Introduction
Ovarian cancer (OC) is a prevalent malignancy of the
female reproductive system. Its incidence rate is second only to
cervical cancer and endometrial cancer (1). Epithelial cancer is the most common
subtype in OC, followed by malignant germ cell tumor. The mortality
of ovarian epithelial cancer ranks first of the various
gynecological tumors, posing a serious threat to females (2). Due to the deep location, small volume
and atypical symptoms, OC is hard to diagnose at early stage
(3). Non-metastatic ovarian
epithelial cancer only accounts for 30% of all OC cases (4). It is prone to metastasize, especially
in the pelvic and abdominal organs. Sufficient diagnosis of OC at
early stage is urgently needed.
As a subtype of non-coding RNAs, lncRNAs are
long-chain RNAs over 200 nucleotides in length (5). They exert crucial roles in biological
processes, and have been extensively explored in genetic researches
(6). Accumulating evidence has shown
the vital functions of lncRNAs in the regulation of cellular
behavior (7). Several lncRNAs are
reported to participate in the occurrence and progression of OC
through interacting with other proteins, microRNAs or mRNAs. These
lncRNAs may serve as diagnostic and therapeutic targets of OC
(8).
It is reported that LINC00460 is involved in
regulation of malignant phenotypes of tumors (9). LINC00460 is abnormally upregulated in
multiple types of malignant tumors (10–16). Its
role in OC, however, has not been fully explored. This study aimed
to uncover the potential function of LINC00460 in the progression
of OC and the underlying mechanism to provide new directions for
developing treatment strategies for OC patients by targeting
LINC00460.
Patients and methods
Sample collection and ethical
statements
OC tissues were harvested from 40 OC patients
undergoing radical surgery in Yuncheng County Hospital of
Traditional Chinese Medicine (Heze, China). Normal ovarian tissues
were collected from healthy controls during the same period.
Samples were immediately placed in liquid nitrogen and preserved at
−80°C. Patients and their families of this study were fully
informed. This study was approved by Ethics Committee of Yuncheng
County Hospital of Traditional Chinese Medicine. Signed informed
consents were obtained from the patients and/or guardians.
Cell culture and transfection
Epithelial ovarian cell line (IOSE-386) and OC cell
lines (HO8910, SKOV-3, A2780 and ES-2) were obtained from American
Type Culture Collection (ATCC). Cell culture was conducted in
Roswell Park Memorial Institute-1640 (RPMI-1640) (HyClone) with 10%
fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific, Inc.) in
an incubator with 5% CO2 at 37°C. For cell transfection,
1.5 ml of serum-free medium and 0.5 ml of Lipofectamine™ 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) containing
transfection vectors were applied to each well of a 6-well plate.
Fresh medium was replaced 4–6 h later. Cells transfected for 24–48
h were collected for determination.
RNA extraction and quantitative
real-time polymerase chain reaction (qRT-PCR)
Cells were lysed and tissues were homogenized using
TRIzol method (Invitrogen; Thermo Fisher Scientific, Inc.). The
extracted RNAs were qualified using spectrometer and reverse
transcribed into cDNAs using PrimeScript RT reagent Kit (Takara).
QRT-PCR was carried out at 94°C for 5 min and 40 cycles at 94°C for
30 sec, 55°C for 30 sec and 72°C for 90 sec following the protocols
of SYBR Premix Ex Taq™ (Takara). The relative gene expression was
calculated using 2−∆Ct method.
Cell counting kit-8 (CCK-8) assay
Cells (1×105/ml) were pre-inoculated in a
96-well plate. At the appointed times, each well was replaced with
10 µl of CCK-8 solution (Dojindo). The absorbance at 450 nm was
measured by a microplate reader (Bio-Rad Laboratories).
Western blot analysis
Cells were lysed by radioimmunoprecipitation assay
(RIPA) (Beyotime Institute of Biotechnology) to extract the total
protein and underwent gel electrophoresis. Protein samples were
separated and incubated with primary antibody (Cell Signaling
Technology) overnight at 4°C. Subsequently, membranes were
incubated with corresponding secondary antibodies. Then, protein
band was exposed and captured by the Tanon detection system using
electrochemiluminescence (ECL) reagent (Thermo Fisher Scientific,
Inc.).
Chromatin immunoprecipitation
(ChIP)
Cells were subjected to cross-link with 1%
formaldehyde for 10 min at room temperature. Subsequently, the
cross-linked cells were lysed and sonicated for 30 min. Finally,
the sonicated lysate was immuno-precipitated with antibodies and
IgG.
Colony formation assay
Cells were seeded in a 6-well plate with 500 cells
per well and cultured for 2 weeks. Subsequently, cells were
subjected to 15-min fixation in 4% paraformaldehyde and 30-min
staining in 0.1% violet crystal. After removing the staining
solution, colonies were air dried and observed under a
microscope.
Cell cycle detection
Cells were digested for preparation of cell
suspension and washed with pre-cooled PBS twice. After pre-cooled
75% alcohol was added, cells were placed at 4°C in a refrigerator
for overnight fixation. Then the ethanol was discarded, and cells
were washed with 1X phosphate-buffered saline (PBS). After
centrifugation at 4°C, 950 × g for 5 min, cells were incubated with
100 µl propidium iodide and 100 µl RNA enzyme. Cell cycle was
detected using flow cytometry (FACSCalibur; BD Biosciences).
In vivo tumorigenesis in nude
mice
Female, 5-week-old, nude mice were provided by
Shandong University Experimental Animal Center, and habituated in
Specific Pathogen Free (SPF) level according to protocols approved
by the Animal Care and Use Committee. A2780 cells were transfected
with sh-NC or sh-LINC00460. Transfected cells (1×107)
were subcutaneously implanted in the right armpit of nude mice
(n=6). Tumor size was recorded every week. Four weeks later, mice
were sacrificed for collecting tumor tissues and weighing. This
study was approved by the Animal Ethics Committee of Yuncheng
County Hospital of Traditional Chinese Medicine Animal Center.
Statistical analysis
Statistical Product and Service Solutions (SPSS)
19.0 statistical software (IBM Corp.) was used for data analysis.
Data were expressed as mean ± standard deviation (mean ± SD).
Intergroup data were compared using the t-test. P<0.05 indicates
the difference is statistically significant.
Results
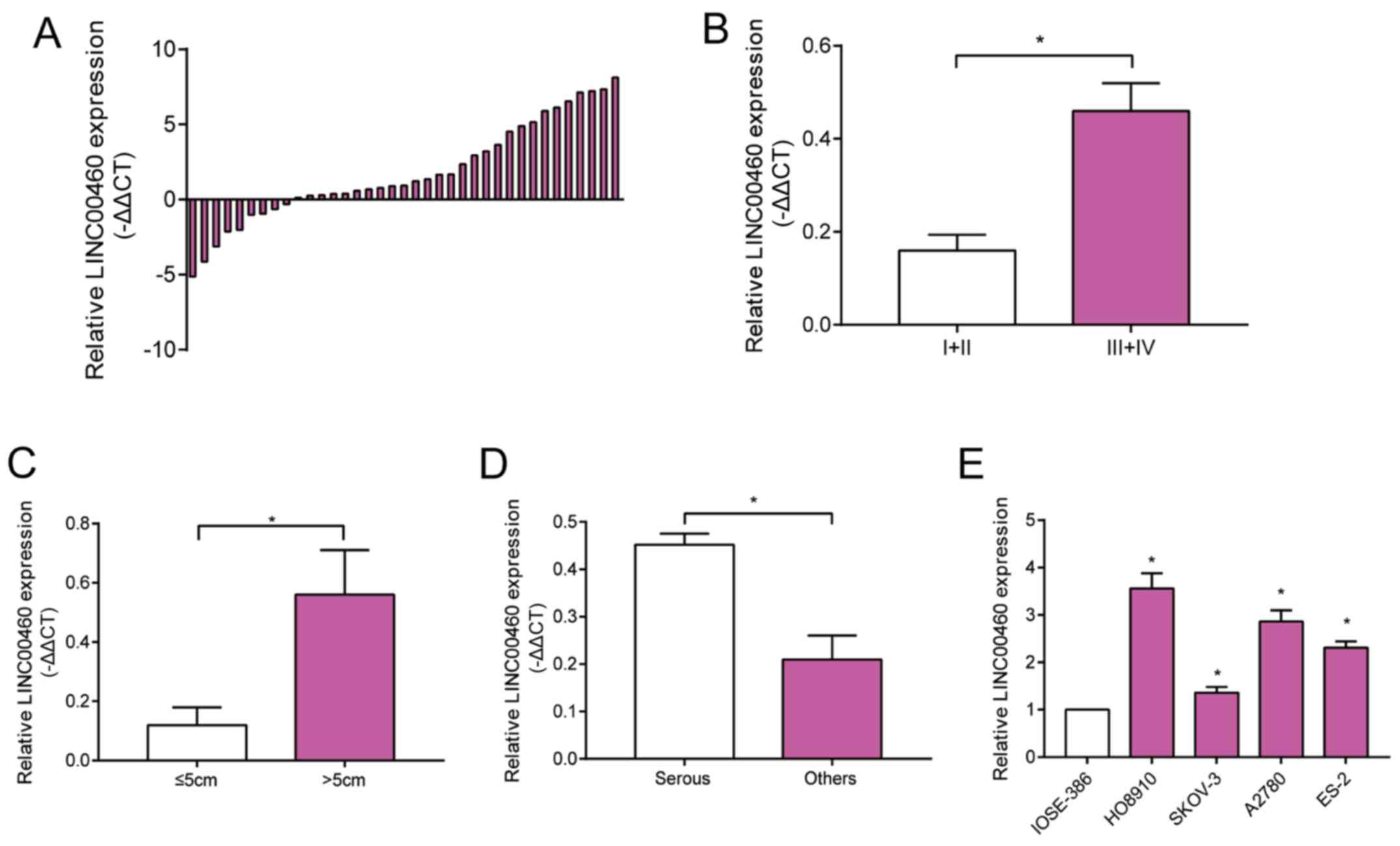
LINC00460 is upregulated in OC tissues
and cell lines
Expression level of LINC00460 was higher in OC
tissues relative to normal ovarian tissues (Fig. 1A). In particular, LINC00460 level
remained higher in OC patients with stage III–IV than those with
stage I–II (Fig. 1B). OC >5 cm in
tumor size presented higher abundance of LINC00460 compared with
those smaller than 5 cm (Fig. 1C).
Taking into consideration pathological subtypes of OC, serous OC
expressed higher level of LINC00460 relative to other subtypes
(Fig. 1D). LINC00460 was upregulated
in OC cell lines compared with that of epithelial ovarian cell line
(Fig. 1E). It is suggested that
LINC00460 was involved in the progression of OC.
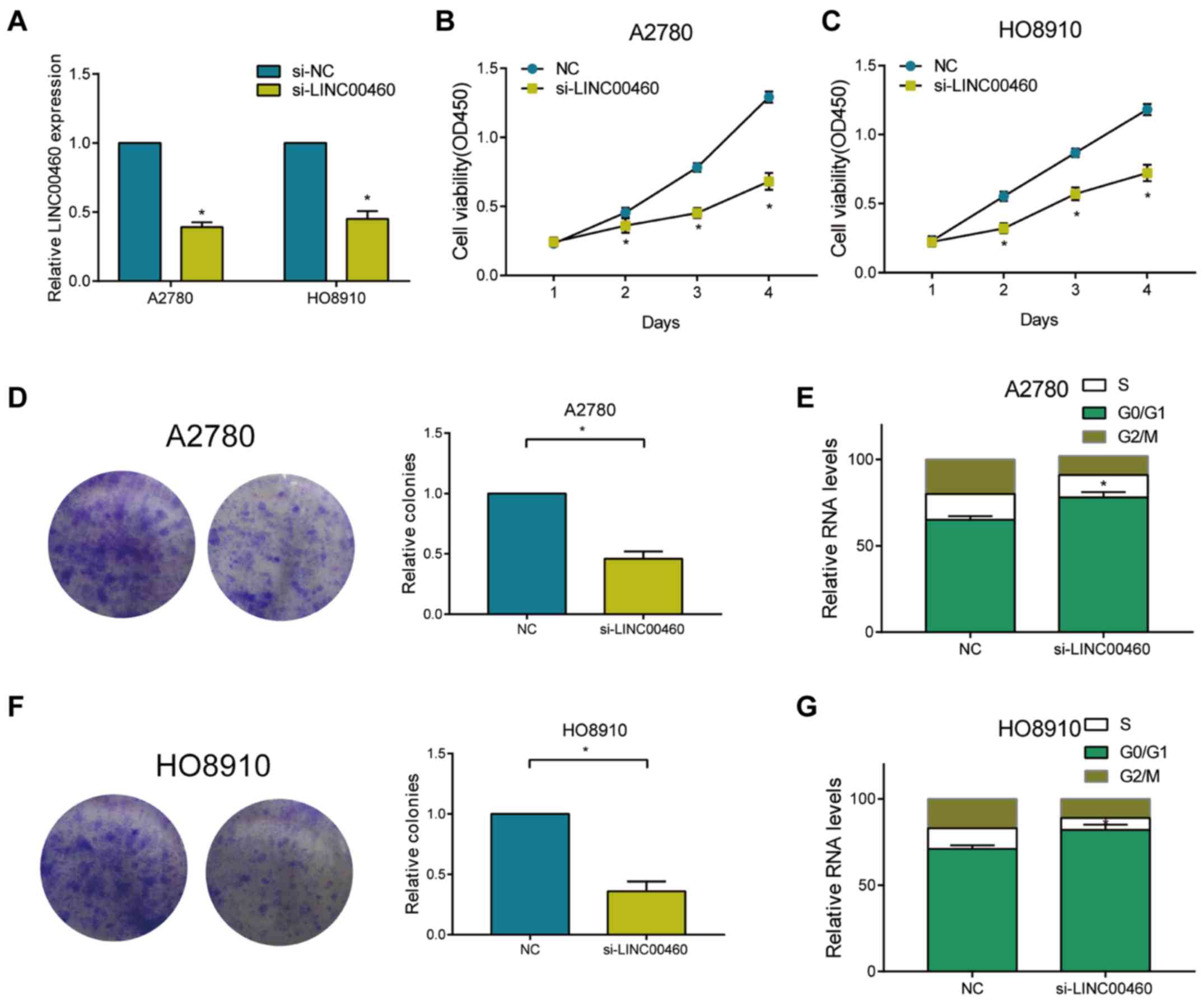
Knockdown of LINC00460 suppresses
proliferative ability and arrests cell cycle of OC cells
Among the four detected OC cell lines, HO8910 and
A2780 cells expressed high abundance of LINC00460. Transfection of
si-LINC00460 in these two cell lines markedly downregulated
LINC00460 level, showing a pronounced transfection efficacy
(Fig. 2A). After transfection of
si-LINC00460, the viability in A2780 and HO8910 markedly decreased
(Fig. 2B and C). Knockdown of
LINC00460 decreased the relative colony number in OC cells
(Fig. 2D and E). Flow cytometry
revealed elevated cell ratio in G0/G1 phase after transfection of
si-LINC00460 in OC cells, suggesting arrested cell cycle
progression (Fig. 2F and G).
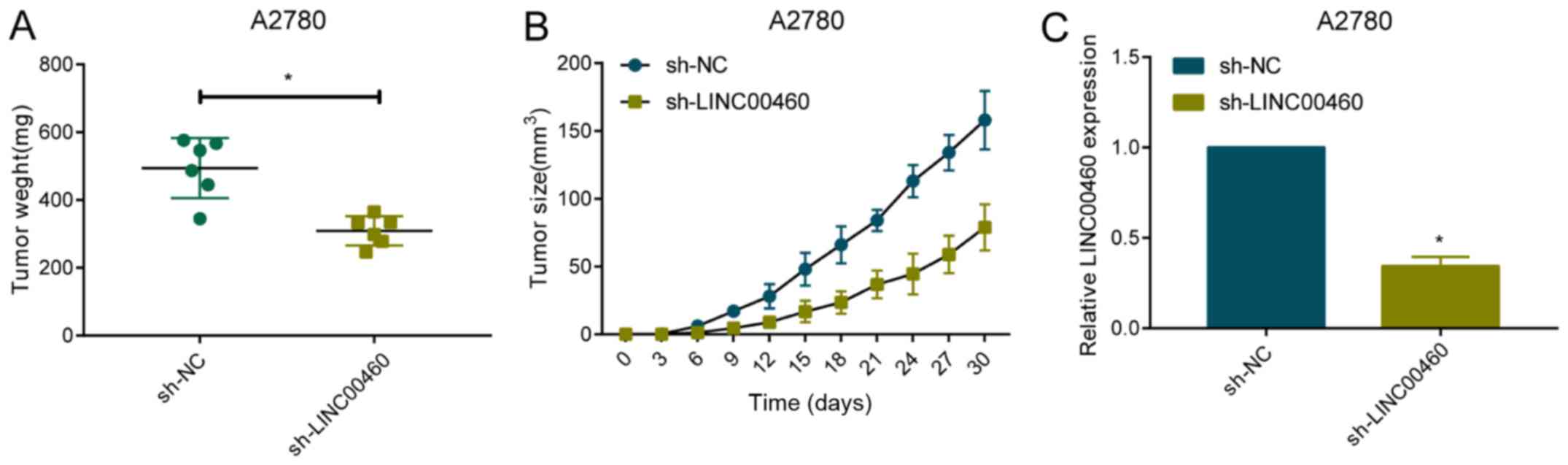
Knockdown of LINC00460 alleviates in
vivo growth of OC
To further analyze the in vivo role of
LINC00460 in the progression of OC, A2780 cells were transfected
with sh-NC or sh-LINC00460. After the sacrifice of mice for
harvesting tumor samples, tumor weight was found to be markedly
lower in mice administered with A2780 cells transfected with
sh-LINC00460 relative to controls (Fig.
3A). Transfected cells were subcutaneously implanted in the
right armpit of nude mice. Tumor growth was regularly observed.
During the experimental period, tumor size was smaller in nude mice
implanted with A2780 cells with downregulated level of LINC00460
(Fig. 3B). Transfection of
sh-LINC00460 stably downregulated LINC00460 level in A2780 cells
(Fig. 3C). It is believed that
knockdown of LINC00460 alleviated tumor growth in OC bearing nude
mice.
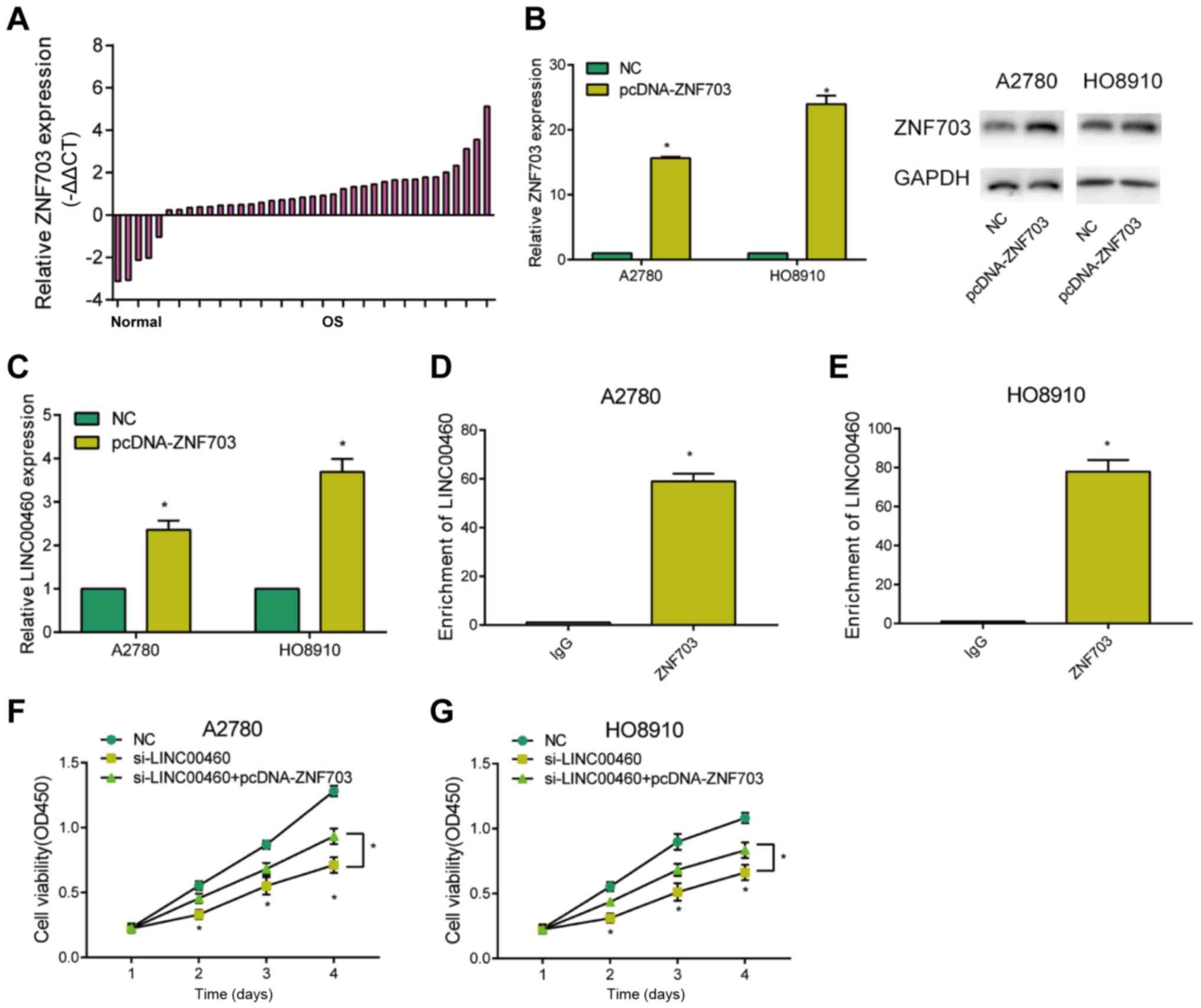
ZNF703 participates in
LINC0046-mediated progression of OC
ZNF703 was upregulated in OC tissues relative to
controls (Fig. 4A). To further
explore its potential function, pcDNA-ZNF703 was constructed.
Transfection of pcDNA-ZNF703 remarkably upregulated ZNF703 at both
mRNA and protein levels in OC cells (Fig. 4B). Overexpression of ZNF703 greatly
upregulated LINC00460 level in OC cells, indicating a potential
correlation between these two genes (Fig. 4C). ChIP assay demonstrated the
pronounced enrichment of LINC00460 in ZNF703 relative to control
IgG, suggesting the interaction between LINC00460 and ZNF703
(Fig. 4D and E). Interestingly, the
inhibited viability in OC cells transfected with si-LINC00460 was
reversed by co-transfection of pcDNA-ZNF703 (Fig. 4F and G). Hence, LINC00460 regulated
the proliferative ability of OC cells via interacting with
ZNF703.
Discussion
Pathological subtypes of OC are diverse owing to the
histological features of the ovaries (17). The specific pathogenesis of OC
remains unclear, and genetic and endocrine factors are considered
to be involved (18). Ovarian
epithelial cancer is more common in postmenopausal women, and
malignant germ cell tumor is prevalent in adolescents or young
women (19). Obvious symptoms of
early-stage OC are lacking. Approximately 70% of OC patients are
diagnosed in advanced stage, manifesting as abdominal distension,
abdominal pain, and weight loss. According to the International
Harmonized Classification developed by the World Health
Organization (WHO), the main histological type of OC is
epithelial-derived tumors. Pathological subtypes of OC include
serous tumors, mucinous tumors, endometrioid tumors, clear cell
tumors, fibro-epitheliomas and mixed type of epitheliomas (20). Although therapeutic approaches for OC
have been advanced, tumor metastasis and chemotherapy-resistance
are treatment difficulties that severely restrict the clinical
outcome of OC (21). Generally
speaking, the occurrence and progression of OC are multi-step
processes, where genomic alterations and genetic mutations are
involved (22).
Dysregulated lncRNAs are crucial in the pathogenesis
of OC (23,24). It is reported that lncRNA NEAT1
stimulates paclitaxel-resistance in OC cells through interacting
with miR-194 to regulate ZEB1 level (25). lncRNA MNX1-AS1 is upregulated in EOC,
posing a positive relationship with tumor staging, grade, distant
metastasis, and poor prognosis (26). lncRNA FAM83H-AS1 promotes
radioresistance, proliferation and metastasis of OC by stabilizing
HuR (27). HOXD-AS1 promotes
proliferative, invasive abilities and epithelial-mesenchymal
transition (EMT) of EOC by activating Wnt/β-catenin pathway and
sponging miR-133a-3p (28). lncRNA
As-SLC7A11 inhibits proliferative, migratory and invasive
abilities, and induces apoptosis of EOC cells through targeting
SLC7A11 (29). Through targeting
miR-186-5p and PIK3R3, HOXD-AS1 promotes EMT of EOC cells (30).
Located on 13q33.2, LINC00460 is upregulated and
exerts a carcinogenic role in many tumors (31,32).
Relative level, regulatory characteristic and clinical significance
of LINC00460 in OC, have not been fully elucidated. This study
showed that LINC00460 was upregulated in OC tissues and cell lines.
LINC00460 level was positively correlated to tumor staging and
tumor size of OC. In vitro experiments proved that knockdown
of LINC00460 attenuated proliferative ability and arrested cell
cycle of OC cells. Furthermore, in vivo function of
LINC00460 was identified. LINC00460 stimulated tumor enlargement
and growth rate in OC bearing nude mice.
Studies have confirmed that transcriptional factors
are able to activate lncRNAs and thus upregulate their expression
levels. The interaction between transcriptional factors and lncRNAs
participates in various biological progresses, including
tumorigenesis and tumor progression. Herein, potential binding
sequences between ZNF703 and LINC00460 were predicted in JASPAR
database. Furthermore, ChIP confirmed the interaction between
ZNF703 and LINC00460. Notably, ZNF703 overexpression reversed the
inhibited viability in OC cells with LINC00460 knockdown.
Collectively, this study is the first to demonstrate
the carcinogenic role of LINC00460 in the progression of OC. The
biological function of LINC00460 in OC was closely regulated to the
transcriptional factor ZNF703. LINC00460 shows promise as a
diagnostic and therapeutic target for OC.
In conclusion, LINC00460 is upregulated in OC
tissues and cell lines, which is closely related to tumor
progression. It accelerates proliferative ability and cell cycle
progression of OC cells via interacting with ZNF703.
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
XW, XG and WZ designed the study and performed the
experiments, XW, and CL collected the data, XG and CL analyzed the
data, XW, XG and WZ prepared the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Yuncheng County Hospital of Traditional Chinese Medicine (Heze,
China). Signed informed consents were obtained from the patients
and/or guardians. This study was approved by the Animal Ethics
Committee of Yuncheng County Hospital of Traditional Chinese
Medicine Animal Center.
Patients consent for publication
Not applicable.
Competing interests
The authors declare they have no competing
interests.
References
|
1
|
Eisenhauer EA: Real-world evidence in the
treatment of ovarian cancer. Ann Oncol. 28 (Suppl 8):viii61–viii65.
2017. View Article : Google Scholar
|
|
2
|
Webb PM and Jordan SJ: Epidemiology of
epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol.
41:3–14. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nammalwar B, Bunce RA, Berlin KD, Benbrook
DM and Toal C: Synthesis and biological evaluation of SHetA2
(NSC-721689) analogs against the ovarian cancer cell line A2780.
Eur J Med Chem. 170:16–27. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Marchetti C, De Felice F, Perniola G,
Palaia I, Musella A, Di Donato V, Cascialli G, Muzii L, Tombolini V
and Benedetti Panici P: Role of intraperitoneal chemotherapy in
ovarian cancer in the platinum-taxane-based era: A meta-analysis.
Crit Rev Oncol Hematol. 136:64–69. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Andersen RE and Lim DA: Forging our
understanding of lncRNAs in the brain. Cell Tissue Res. 371:55–71.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peng WX, Koirala P and Mo YY:
LncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu X, Wen J, Wang H and Wang Y: Long
non-coding RNA LINC00460 promotes epithelial ovarian cancer
progression by regulating microRNA-338-3p. Biomed Pharmacother.
108:1022–1028. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ye JJ, Cheng YL, Deng JJ, Tao WP and Wu L:
LncRNA LINC00460 promotes tumor growth of human lung adenocarcinoma
by targeting miR-302c-5p/FOXA1 axis. Gene. 685:76–84. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Y, Liu X, Li Q and Zhang Y: lncRNA
LINC00460 promoted colorectal cancer cells metastasis via
miR-939-5p sponging. Cancer Manag Res. 11:1779–1789. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ma G, Zhu J, Liu F and Yang Y: Long
Noncoding RNA LINC00460 promotes the gefitinib resistance of
nonsmall cell lung cancer through epidermal growth factor receptor
by sponging miR-769-5p. DNA Cell Biol. 38:176–183. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang F, Liang S, Liu X, Han L, Wang J and
Du Q: LINC00460 modulates KDM2A to promote cell proliferation and
migration by targeting miR-342-3p in gastric cancer. OncoTargets
Ther. 11:6383–6394. 2018. View Article : Google Scholar
|
|
13
|
Ge SS, Wu YY, Gao W, Zhang CM, Hou J, Wen
SX and Wang BQ: Expression of long non-coding RNA LINC00460 in
laryngeal squamous cell carcinoma tissue and its clinical
significance. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi.
32:18–22. 2018.(In Chinese). PubMed/NCBI
|
|
14
|
Kong YG, Cui M, Chen SM, Xu Y, Xu Y and
Tao ZZ: LncRNA-LINC00460 facilitates nasopharyngeal carcinoma
tumorigenesis through sponging miR-149-5p to up-regulate IL6. Gene.
639:77–84. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liang Y, Wu Y, Chen X, Zhang S, Wang K,
Guan X, Yang K, Li J and Bai Y: A novel long noncoding RNA
linc00460 up-regulated by CBP/P300 promotes carcinogenesis in
esophageal squamous cell carcinoma. Biosci Rep. 37:372017.
View Article : Google Scholar
|
|
16
|
Zhao G, Fu Y, Su Z and Wu R: How long
non-coding RNAs and MicroRNAs mediate the endogenous RNA Network of
head and neck squamous cell carcinoma: A comprehensive analysis.
Cell Physiol Biochem. 50:332–341. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vlad C, Dina C, Kubelac P, Vlad D, Pop B
and Achimas Cadariu P: Expression of toll-like receptors in ovarian
cancer. J BUON. 23:1725–1731. 2018.PubMed/NCBI
|
|
18
|
Zhou H, Zhao H, Liu H, Xu X, Dong X and
Zhao E: Influence of carboplatin on the proliferation and apoptosis
of ovarian cancer cells through mTOR/p70s6k signaling pathway. J
BUON. 23:1732–1738. 2018.PubMed/NCBI
|
|
19
|
Yang X, Guo F, Peng Q, Liu Y and Yang B:
Suppression of in vitro and in vivo human ovarian cancer growth by
isoacteoside is mediated via sub-G1 cell cycle arrest, ROS
generation, and modulation of AKT/PI3K/m-TOR signalling pathway. J
BUON. 24:285–290. 2019.PubMed/NCBI
|
|
20
|
Ottevanger PB: Ovarian cancer stem cells
more questions than answers. Semin Cancer Biol. 44:67–71. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ding SS, Li L and Yu CX: Systematic
evaluation of bevacizumab in recurrent ovarian cancer treatment. J
BUON. 19:965–972. 2014.PubMed/NCBI
|
|
22
|
Maduro MR: Potential biomarkers for
ovarian cancer. Reprod Sci. 26:4492019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Worku T, Bhattarai D, Ayers D, Wang K,
Wang C, Rehman ZU, Talpur HS and Yang L: Long non-coding RNAs: The
new horizon of gene regulation in ovarian cancer. Cell Physiol
Biochem. 44:948–966. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fu LL, Li CJ, Xu Y, Li LY, Zhou X, Li DD,
Chen SX, Wang FG, Zhang XY and Zheng LW: Role of lncRNAs as novel
biomarkers and therapeutic targets in ovarian cancer. Crit Rev
Eukaryot Gene Expr. 27:183–195. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
An J, Lv W and Zhang Y: LncRNA NEAT1
contributes to paclitaxel resistance of ovarian cancer cells by
regulating ZEB1 expression via miR-194. OncoTargets Ther.
10:5377–5390. 2017. View Article : Google Scholar
|
|
26
|
Li AH and Zhang HH: Overexpression of
lncRNA MNX1-AS1 is associated with poor clinical outcome in
epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 21:5618–5623.
2017.PubMed/NCBI
|
|
27
|
Dou Q, Xu Y, Zhu Y, Hu Y, Yan Y and Yan H:
LncRNA FAM83H-AS1 contributes to the radioresistance,
proliferation, and metastasis in ovarian cancer through stabilizing
HuR protein. Eur J Pharmacol. 852:134–141. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Y, Dun Y, Zhou S and Huang XH:
LncRNA HOXD-AS1 promotes epithelial ovarian cancer cells
proliferation and invasion by targeting miR-133a-3p and activating
Wnt/β-catenin signaling pathway. Biomed Pharmacother. 96:1216–1221.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yuan J, Liu Z and Song R: Antisense lncRNA
As-SLC7A11 suppresses epithelial ovarian cancer progression mainly
by targeting SLC7A11. Pharmazie. 72:402–407. 2017.PubMed/NCBI
|
|
30
|
Dong S, Wang R, Wang H, Ding Q, Zhou X,
Wang J, Zhang K, Long Y, Lu S, Hong T, et al: HOXD-AS1 promotes the
epithelial to mesenchymal transition of ovarian cancer cells by
regulating miR-186-5p and PIK3R3. J Exp Clin Cancer Res.
38:1102019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li K, Sun D, Gou Q, Ke X, Gong Y, Zuo Y,
Zhou JK, Guo C, Xia Z, Liu L, et al: Long non-coding RNA linc00460
promotes epithelial-mesenchymal transition and cell migration in
lung cancer cells. Cancer Lett. 420:80–90. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yue QY and Zhang Y: Effects of Linc00460
on cell migration and invasion through regulating
epithelial-mesenchymal transition (EMT) in non-small cell lung
cancer. Eur Rev Med Pharmacol Sci. 22:1003–1010. 2018.PubMed/NCBI
|