Introduction
Hepatocellular carcinoma (HCC) is a leading cause of
cancer-associated mortality, with >700,000 new cases being
diagnosed annually throughout the world (1). HCC is the result of complex
interactions between genetic and non-genetic host factors,
including exposure to environmental chemicals and viruses. In
>90% of cases, a primitive chronic liver disease, e.g.,
cirrhosis, usually develops and creates a procarcinogenic
environment in its final stage (2).
The yhwaz gene encodes 14-3-3ζ, which belongs
to a family of highly conserved dimeric proteins that are
considered master regulators of intracellular signaling (3–5). 14-3-3ζ
has been shown to interact with numerous protein kinases, enzymes,
receptor proteins, structural and cytoskeletal proteins, proteins
associated with cell cycle and transcriptional control and proteins
involved in apoptosis (6).
Upregulated expression of yhwaz is frequently observed in
several types of cancer (including lung and liver cancer) (7–9). For
example, yhwaz is upregulated in >40% of cases of breast
cancer and is able to activate transforming growth factor-β/SMAD
signaling during the epithelial-to-mesenchymal-transition, thus
promoting tumor progression (10).
14-3-3ζ interacts with β-catenin, GLI family zinc finger 2,
Yes-associated protein 1 and the downstream effectors of the Wnt,
Hedgehog and Hippo signaling pathways, all of which are involved to
varying degrees in a number of different types of cancer (such as
breast cancer or lymphocytic leukemia) (11–14).
Therefore, 14-3-3ζ may potentially be used as a prognostic
biomarker for the diagnosis of certain types of cancer.
The upregulation of yhwaz in patients with
HCC has been frequently examined (15) and 14-3-3ζ protein expression levels
have been found to be significantly upregulated in hepatoma cell
lines. In HCC cell lines with upregulated 14-3-3ζ expression,
knockdown of 14-3-3ζ using RNA interference was reported to inhibit
cell proliferation by activating the c-jun N-terminal kinase and
p38/mitogen-activated protein kinase (MAPK) (16). Additionally, knockdown of 14-3-3ζ was
observed to enhance radio-induced apoptosis in liver cancer
stem-like cells (17). Wang et
al (18) demonstrated that
14-3-3ζ was upregulated in the hepatocarcinomatous environment,
attenuated the anti-tumor function of tumor-infiltrating T cells
and may have partially been transferred from liver cancer cells to
T cells via exosomes. A recent study revealed that 14-3-3ζ
regulated the stability of heme oxygenase 1, which in turn promoted
cancer cell proliferation via activation of STAT3 signaling in HCC
(19). Nonetheless, the prognostic
roles of yhwaz at the mRNA level in HCC remain to be clearly
elucidated. In the present study, bioinformatics tools were used to
analyze the functional effects of yhwaz expression in liver
cancer. The expression of yhwaz was shown to be elevated in
liver cancer tissues and cell lines and this upregulation was
associated with poor prognosis. Additionally, it was demonstrated
that mutations in yhwaz may have important implications in
the survival rates of patients with liver cancer. By contrast,
upregulated expression levels of microRNAs (miRNAs/miRs) that
targeted yhwaz were associated with improved prognosis. The
results demonstrated that the regulatory network of 14-3-3ζ and its
interacting proteins serve an important role in the development and
prognosis of HCC, suggesting the potential application of 14-3-3ζ
as a therapeutic target for liver cancer. Additionally, the results
of the present study support additional in vitro and in
vivo studies on the functional effects of 14-3-3ζ expression in
HCC.
Materials and methods
Bioinformatics tools and
databases
Several bioinformatics tools were used to analyze
yhwaz expression in patients with HCC. The specific tools
that were used are listed in Table
I.
 | Table I.Bioinformatics tools used for
analysis in the present study. |
Table I.
Bioinformatics tools used for
analysis in the present study.
| Databases | Samples | (Refs.) |
|---|
| HCCDB | Tissues | (20) |
| TCGA | Tissues | (21) |
| GTEx | Tissues | (22) |
| UALCAN | Tissues | (23) |
| Oncomine | Tissues | (24) |
| KEGG | N/A | (25) |
| Kaplan-Meier
plotter | Tissues | (26) |
| cBioPortal | N/A | (27,28) |
| TargetScan | N/A | (29) |
| GSEA | N/A | (30,31) |
| GeneMANIA | N/A | (32) |
| GEPIA | Tissues | (33) |
Integrative molecular database of
hepatocellular carcinoma (HCCDB)
The HCCDB is a database that contains information on
HCC expression (20). In the current
database release, HCCDB archived 15 public HCC gene expression
datasets (HCCDB1, 3, 4, 6–9, 11–18) containing a total of 3917
samples from The Cancer Genome Atlas (TCGA) (21).
Genotype-tissue expression (GTEx)
Among the 15 datasets from GTEx (https://www.gtexportal.org) (22), 12 datasets contain both tumor and the
adjacent normal samples, whereas only tumor samples are available
for the three remaining datasets.
UALCAN
UALCAN is an interactive web resource for studying
cancer transcriptome data (23). In
particular, 50 healthy individuals and 371 patients with primary
HCC were used to analyze the association among yhwaz levels
and their clinical characteristics.
Oncomine
Oncomine database is a bioinformatics initiative
aimed at collecting, standardizing, analyzing, and delivering
cancer transcriptome data to the biomedical research community
(24). Two microarray datasets
Roessler Liver (43 samples) and Roessler Liver 2 (445
samples) were used for analysis in this study. Based on these open
source bioinformatics platforms, yhwaz expression profiles
were obtained for both human HCC tissues and cell lines.
Kyoto encyclopedia of genes and
genomes (KEGG) pathway analysis
KEGG (25) is a
database resource for understanding high-level functions and
utilities of the biological system, such as the cell, the organism
and the ecosystem, from molecular-level information, especially
large-scale molecular datasets generated by genome sequencing and
other high-throughput experimental technologies.
Kaplan-Meier plotter
The Kaplan-Meier plotter (26) with the log-rank test was used to
determine disease prognosis, including overall survival (OS) time
and post-progression survival (PPS) time. Clinical data from TCGA
were used for Kaplan-Meier plotter analysis in order to evaluate
the clinical relevance of yhwaz mRNA expression levels in
HCC. In this project 364 (male, 250; female, 121) HCC RNA-seq
samples were used.
CBioPortal
CBioPortal (27,28) can
be used to download and analyze large-scale cancer genomics
datasets and was used in the present study to analyze mutations in
yhwaz. In this project 1065 HCC samples were used for
analysis.
TargetScan (version 7.1)
TargetScan (29) was
used to predict the biological targets of miRNAs by searching for
the presence of conserved 8mer and 7mer sites that matched the seed
region of each miRNA.
Gene set enrichment analysis
(GSEA)
GSEA (30,31) is a computational method that
determines whether an a priori defined set of genes shows
statistically significant, concordant differences between two
biological states.
GeneMANIA
GeneMANIA (32) is a
flexible, user-friendly web interface for constructing
protein-protein interaction (PPI) network, generating hypotheses
about gene function, analyzing gene lists, and prioritizing genes
for functional assays.
Gene expression profiling interactive
analysis (GEPIA)
GEPIA (33) is a
newly developed interactive web server for analyzing the RNA
sequencing expression data of 9,736 tumors and 8,587 normal samples
from the TCGA and the GTEx projects, using a standard processing
pipeline.
Tumor grade and tumor stages
The tumor grade is divided into four subtypes: Grade
1, well differentiated (low grade); grade 2, moderately
differentiated (intermediate grade); grade 3, poorly differentiated
(high grade); and grade 4, undifferentiated (high grade). The tumor
stage class depends on the tumor-node-metastasis staging system
(AJCC 8th edition) (34).
Statistical analysis
Data are expressed as mean ± SD and analyzed for
significance using GraphPad Prism 6.00 software (IBM Corp.).
Difference between two-groups was assessed using Student's t-test.
One-way ANOVA followed by Newman-Keuls post hoc testing (95%
confidence) was used to determine difference among more than two
groups. The survival analysis was illustrated by Kaplan-Meier
curves with log-rank test. Univariate and multivariate survival
analyses were performed using the likelihood ratio test of the
stratified Cox proportional hazards model of SPSS 17.0 (IBM Corp.).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Yhwaz is upregulated in HCC cell lines
and tissues
The results from the analysis of the data in the
GTEx portal revealed that ywhaz is expressed in multiple
organs and tissues (Fig. S1). As
yhwaz has been reported to affect a number of different
types of cancer, such as liver, lung and breast (3), HCCDB, an analytical tool for gene
expression profiling, was used to determine yhwaz expression
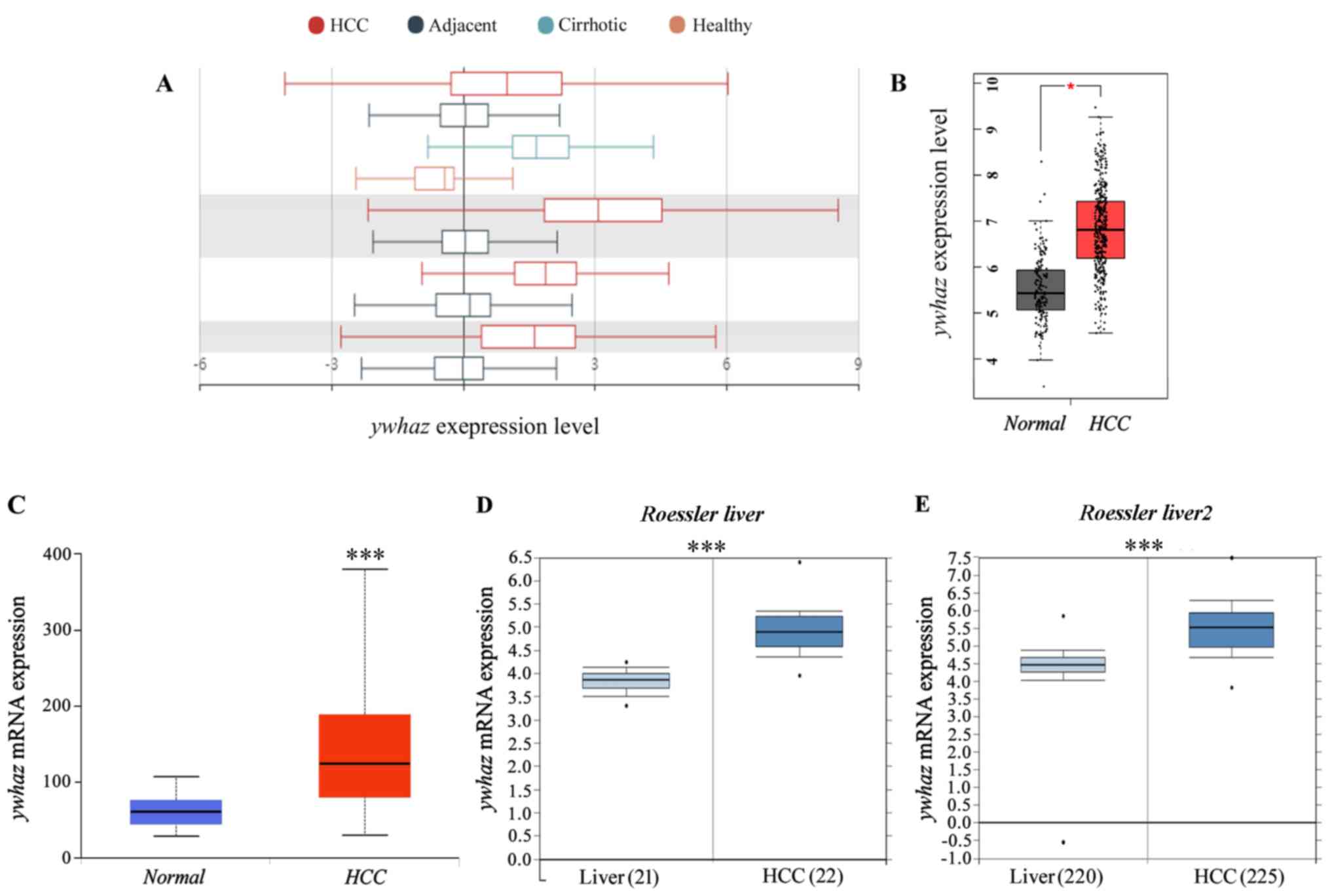
in tumor and other tissues. As presented in Fig. 1A and Table II, yhwaz expression was
significantly higher in tumor tissues compared with tissue adjacent
to HCC tissues. Additionally, in patients with cirrhosis,
yhwaz expression was higher compared with healthy human
liver tissues. Liver HCC tissues exhibited upregulated expression
levels of yhwaz compared with healthy liver tissue (Fig. 1B). In order to investigate the
changes in yhwaz expression between HCC and adjacent
non-tumor tissues, yhwaz expression profiles were used from
different independent bioinformatics databases. Based on the gene
expression profiles obtained from the UALCAN database, yhwaz
was determined to be upregulated in the majority of HCC tissues
(Fig. 1C). To confirm this finding,
two microarray datasets from the Oncomine database (24) were analyzed (Fig. 1D and E). yhwaz expression
levels were significantly increased in HCC tissues compared with in
healthy liver tissues. Taken together, these data demonstrate that
increased mRNA expression of yhwaz contributed to HCC,
indicating its potential role in liver cancer.
 | Table II.Summary of yhwaz expression
profiles based on HCCDB datasets. |
Table II.
Summary of yhwaz expression
profiles based on HCCDB datasets.
| Datasets | P-value | Type | Nums | Mean | SD | IQR |
|---|
| HCCDB3 |
7.20×10−15 | HCC | 268 | 10.43 | 3.269 | 4.115 |
|
|
| Adjacent | 243 | 8.609 | 1.632 | 1.771 |
|
|
| Cirrhotic | 40 | 11.55 | 1.868 | 2.095 |
|
|
| Healthy | 6 | 7.790 | 1.401 | 1.458 |
| HCCDB4 |
7.58×10−66 | HCC | 240 | 8.692 | 0.5139 | 0.6828 |
|
|
| Adjacent | 193 | 7.886 | 0.2552 | 0.2674 |
| HCCDB6 |
2.62×10−52 | HCC | 225 | 9.271 | 0.8106 | 0.9875 |
|
|
| Adjacent | 220 | 8.014 | 0.7027 | 0.8710 |
| HCCDB18 |
5.66×10−28 | HCC | 212 | 6.626 | 0.8582 | 1.135 |
|
|
| Adjacent | 177 | 5.774 | 0.5318 | 0.5900 |
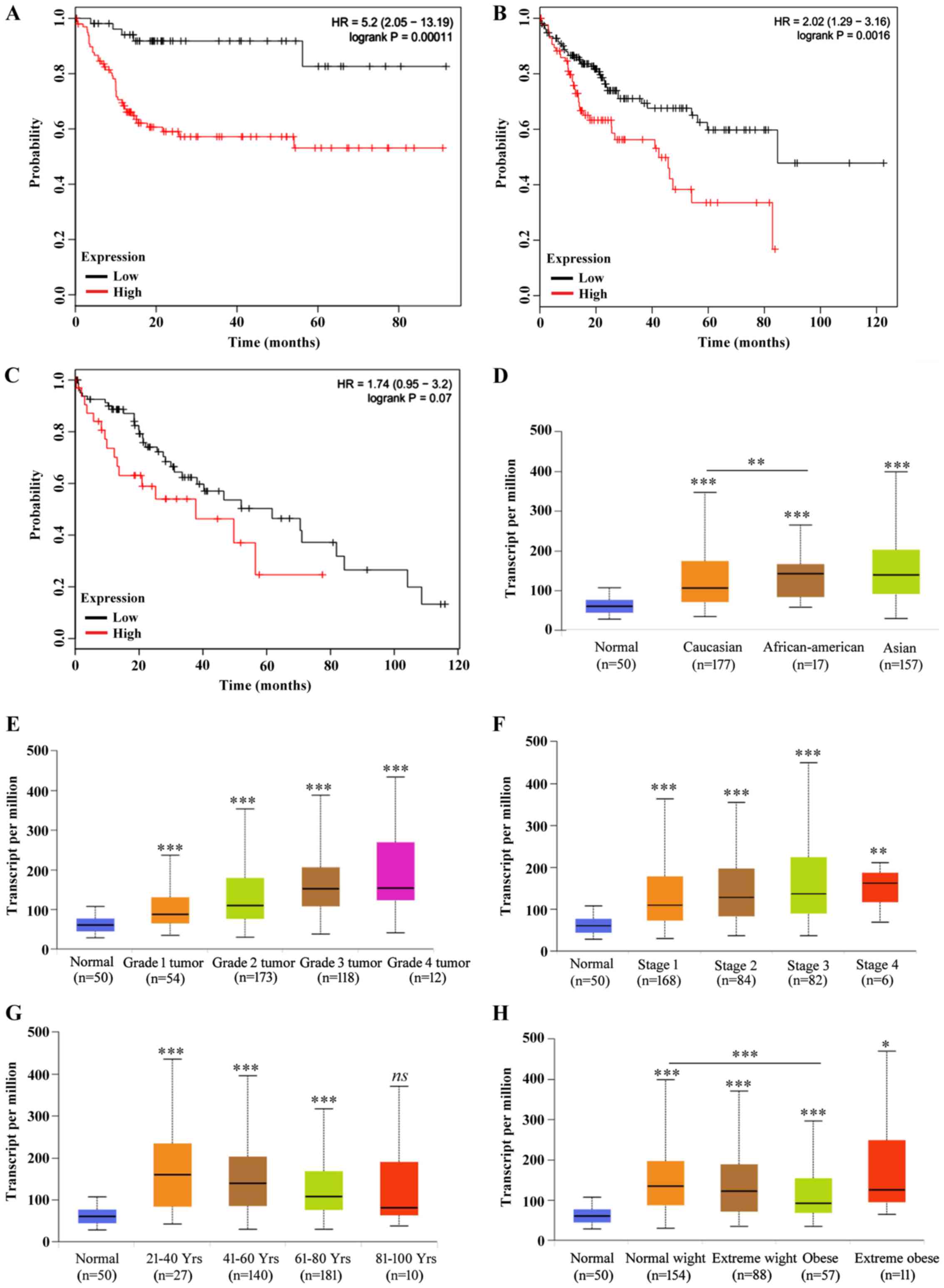
Yhwaz expression may be a prognostic
factor in HCC
Although yhwaz has been demonstrated to
participate in the development of multiple types of cancer, there
are no clear reports on the association between its expression and
clinical prognosis in HCC. The prognostic efficacy of the mRNA
expression of ywhaz in patients with HCC was analyzed using
the Kaplan-Meier plotter (26). As
presented in Fig. 2A-C and Table II, increased expression of
yhwaz was found to increase the risk of death and decrease
survival probability in female (Fig.
2A) and male (Fig. 2B) patients
with HCC alone or all both sex combined (Fig. 2C; Table
III). Therefore, yhwaz expression analysis was performed
with regard to the clinicopathological characteristics of patients.
As presented in Fig. 2D,
yhwaz expression in African-American patients with HCC was
significantly higher compared with Caucasian patients. The
expression levels of yhwaz were shown to increase with tumor
grade (Fig. 2E). Based on tumor
stages (34), the ywhaz
expression level was significant higher in patients of all tumor
stages compared with healthy individuals, but yhwaz
expression levels in stage 3 cancer were significantly higher
compared with stage 1 (P<0.01; Fig.
2F), and there were no other significant difference amongst the
other stages. The expression levels of yhwaz decreased with
age in patients with HCC (Fig. 2G).
The weight of patients seemed to have less influence on the
expression of yhwaz, with the only significant difference
observed between normal weight patients and obese patients with HCC
(Fig. 2H), and Together, these data
suggest that high expression levels of yhwaz may predict
tumors with increased malignancy.
 | Table III.Association between yhwaz
expression and sex in patients with hepatocellular carcinoma. |
Table III.
Association between yhwaz
expression and sex in patients with hepatocellular carcinoma.
| Sex | Gene IDs | HR | 95% CI | P-value |
|---|
| Female | 7534 | 5.20 | 2.05–13.19 | 0.00011 |
| Male | 7534 | 2.02 | 1.29–3.16 | 0.0016 |
| All | 7534 | 1.74 | 0.95–3.20 | 0.07 |
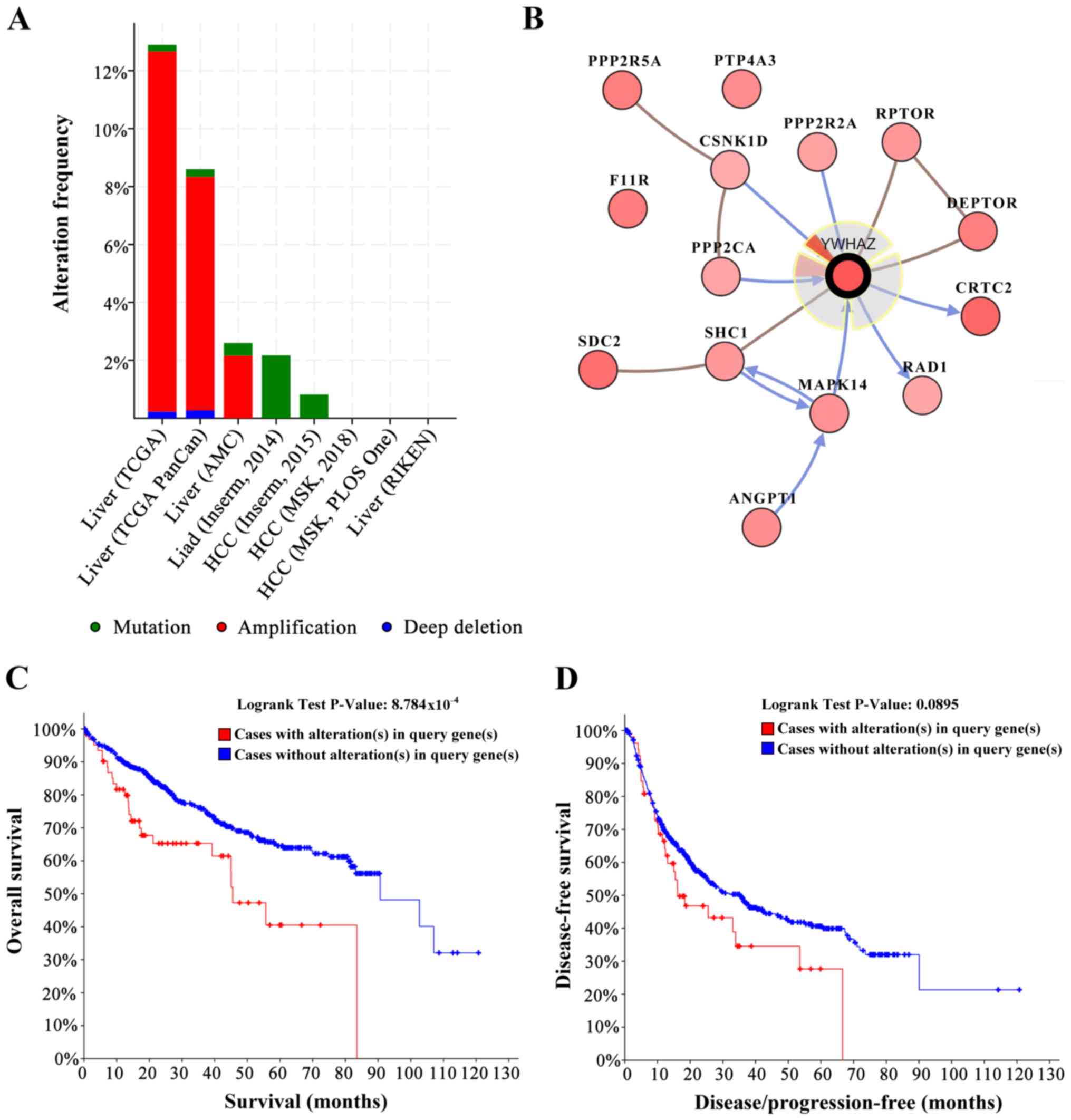
Yhwaz mutations affect the outcome of
patients with HCC
Mutations in a gene may not always affect the
function of the coded proteins. In the cBioPortal data sets, based
on the TCGA, there were 8 studies (35–40) on
HCC, and mutations, amplification and deep deletions were observed
in the yhwaz sequence in these datasets (Fig. 3A). yhwaz was mutated or
otherwise altered in 98 (7.00%) of the 1,487 patients. These
alterations were amplified in 90 cases (6.00%), deep deletion in 2
cases (0.13%) and mutations in 6 cases (0.40%). Therefore,
amplification was the most common type of yhwaz alteration
in HCC. The 6 mutations identified are shown in Table IV, and the single nucleotide
polymorphisms seen were S207I, N108S, R127G and C94S. The
biological interaction network of yhwaz was next identified
in HCC. The tab Network in cBioPortal was used to show yhwaz
neighboring genes that were altered with a frequency >15%
(Fig. 3B; Table V). The neighbor genes of yhwaz
with highest frequency of alterations were CRTC2 (38.1%), SDC2
(34.4%) and F11R (29.2%). OS and disease-free survival were
significantly decreased in patients with yhwaz alterations
compared with individuals without alterations (Fig. 3D), suggesting that alterations in
yhwaz may affected the survival of patients with HCC.
 | Table IV.Known mutations in the yhwaz
gene. |
Table IV.
Known mutations in the yhwaz
gene.
| Sample ID | Protein change | Type of
mutation | Copy no. | COSMIC | Allele frequency
(T) | No. of
mutations |
|---|
| CHC1915T | N108S | Missense |
| 1 |
| 39 |
| CHC703T | R127G | Missense |
|
|
| 104 |
| CHC1915T | N108S | Missense |
| 1 |
| 36 |
| H093892 | S207I | Missense | Diploid |
| 0.04 | 74 |
|
TCGA-DD-A115-01 | C94S | Missense | ShallowDel |
| 0.08 | 66 |
|
TCGA-DD-A115-01 | C94S | Missense | ShallowDel |
| 0.07 | 80 |
 | Table V.Type and frequency of yhwaz
neighbor gene alterations in patients with hepatocellular
carcinoma. |
Table V.
Type and frequency of yhwaz
neighbor gene alterations in patients with hepatocellular
carcinoma.
| Gene symbol | Amplification | Homozygous
deletion | Upregulation | Downregulation | Mutation | Total
alteration |
|---|
| YWHAZ | 15.3 | 0.3 | 23.6 | 0 | 0.3 | 30.8 |
| AGPT1 | 16.7 | 0.3 | 7.5 | 0 | 1.4 | 23.1 |
| CRTC2 | 12.2 | 0 | 34.2 | 0 | 0.8 | 38.1 |
| CSNK1D | 6.9 | 0.6 | 11.1 | 0.6 | 0 | 15.8 |
| DEPTOR | 17.5 | 0.3 | 14.7 | 0 | 0.3 | 28.3 |
| F11R | 11.4 | 0 | 24.2 | 0 | 0.3 | 29.2 |
| MAPK14 | 3.1 | 0.3 | 20.3 | 1.1 | 0.6 | 22.5 |
| NTRK1 | 11.9 | 0 | 1.9 | 0 | 1.1 | 15.0 |
| PPP2CA | 0.6 | 0 | 13.9 | 3.1 | 0.6 | 17.5 |
| PPP2R2A | 0 | 6.1 | 2.5 | 9.7 | 0.8 | 18.1 |
| PPP2R5A | 8.9 | 0 | 24.2 | 0 | 0 | 28.3 |
| PTP4A3 | 16.4 | 0.3 | 11.7 | 0 | 0.8 | 23.9 |
| SDC2 | 15.3 | 0.3 | 21.9 | 2.5 | 0.3 | 34.4 |
| SHC1 | 13.6 | 0 | 8.9 | 0 | 0.8 | 20.6 |
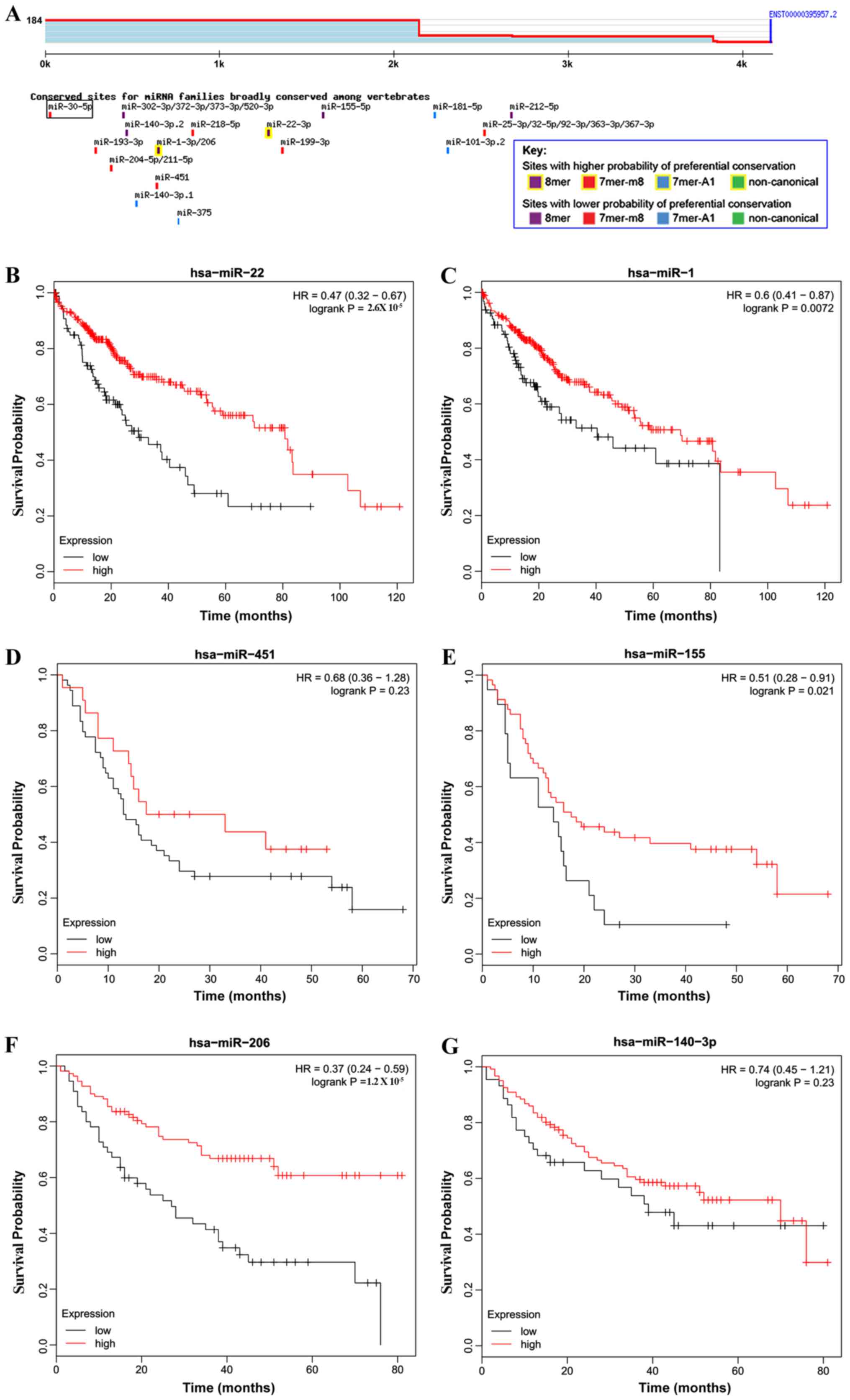
Increased expression of miRNAs that
target yhwaz are associated with improved survival time in patients
with HCC
miR-22 has been reported to target yhwaz and
inhibit its expression in HCC cells (41). miRNAs that targeted yhwaz were
predicted using TargetScan. As presented in Fig. 4A, miR-22 and miR-1/miR-206 were
predicted to exhibit a higher probability of binding with
yhwaz. miR-451 negatively regulated the expression of
yhwaz through binding with the 3′untranslated region of
yhwaz (42). miR-155 and
miR-140 were also predicted to exhibit a high probability of
binding with yhwaz. For reasons not yet known, the survival
curves for miR-140-3p (Fig. 4G)
exhibited a late stage crossover. The most mentioned miRNAs that
were predicted to bind with yhwaz, were associated with improved
survival time in patients with HCC when expression was upregulated
(Fig. 4B-G; Table VI). Together, these data show that
miRNAs that were predicted to target yhwaz were associated
with improved survival time in patients with HCC.
 | Table VI.Association between yhwaz
targeting miRNAs and survival time in patients with hepatocellular
carcinoma. |
Table VI.
Association between yhwaz
targeting miRNAs and survival time in patients with hepatocellular
carcinoma.
| miRNAs | Gene IDs | HR | 95% CI | P-value |
|---|
| miR-22 | Has-miR-22 | 0.47 | 0.32–0.67 |
2.6×10−5 |
| miR-1 | has-miR-1 | 0.6 | 0.41–0.87 | 0.0072 |
| miR−451 | has-miR-451 | 0.68 | 0.36–1.28 | 0.23 |
| miR−155 | has-miR-155 | 0.51 | 0.28–0.91 | 0.021 |
| miR−206 | has-miR-206 | 0.37 | 0.24–0.59 |
1.2×10−5 |
| miR−140 | has-miR-140-3p | 0.74 | 0.45–1.21 | 0.23 |
KEGG pathway analysis of co-expression
genes correlated with yhwaz and its networks of kinase and miRNA
targets in HCC
To further examine the targets of yhwaz in
HCC, the KEGG pathway analysis was used to determine the kinase and
miRNA target networks of associated gene sets generated by Gene Set
Enrichment Analysis (GSEA) (30,31).
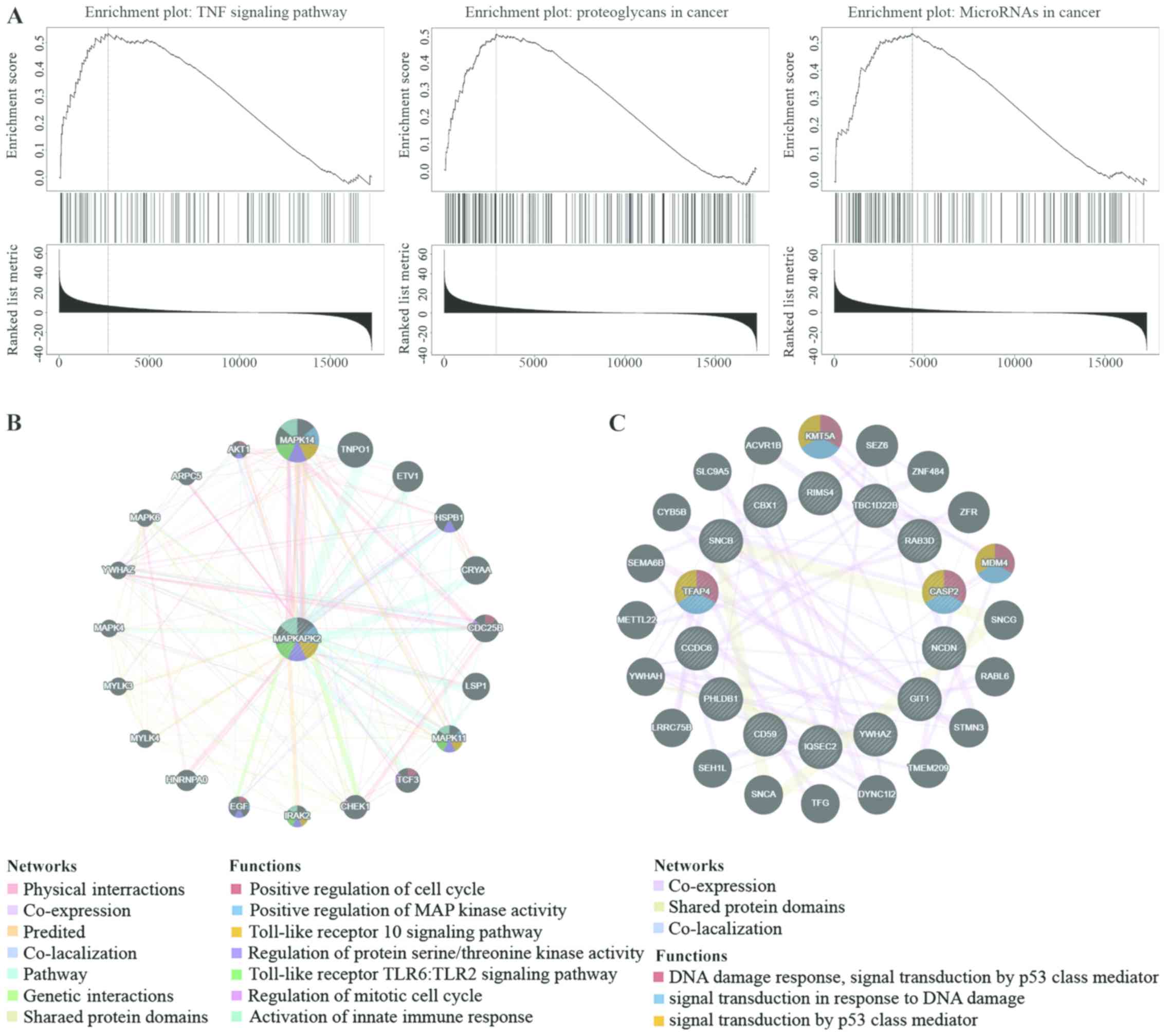
KEGG pathway analysis showed enrichment in tumor necrosis factor,
proteoglycans in cancer and miRNAs in cancer pathways (Fig. 5A). The miRNA-target network was
associated with miR-149 (GAG CCA G), miR-133A/miR-133B (GGG ACC A)
and miR-296 (GGG GCC C). The most significant target networks were
the kinase-target networks associated primarily with the MAPK2, Rho
Associated Coiled-Coil Containing Protein Kinase 1 and Polo-like
Kinase 1 (Table VII). The
protein-protein interaction network constructed by GeneMANIA
(32) revealed associations amongst
genes for MAPKAPK2 and miRNA-149. The GSEA for MAPK2 is responsible
primarily for regulating the cell cycle, kinase activity and the
immune response (Fig. 5B), and the
GSEA for miRNA-527 (Fig. 5C) was
involved primarily in the regulation of cell activation, kinase
activity and leukocyte activation.
 | Table VII.Kinase and miRNA target networks of
yhwaz in hepatocellular carcinoma. |
Table VII.
Kinase and miRNA target networks of
yhwaz in hepatocellular carcinoma.
| Enriched
category | Geneset | Leading
EdgeNum | FDR |
|---|
| miRNA target | GAGCCAG,
MIR-149 | 46 | 0.039256 |
|
| GGGACCA, MIR-133A,
MIR-133B | 52 | 0.038166 |
|
| GGGGCCC,
MIR-296 | 21 | 0.037802 |
| Kinase target |
Kinase_MAPKAPK2 | 12 | 0.043584 |
|
| Kinase_ROCK1 | 19 | 0.035556 |
|
| Kinase_PLK1 | 41 | 0.030585 |
Discussion
The 14-3-3 family of proteins serve various roles in
signaling and interact with several protein partners, including
their function as adaptors that stimulate protein-protein
interactions (3,41). Among the 14-3-3 proteins, the 14-3-3ζ
isotype is one of the most studied members of this family (42). 14-3-3ζ interacts with numerous key
cellular proteins involved in tumor development and progression
(43). The importance of 14-3-3ζ in
the development and progression of cancer has been demonstrated in
a number of different types of cancer (44). In the present study, bioinformatics
analysis was used to determine the prognostic value of yhwaz
in liver cancer, and the results revealed that yhwaz
expression was upregulated in HCC tissues and cell lines.
Upregulated levels of yhwaz were significantly associated
with a poor prognosis. Mutations in yhwaz significantly
affected the survival time of patients with HCC, and miRNAs that
targeted yhwaz were associated with outcomes of HCC.
14-3-3ζ has been suggested to be a potential
prognostic marker and therapeutic target in a number of different
types of cancer (43). 14-3-3ζ
overexpression increases Akt phosphorylation and further increased
hypoxia-inducible factor-1α expression in HCC cells (45,46). In
the present study, yhwaz expression was upregulated in the
major and different sub-types of HCC. 14-3-3ζ has been hypothesized
to bind with the hepatitis B virus (HBV) protein X (HBx) and
maintain its protein stability in HCC cells. In cancer cells where
14-3-3ζ was silenced, the cells exhibited decreased migratory and
invasive capacities, and this was accompanied by decreased
expression of HBx (47). Together,
these data suggest that upregulated expression of 14-3-3ζ may
increase the risk of HBV infection.
14-3-3ζ targets and affects the phosphorylation of a
number of proteins, a number of which are involved in
tumor-promoting processes, such as regulation of autophagy and
tumor suppressor pathways (48).
Thus, alterations in yhwaz expression may result in cancer
of the liver through regulation of these pathways and processes.
The most common type of alteration observed in HCC was
amplification, which was observed in 6% of cases. Multiple
yhwaz mutations have been observed in patients with liver
cancer, the majority of which are primarily phosphorylation sites.
p-AKT is the one of the targets of 14-3-3ζ, upregulation of both
14-3-3ζ. p-Akt in patients with HCC predicts a poor prognosis, and
14-3-3ζ triggers activation of the Akt signaling pathway, thus
contributing to the development of HCC (49). However, in patients with upregulated
expression of miRNAs that target ywhaz, prognosis was
predicted to be improved compared with patients with lower
expression levels of the same miRNAs. Enrichment analysis of
yhwaz co-expression genes determined that the majority of
the interacting genes were involved in processes associated with
cancer progression, and yhwaz target GSEA may assist in
identifying important networks of target kinases and miRNAs.
The present study used the online tools to perform
target gene analysis on tumor data from public databases. At the
same time, the limitation is that transcriptome sequencing can only
detect static mutations; it does not directly provide information
on protein activity or expression levels. These issues should be
addressed in subsequent research using molecular biology
techniques. The other one limitation is that the LIHC samples
contained a relatively small number of stage 4 patients, but the
clinical reality is that most patients with HCC are diagnosed for
the first time when the disease progresses, and thus, the prognosis
is extremely poor. Therefore, the results of the present study
should be validated in clinical samples including a full coverage
of different ethnic groups and HCC stages.
The present study provided theoretical evidence of
the importance of yhwaz expression in hepatocarcinogenesis
and highlighted its potential as a marker in HCC. Based on the
results of the present study, it may be hypothesized that targeting
14-3-3ζ in patients with liver cancer may effectively improve the
survival rates and prognosis of patients.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by Zhejiang Provincial
Natural Science Foundation of China (grant no: LQ18C070001), the
Project Grant from Wenzhou Science and Technology Bureau (grant no:
Y20180722) and the Program of Science and Technology Development in
Yancheng (grant no: YK2016053).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL, LS and XY designed and conducted the
experiments. YL and LS wrote and revised the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
European Association For The Study Of The
Liver1; European Organisation For Research And Treatment Of Cancer,
. EASL-EORTC clinical practice guidelines: Management of
hepatocellular carcinoma. J Hepatol. 56:908–943. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fan X, Cui L, Zeng Y, Song W, Gaur U and
Yang M: 14-3-3 proteins are on the crossroads of cancer, aging, and
age-related neurodegenerative disease. Int J Mol Sci. 20(pii):
E35182019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sluchanko NN and Gusev NB: Moonlighting
chaperone-like activity of the universal regulatory 14-3-3
proteins. FEBS J. 284:1279–1295. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
van Heusden GP: 14-3-3 proteins:
Regulators of numerous eukaryotic proteins. IUBMB Life. 57:623–629.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yaffe MB: How do 14-3-3 proteins
work?-Gatekeeper phosphorylation and the molecular anvil
hypothesis. FEBS Lett. 513:53–57. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lin M, Morrison CD, Jones S, Mohamed N,
Bacher J and Plass C: Copy number gain and oncogenic activity of
YWHAZ/14-3-3zeta in head and neck squamous cell carcinoma. Int J
Cancer. 125:603–611. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bajpai U, Sharma R, Kausar T, Dattagupta
S, Chattopadhayay TK and Ralhan R: Clinical significance of 14-3-3
zeta in human esophageal cancer. Int J Biol Markers. 23:231–237.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Matta A, Siu KW and Ralhan R: 14-3-3 zeta
as novel molecular target for cancer therapy. Expert Opin Ther
Targets. 16:515–523. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lu J, Guo H, Treekitkarnmongkol W, Li P,
Zhang J, Shi B, Ling C, Zhou X, Chen T, Chiao PJ, et al: 14-3-3zeta
Cooperates with ErbB2 to promote ductal carcinoma in situ
progression to invasive breast cancer by inducing
epithelial-mesenchymal transition. Cancer Cell. 16:195–207. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu J, Acharya S, Sahin O, Zhang Q, Saito
Y, Yao J, Wang H, Li P, Zhang L, Lowery FJ, et al: 14-3-3ζ turns
TGF-β's function from tumor suppressor to metastasis promoter in
breast cancer by contextual changes of Smad partners from p53 to
Gli2. Cancer Cell. 27:177–192. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cornell B and Toyo-Oka K: Deficiency of
14-3-3ε and 14-3-3ζ by the Wnt1 promoter-driven Cre recombinase
results in pigmentation defects. BMC Res Notes. 9:1802016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu J, Chen L, Chen Y, Hasan MK, Ghia EM,
Zhang L, Wu R, Rassenti LZ, Widhopf GF, Shen Z, et al: Wnt5a
induces ROR1 to associate with 14-3-3ζ for enhanced chemotaxis and
proliferation of chronic lymphocytic leukemia cells. Leukemia.
31:2608–2614. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang B, Shi Y, Gong A, Pan Z, Shi H, Yang
H, Fu H, Yan Y, Zhang X, Wang M, et al: HucMSC Exosome-delivered
14-3-3ζ orchestrates self-control of the Wnt response via
modulation of YAP during cutaneous regeneration. Stem Cells.
34:2485–2500. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Choi JE, Hur W, Jung CK, Piao LS, Lyoo K,
Hong SW, Kim SW, Yoon HY and Yoon SK: Silencing of 14-3-3ζ
over-expression in hepatocellular carcinoma inhibits tumor growth
and enhances chemosensitivity to Cis-diammined dichloridoplatium.
Cancer Lett. 303:99–107. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Davidson CE, Reese BE, Billingsley ML and
Yun JK: The protein stannin binds 14-3-3zeta and modulates
mitogen-activated protein kinase signaling. Brain Res Mol Brain
Res. 138:256–263. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee YK, Hur W, Lee SW, Hong SW, Kim SW,
Choi JE and Yoon SK: Knockdown of 14-3-3ζ enhances radiosensitivity
and radio-induced apoptosis in CD133(+) liver cancer stem cells.
Exp Mol Med. 46:e772014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang X, Shen H, Zhangyuan G, Huang R,
Zhang W, He Q, Jin K, Zhuo H, Zhang Z, Wang J, et al: 14-3-3ζ
delivered by hepatocellular carcinoma-derived exosomes impaired
anti-tumor function of tumor-infiltrating T lymphocytes. Cell Death
Dis. 9:1592018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Song J, Zhang X, Liao Z, Liang H, Chu L,
Dong W, Zhang X, Ge Q, Liu Q, Fan P, et al: 14-3-3ζ inhibits heme
oxygenase-1 (HO-1) degradation and promotes hepatocellular
carcinoma proliferation: Involvement of STAT3 signaling. J Exp Clin
Cancer Res. 38:32019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lian Q, Wang S, Zhang G, Wang D, Luo G,
Tang J, Chen L and Gu J: HCCDB: A database of hepatocellular
carcinoma expression atlas. Genomics Proteomics Bioinformatics.
16:269–275. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hutter C and Zenklusen JC: The cancer
genome atlas: Creating lasting value beyond its data. Cell.
173:283–285. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
GTEx Consortium: The genotype-tissue
expression (GTEx) project. Nat Genet. 45:580–585. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rhodes DR, Kalyana-Sundaram S, Mahavisno
V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ,
Kincead-Beal C, Kulkarni P, et al: Oncomine 3.0: Genes, pathways,
and networks in a collection of 18,000 cancer gene expression
profiles. Neoplasia. 9:166–180. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Altermann E and Klaenhammer TR:
PathwayVoyager: Pathway mapping using the Kyoto Encyclopedia of
Genes and Genomes (KEGG) database. BMC Genomics. 6:602005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lánczky A, Nagy Á, Bottai G, Munkácsy G,
Szabó A, Santarpia L and Győrffy B: miRpower: A web-tool to
validate survival-associated miRNAs utilizing expression data from
2178 breast cancer patients. Breast Cancer Res Treat. 160:439–446.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 42015.doi: 10.7554/eLife.05005.
|
|
30
|
Mootha VK, Lindgren CM, Eriksson KF,
Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E,
Ridderstråle M, Laurila E, et al: PGC-1alpha-responsive genes
involved in oxidative phosphorylation are coordinately
downregulated in human diabetes. Nat Genet. 34:267–273. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38:W214–W220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Amin MB, Edge S, Greene F, Byrd DR,
Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR,
Sullivan DC, et al: AJCC cancer staging manual. 8th. New York:
Springer; 2017, View Article : Google Scholar
|
|
35
|
Pilati C, Letouze E, Nault JC, Imbeaud S,
Boulai A, Calderaro J, Poussin K, Franconi A, Couchy G, Morcrette
G, et al: Genomic profiling of hepatocellular adenomas reveals
recurrent FRK-activating mutations and the mechanisms of malignant
transformation. Cancer Cell. 25:428–441. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Harding JJ, Nadakumar S, Armenia J, Khalil
DN, Albano M, Ly M, Shia J, Hechtman JF, Kundra R, El Dika I, et
al: Prospective genotyping of hepatocellular carcinoma: Clinical
Implications of next-generation sequencing for matching patients to
targeted and immune therapies. Clin Cancer Res. 25:2116–2126. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ahn SM, Jang SJ, Shim JH, Kim D, Hong SM,
Sung CO, Baek D, Haq F, Ansari AA, Lee SY, et al: Genomic portrait
of resectable hepatocellular carcinomas: Implications of RB1and
FGF19 aberrations for patient stratifications. Hepatology.
60:1972–1982. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Schulze K, Imbeaud S, Letouze E,
Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C,
Shinde J, Soysouvanh F, et al: Exome sequencing of hepatocellular
carcinomas identifies new mutational signatures and potential
therapeutic targets. Nat Genet. 47:505–511. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fujimoto A, Totoku Y, Abe T, Boroevich KA,
Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, et al:
Whole-genome sequencing of liver cancers identifies etiological
influences of mutation patterns and recurrent mutations in
chromatin regulators. Nat Genet. 7:760–764. 2012. View Article : Google Scholar
|
|
40
|
Hoadley KA, Yau C, Hinoue T, Wolf DM,
Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, et
al: Cell-of-origin patterns dominate the molecular classification
of 10,000 tumors from 33 types of cancer. Cell. 173:291–304. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kaplan A, Ottmann C and Fournier AE:
14-3-3 adaptor protein-protein interactions as therapeutic targets
for CNS diseases. Pharmacol Res. 125:114–121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Woodcock JM, Goodwin KL, Sandow JJ, Coolen
C, Perugini MA, Webb AI, Pitson SM, Lopez AF and Carver JA: Role of
salt bridges in the dimer interface of 14-3-3ζ in dimer dynamics,
N-terminal α-helical order, and molecular chaperone activity. J
Biol Chem. 293:89–99. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Neal CL and Yu D: 14-3-3ζ as a prognostic
marker and therapeutic target for cancer. Expert Opin Ther Targets.
14:1343–1354. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Aghazadeh Y and Papadopoulos V: The role
of the 14-3-3 protein family in health, disease, and drug
development. Drug Discov Today. 21:278–287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tang Y, Lv P, Sun Z, Han L, Luo B and Zhou
W: 14-3-3ζ up-regulates hypoxia-inducible factor-1α in
hepatocellular carcinoma via activation of PI3K/Akt/NF-κB signal
transduction pathway. Int J Clin Exp Pathol. 8:15845–15853.
2015.PubMed/NCBI
|
|
46
|
Tang Y, Liu S, Li N, Guo W, Shi J, Yu H,
Zhang L, Wang K, Liu S and Cheng S: 14-3-3ζ promotes hepatocellular
carcinoma venous metastasis by modulating hypoxia-inducible
factor-1α. Oncotarget. 7:15854–15867. 2016.PubMed/NCBI
|
|
47
|
Tang Y, Zhang Y, Wang C, Sun Z, Li L, Dong
J and Zhou W: 14-3-3ζ binds to hepatitis B virus protein X and
maintains its protein stability in hepatocellular carcinoma cells.
Cancer Med. 7:5543–5553. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tzivion G, Gupta VS, Kaplun L and Balan V:
14-3-3 proteins as potential oncogenes. Semin Cancer Biol.
16:203–213. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tang Y, Wang R, Zhang Y, Lin S, Qiao N,
Sun Z, Cheng S and Zhou W: Co-Upregulation of 14-3-3ζ and P-Akt is
associated with oncogenesis and recurrence of hepatocellular
carcinoma. Cell Physiol Biochem. 45:1097–1107. 2018. View Article : Google Scholar : PubMed/NCBI
|