene expression profiling allows distinction between
primary and metastatic squamous cell carcinomas in the lung. Cancer
Res. 65:3063–3071. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hou J, Aerts J, den Hamer B, van Ijcken W,
den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens
JA, Hoogsteden HC, et al: Gene expression-based classification of
non-small cell lung carcinomas and survival prediction. PLoS One.
5:e103122010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Su LJ, Chang CW, Wu YC, Chen KC, Lin CJ,
Liang SC, Lin CH, Whang-Peng J, Hsu SL, Chen CH and Huang CY:
Selection of DDX5 as a novel internal control for Q-RT- PCR from
microarray data using a block bootstrap re-sampling scheme. BMC
Genomics. 8:1402007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Selamat SA, Chung BS, Girard L, Zhang W,
Zhang Y, Campan M, Siegmund KD, Koss MN, Hagen JA, Lam WL, et al:
Genome-scale analysis of DNA methylation in lung adenocarcinoma and
integration with mRNA expression. Genome Res. 22:1197–1211. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Okayama H, Kohno T, Ishii Y, Shimada Y,
Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S,
et al: Identification of genes upregulated in ALK-positive and
EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res.
72:100–111. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Alhopuro P, Karhu A, Winqvist R, Waltering
K, Visakorpi T and Aaltonen LA: Somatic mutation analysis of MYH11
in breast and prostate cancer. BMC Cancer. 8:2632008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Deng Z, Liu P, Marlton P, Claxton DF, Lane
S, Callen DF, Collins FS and Siciliano MJ: Smooth muscle myosin
heavy chain locus (MYH11) maps to 16p13.13-p13.12 and establishes a
new region of conserved synteny between human 16p and mouse 16.
Genomics. 18:156–159. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Trybus KM: Regulation of smooth muscle
myosin. Cell Motil Cytoskeleton. 18:81–85. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Issouf M, Vargas A, Boivin R and Lavoie
JP: SRSF6 is upregulated in asthmatic horses and involved in the
MYH11 SMB expression. Physiol Rep. 6:e138962018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Goyama S and Mulloy JC: Molecular
pathogenesis of core binding factor leukemia: Current knowledge and
future prospects. Int J Hematol. 94:126–133. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu P, Tarlé SA, Hajra A, Claxton DF,
Marlton P, Freedman M, Siciliano MJ and Collins FS: Fusion between
transcription factor CBF beta/PEBP2 beta and a myosin heavy chain
in acute myeloid leukemia. Science. 261:1041–1044. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ning X and Deng Y: Identification of key
pathways and genes influencing prognosis in bladder urothelial
carcinoma. Onco Targets Ther. 10:1673–1686. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Seitz S, Korsching E, Weimer J, Jacobsen
A, Arnold N, Meindl A, Arnold W, Gustavus D, Klebig C, Petersen I
and Scherneck S: Genetic background of different cancer cell lines
influences the gene set involved in chromosome 8 mediated breast
tumor suppression. Genes Chromosomes Cancer. 45:612–627. 2010.
View Article : Google Scholar
|
|
39
|
Alhopuro P, Phichith D, Tuupanen S,
Sammalkorpi H, Nybondas M, Saharinen J, Robinson JP, Yang Z, Chen
LQ, Orntoft T, et al: Unregulated smooth-muscle myosin in human
intestinal neoplasia. Proc Natl Acad Sci USA. 105:5513–5518. 2008.
View Article : Google Scholar : PubMed/NCBI
|