Introduction
As a leading cause of cancer-associated mortalities
globally (1), lung cancer is an
important and highly investigated area in cancer research.
Moreover, non-small cell lung cancer (NSCLC) accounts for 80% of
all lung cancer cases (2,3). Despite improvements in diagnosis and
standard therapy, the 5-year survival rate of NSCLC patients is
only 18% (4). A possible reason for
this low survival rate is the uncontrolled proliferation and
metastatic potential of NSCLC cells (5). While numerous molecular genetic studies
have been conducted in NSCLC, the precise molecular mechanisms
underlying NSCLC progression remain unknown (6). Thus, the aim of the present study was
to identify valuable biomarkers for NSCLC.
Epididymal protein 3A (EDDM3A) is located on human
chromosome (chr) 14q11.2 and there is only one transcript
(NC_000014.9) for EDDM3A (7).
Damyanova et al (8) reported
that the loss of chr14q11.2 affects proteins that are synthesized
and secreted by epididymal epithelial cells, which are upregulated
in the epididymis of male patients with non-obstructive
azoospermia. Despite these previous findings, there is an overall
lack of evaluation of the clinicopathological significance of
EDDM3A in NSCLC, and the molecular mechanisms underlying its role
are not fully understood.
In the present study, it was demonstrated that
EDDM3A expression is significantly upregulated in NSCLC by using
human NSCLC tissues and data from The Cancer Genome Atlas (TCGA).
Moreover, the aim of the present study was to identify whether
EDDM3A was significantly associated with cell proliferation, cell
cycle progression and apoptosis in the A549 lung cancer cell line.
It was indicated that EDDM3A is an oncogene in NSCLC, which may
represent a novel diagnostic and therapeutic target for the
treatment of NSCLC. Therefore, the present study may have
identified a potential new therapeutic and prognostic target for
NSCLC.
Materials and methods
Gene expression datasets
TCGA datasets TCGA_LUNG_exp_HiSeq V2-2015-02-24,
TCGA_LUAD_exp_HiSeqV2-2015-02-24 and
TCGA_LUSC_exp_HiSeqV2-2015-02-24 were downloaded from TCGA
(cancergenome.nih.gov/) database and
these contained 51 pairs of NSCLC tissues and matched non-tumor
tissues (9).
Patients and tissues
A total of 30 patients (46.6% females and 53.4%
males; mean age 59.8 and age range 46–82) diagnosed with NSCLC at
The First Affiliated Hospital of Soochow University (Suzhou, China)
were enrolled between July 2018 and June 2019. The
paraffin-embedded slides, including 30 pairs of NSCLC tissues and
adjacent healthy lung tissues, were obtained from the Department of
Pathology, The First Affiliated Hospital of Soochow University. The
current study was approved by The Institutional Ethical Committee
of The First Affiliated Hospital of Soochow University (approval
no. 2018011), and a signed informed consent form was obtained from
each participant prior to the study.
Immunohistochemistry (IHC)
5-µm continuous slides were incubated with
anti-EDDM3A antibody produced in rabbit (1:25; Sigma-Aldrich; Merck
KGaA) at 4°C overnight, followed by incubation for 1 h with
horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG
(1:500; cat. no. ab7090; Abcam) at room temperature, then
3,3′-diaminobenzidine staining solution (1:25, cat. no. 070004-D;
Beijing CellChip Biotechnology Co., Ltd.) for 10 min at room
temperature. Slides were blocked using 10% goat serum (Thermo
Fisher Scientific, Inc.) for 10 min at room temperature, and then
treated according to the manufacturer's instructions for the
Rabbit/Rabbit Streptomyces vitellogenin-Biotin Detection system
(OriGene Technologies, Inc.). Then, the slides were mounted and
imaged with an optical microscope at ×400 magnification.
Cell lines and culture conditions
The human NSCLC cell line A549 was obtained from the
American Type Culture Collection and was assessed by short tandem
repeat analysis. All cells were cultured in RPMI-1640 medium
(Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Thermo
Fisher Scientific, Inc.) and 100 IU/ml penicillin/streptomycin and
cultured at 37°C in a humidified incubator containing 5%
CO2.
Reverse transcription-quantitative PCR
(RT-qPCR)
Analysis of relative gene expression was conducted
using RT-qPCR. RNeasy Mini Kit was utilized (Qiagen, USA) to
isolate RNA according to the manufacturer's protocol and the
concentration of RNA was identified by spectrophotometer Nanodrop
2000 (Thermo Fisher Scientific, USA). 1 µg RNA was reverse
transcribed to cDNA using PrimeScriptTM RT Reagent Kit (Takara,
Tokyo, Japan) according to the manufacturer's instructions. EDDM3A
and GAPDH primer sequences were as follows: EDDM3A forward,
5′-CATTGTGGCGTAGATGGATA-3′ and reverse, 5′-ATAAATGTAAGCGGGGAGTG-3′;
and GAPDH forward, 5′-TGACTTCAACAGCGACACCCA-3′ and reverse,
5′-CACCCTGTTGCTGTAGCCAAA-3′. The Cq values were normalized to that
of β-actin, which was used as a reference to estimate the different
gene expression levels. The expression of EDDM3A was calculated by
the 2−ΔΔCq method (10).
To ensure quantitative accuracy, each sample was assayed three
times.
Lentiviral short hairpin (sh)RNA
vector construction
C-terminal HA-tagged full-length cDNA encoding human
EDDM3A (ID: 16978) and the deletion plasmids (UBA and CULLIN) were
amplified by PCR using Phusion DNA Polymerases (Thermo Fisher
Scientific, Inc.). The thermocycling were as follows: Initiation at
94°C for 30 sec denaturation at 95°C for 6 min, annealing at 60°C
for 30 sec and elongation at 73°C for 1.5 min for 40 cycles. The
sequences of forward and reference primers: Forward,
5′-TGTGGCGTAGATGG-3′ and reverse, 5′-ATGTAAGCGGGGAG-3′. Then the
amplified cDNA was cloned into the lentiviral expression vector
pGVX115-GFP (Shanghai GeneChem Co., Ltd.). Primers for PCR were
designed to include BamHI and XhoI restriction sites. Overall, 10
µg pGV115-shControl (Ctrl)/pGV115-shEDDM3A were transfected into
293T cells (Thermo Fisher Scientific, Inc.) with the pHelper system
(Shanghai GeneChem Co., Ltd.) to produce lentiviral particles.
Lentiviruses were harvested after 48 h and filtered using a 0.45 mm
filter.
Western blotting
Western blotting was performed to measure the
protein expression in transfected A549 cells. Cells were lysed with
RIPA buffer (Boster Biological Technology Co., Ltd.) and the total
protein concentration was measured with a bicinchoninic acid kit
(Abbkine Scientific Co., Ltd.). GAPDH was used as the internal
reference. Overall, 5 µg proteins per lane were loaded on a 10%
SDS-PAGE and then transferred to PVDF membranes. Subsequently, the
PVDF membranes were blocked with 5% BSA (Abbkine Scientific Co.,
Ltd.) for 2 h at room temperature. The primary antibodies,
including anti-EDDM3A (1:500; cat. no. ab151083; Abcam) and
anti-GAPDH (1:500; cat. no. ab8245; Abcam), were incubated for 1 h
at room temperature. The blots were then incubated with the
secondary goat anti-rabbit antibody (HRP goat anti-mouse IgG
H&L, 1:1,000, cat. no. ab6708, Abcam; or goat anti-rabbit IgG;
1:1,500, cat. no. bs-0295G, BIOSS) for 1 h at room temperature and
developed with an ECL reagent (Abcam). Densitometry was performed
using a bio-imaging system (DNR Bio-Imaging Systems, Ltd.).
Experiments were performed in triplicate.
Cell proliferation
Transfected A549 cells reached 80% confluency, the
fluorescence of these cells was detected and the number of cells
was automatically calculated using the Celigo Imaging Cytometry
system version 2.0 (Nexcelom Bioscience LLC). The cell viability
was monitored using a Cell Proliferation kit I (MTT)
(Sigma-Aldrich; Merck KGaA) according to the manufacturer's
instructions and 0.25% trypsin was used to dissolve the purple
formazan. Cells were seeded at a density of 3×103
cells/well and cultured for 5 days. Subsequently, the optical
density (OD) was measured by microplate reader under 450 nm. For
each experimental condition, 5 parallel wells were assigned to each
group. The experiments were performed in triplicate.
Colony formation assays
A549 cells transfected with shEDDM3A or shCtrl were
plated in 6-well plates at a density of 1,200 cells/well in culture
medium 24 h after transfection, and the medium was changed every 3
days. After 2 weeks, cells were fixed with 4% paraformaldehyde for
15 min and stained with 0.1% crystal violet for 15 min at (both at
room temperature). Colonies with >50 cells per colony were
counted under the light microscope at ×40 magnification manually
and each experiment was repeated three times.
Apoptosis assay
A total of 1×105 cells/well were
incubated in 6-well plates for 24 h and treated with shEDDM3A and
shCtrl for 48 h. Then cells were collected using centrifuge at
1,000 × g. After washing the cells with PBS, an Annexin V-APC
Apoptosis Detection kit (eBioscience; Thermo Fisher Scientific,
Inc.) was used to assess apoptosis using a flow cytometer
(FACScalibur; BD Biosciences) following the manufacturer's
instructions and the percentage of apoptotic cells was determined
using ModFit LT version 5.0 software (Verity Software House,
Inc.).
Statistical analysis
SPSS software (version 19.0; IBM Corp.) was used to
analyze data for statistical significance. Data are presented as
the mean ± SEM from one representative experiment out of three
independent experiments, and every representative experiment had
three replicates. Unpaired two-tailed Student's t-test or one-way
analysis of variance (ANOVA) followed by Bonferroni's post hoc test
were performed to assess differences among the variables. P<0.05
was considered to indicate a statistically significant difference.
GraphPad Prism 5 (GraphPad Software) was used for image
editing.
Results
EDDM3A expression is significantly
increased in NSCLC tissues
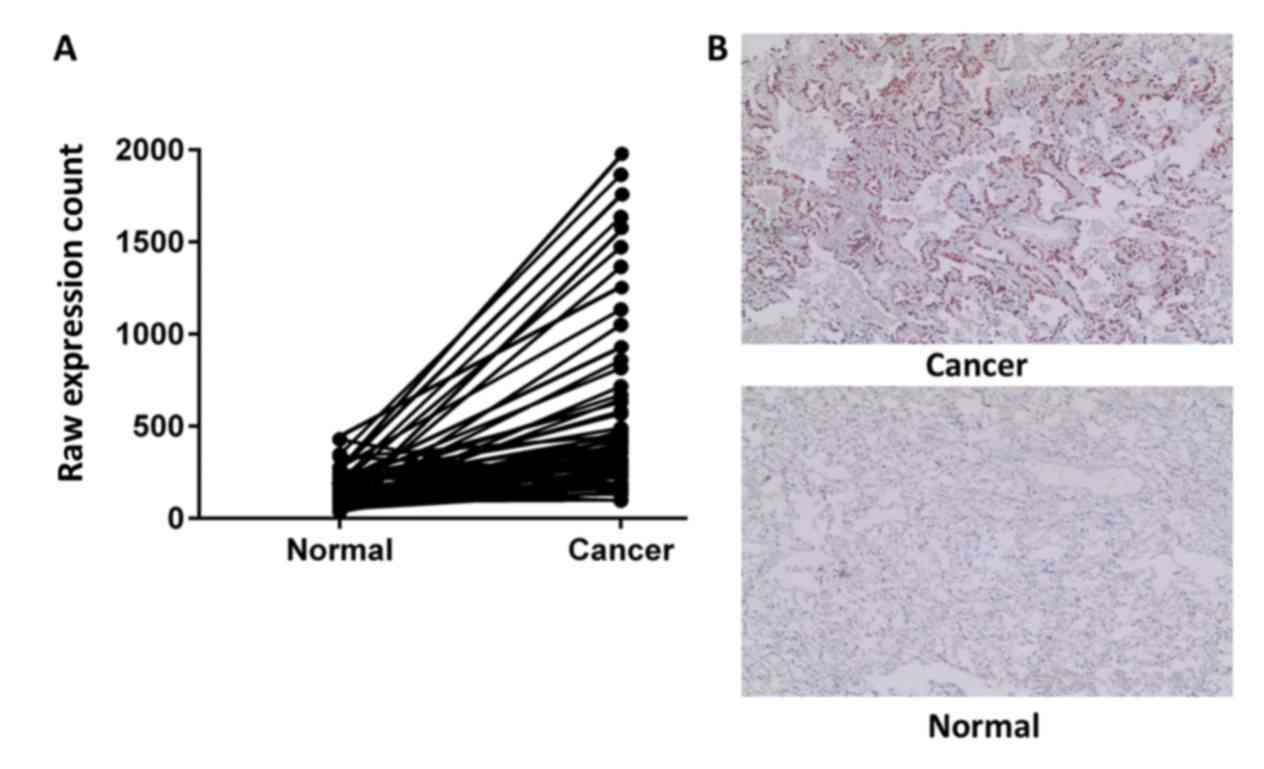
To investigate whether EDDM3A expression was altered
in NSCLC tissues, the mRNA expression profiles of NSCLC tissues and
healthy tissues were retrieved from TCGA database. It was
demonstrated that EDDM3A expression was significantly increased
(P=4.19×10−2) in NSCLC tissues compared with healthy
tissues (Fig. 1A). Furthermore, the
expression of EDDM3A was detected in 30 pairs of NSCLC tissues and
adjacent healthy lung tissues via IHC. The results indicated that
expression of EDDM3A was increased in NSCLC tissues compared with
healthy lung tissues (Fig. 1B),
which was consistent with the results from TCGA database.
Knockdown efficiency of EDDM3A via
shRNA infection in A549 cells
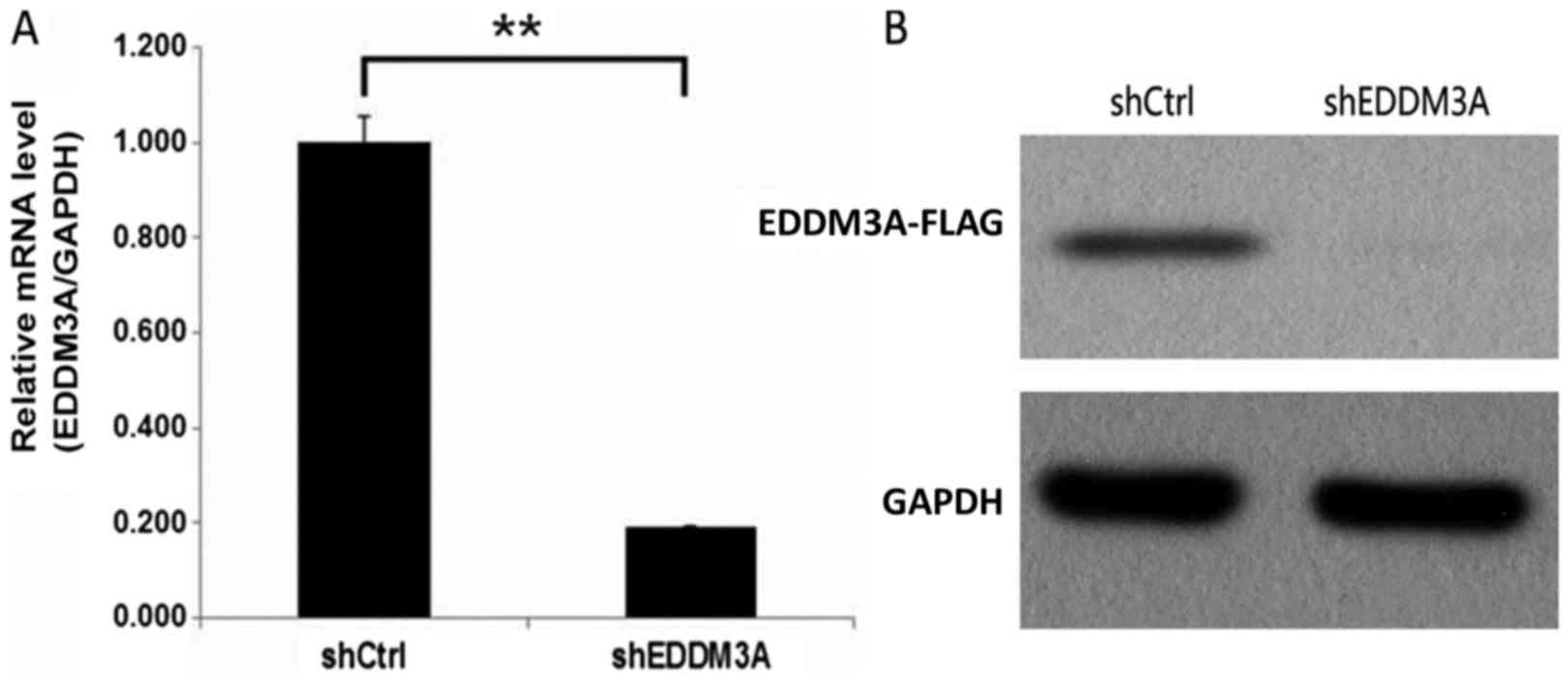
To investigate the role of EDDM3A in A549 cells,
lentiviruses expressing shEDDM3A or shCtrl were used to infect A549
cells. The transfection efficiency was evaluated using RT-qPCR, and
it was revealed that EDDM3A mRNA expression was significantly
reduced by 80% in the shEDDM3A group (Fig. 2A). Moreover, compared with the shCtrl
group, EDDM3A protein expression was knocked down in the shEDDM3A
lentivirus group of A549 cells (Fig.
2B).
Knockdown of EDDM3A inhibits the
proliferation of A549 cells
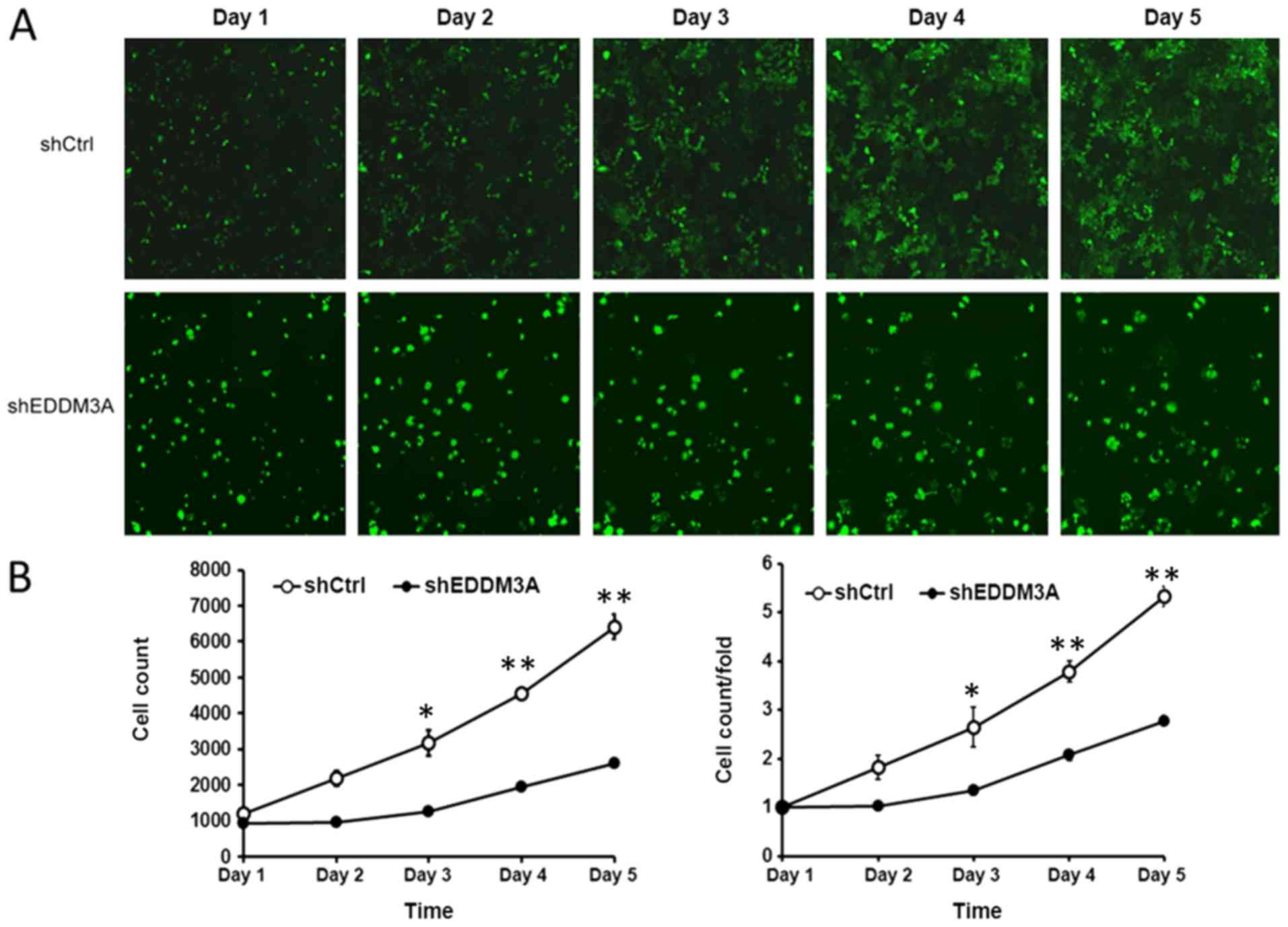
The present study examined the role of EDDM3A in the
proliferation and colony formation of A549 cells after infection
with shEDDM3A or shCtrl in a 5-day study. It was identified that
the proliferation of shEDDM3A-infected cells was inhibited compared
with the shCtrl-infected cells, as demonstrated by the Celigo cell
imaging system (Fig. 3).
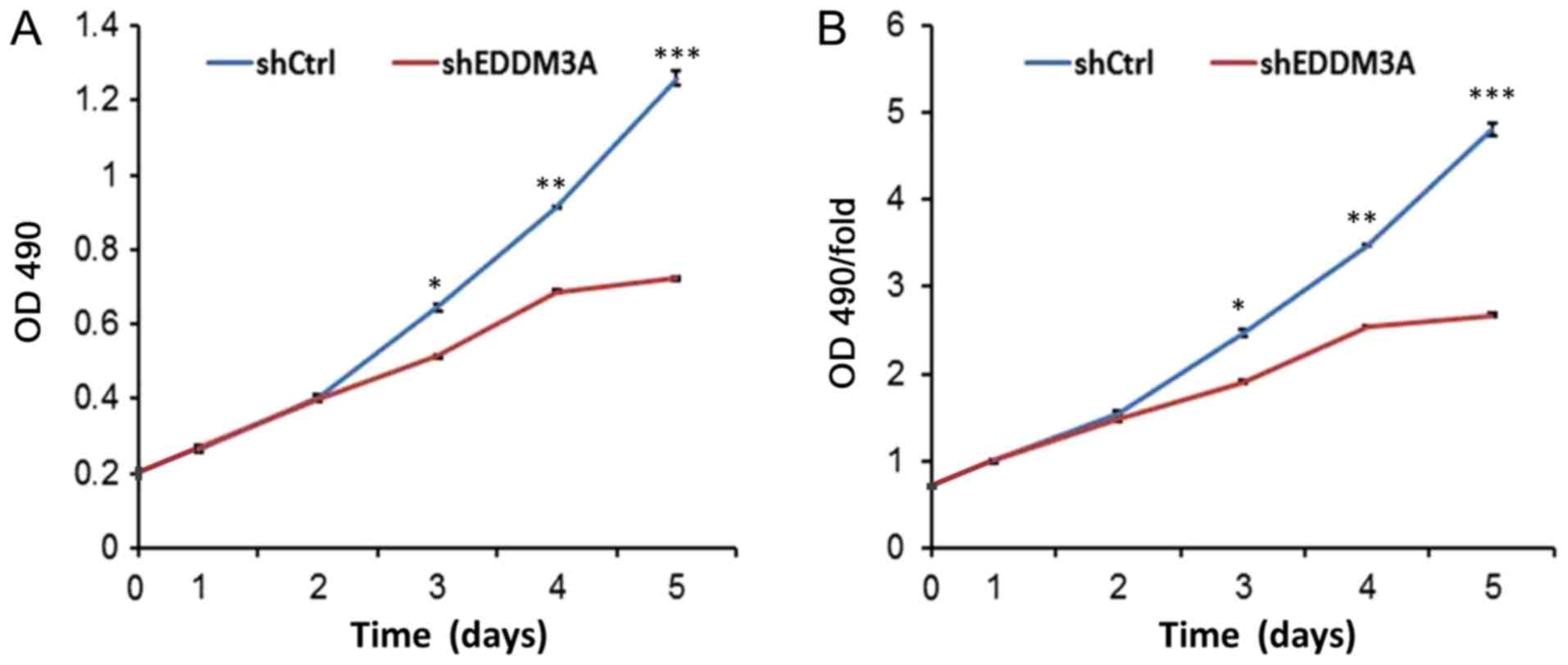
MTT assay results indicated that EDDM3A knockdown
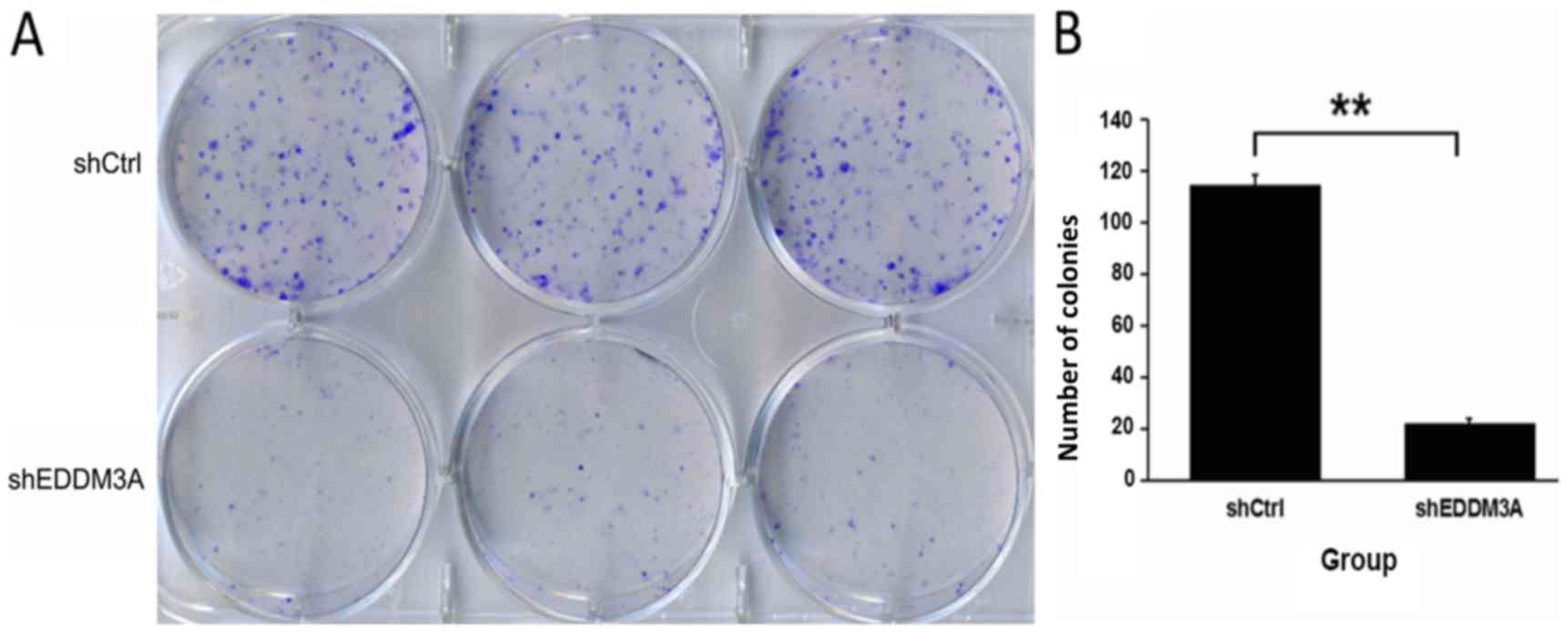
inhibited the proliferative rate of A549 cells (Fig. 4). Moreover, a colony formation assay
was performed to evaluate the role of EDDM3A in long-term survival,
and it was found that knockdown of EDDM3A significantly attenuated
the colony-forming ability of the cellular population (Fig. 5).
Knockdown of EDDM3A accelerates A549
cell apoptosis
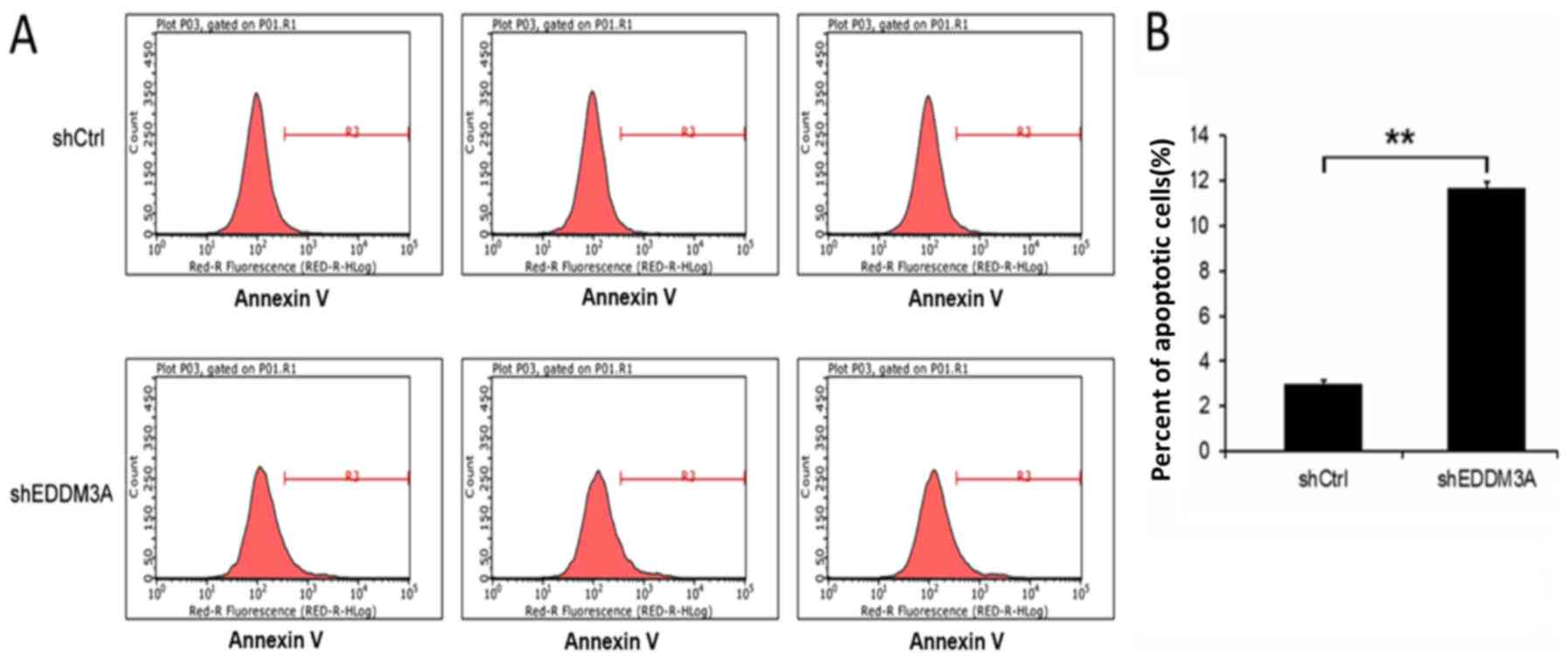
The effect of EDDM3A knockdown on A549 cell
apoptosis was investigated by FACS, which identified that the
apoptotic rate of the shEDDM3A group was elevated compared with the
shCtrl group (P<0.01; Fig. 6).
Therefore, it was suggested that shEDDM3A promoted A549 cell
apoptosis.
Discussion
Lung cancer is the leading cause of
cancer-associated mortality and has led advances in cancer therapy
worldwide (11,12). The development of NSCLC results from
both environmental and genetic changes (13), and the activation of oncogenes
influences the proliferation, adhesion, motility and invasiveness
of cancer cells and reconstructs the contact of tumor cells with
the surrounding extracellular matrix, thus promoting
epithelial-mesenchymal transition (14,15).
EDDM3A is a protein-coding gene located at
chr14q11.2 and it has been shown that loci on chr14q11.2 are
associated with a risk of childhood acute lymphoblastic leukemia
(16). Moreover, double homeobox A
pseudogene 10, located in chr14q11.2, promotes an aggressive
phenotype by binding lysine-specific histone demethylase 1 and
repressing large tumor suppressor kinase 2 and Ras-related
glycolysis inhibitor and calcium channel regulator expression
levels in NSCLC (17). Furthermore,
previous studies have revealed that chr14q11.2 may participate in
cancer development (18–20).
The present study examined the function of EDDM3A in
NSCLC. EDDM3A expression levels in NSCLC tissues compared with
healthy tumor tissues were assessed using TCGA database, and it was
identified that EDDM3A was upregulated in NSCLC tissues. Next,
lentiviruses harboring EDDM3A shRNA were constructed and
transfected into A549 cells. The present results demonstrated that
cell proliferation was inhibited in EDDM3A-silenced A549 cells, as
demonstrated by the Celigo imaging cytometry system and MTT assay.
Moreover, the apoptotic rate of the EDDM3A-shRNA group was
significantly higher compared with the control group. Therefore, it
was speculated that EDDM3A may regulate NSCLC cell proliferation
and apoptosis in vitro.
However, certain limitations of the present study
must be considered. For example, only the A549 cell line was used
in the present study; therefore, in the future the present results
should be assessed in other lung cancer cell lines, such as
NCI-H1299, NCI-H460 and 95D, and healthy cell lines. Future studies
will also perform additional downstream experiments to reveal the
underlying molecular mechanism behind the observed effects.
In conclusion, the present study identified the
functional role of EDDM3A in lung cancer progression. It was
revealed that EDDM3A knockdown may promote cell apoptosis and
inhibit cell proliferation and colony-forming abilities in the
human NSCLC A549 cell line. Thus, it was speculated that EDDM3A
expression in lung cancer may be a valuable biomarker for assessing
aggressive features and poor prognosis. Moreover, the present
results suggested that EDDM3A may represent a potential therapeutic
target in NSCLC.
Acknowledgements
Not applicable.
Funding
The current study was supported by The National
Natural Science Foundation of China (grant no. 81800279), the
Natural Science Foundation of Jiangsu Province (grant no.
BK20180197) and the Science and Education Youth Project of Suzhou
(grant no. kjxw2018002).
Authors' contributions
HM and YF designed the experiments. GL and XT
performed the experiments and wrote the manuscript. LP and HH were
responsible for data analysis. All authors read and approved the
final manuscript.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Ethics approval and consent to
participate
Approval was obtained to review the data from The
Institutional Ethical Committee of The First Affiliated Hospital of
Soochow University (approval no. 2018011). Written informed consent
was provided by each participant prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
EDDM3A
|
epididymal protein 3A
|
|
NSCLC
|
non-small cell lung cancer
|
|
TCGA
|
The Cancer Genome Atlas
|
|
shRNA
|
short hairpin RNA
|
|
IHC
|
immunohistochemistry
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sharma SV, Bell DW, Settleman J and Haber
DA: Epidermal growth factor receptor mutations in lung cancer. Nat
Rev Cancer. 7:169–181. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Noone AM, Cronin KA, Altekruse SF,
Howlader N, Lewis DR, Petkov VI and Penberthy L: Cancer incidence
and survival trends by subtype using data from the surveillance
epidemiology and end results program, 1992–2013. Cancer Epidemiol
Biomarkers Prev. 26:632–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang X and Adjei AA: Lung cancer and
metastasis: New opportunities and challenges. Cancer Metastasis
Rev. 34:169–171. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Koudelakova V, Kneblova M, Trojanec R,
Drabek J and Hajduch M: Non-small cell lung cancer-genetic
predictors. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub.
157:125–136. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chalela R, Curull V, Enriquez C, Pijuan L,
Bellosillo B and Gea J: Lung adenocarcinoma: From molecular basis
to genome-guided therapy and immunotherapy. J Thorac Dis.
9:2142–2158. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ota T, Suzuki Y, Nishikawa T, Otsuki T,
Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al:
Complete sequencing and characterization of 21,243 full-length
human cDNAs. Nat Genet. 36:40–45. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Damyanova V, Dimitrova-Dikanarova D,
Hadjidekova S, Savov A, Nesheva D, Rukova B, Vatev I and Toncheva
D: Genomic study in patients with idiopathic azoospermia and
oligoasthenoteratozoospermia. Akush Ginekol (Sofiia). 52:27–34.
2013.PubMed/NCBI
|
|
9
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Livak JK and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Torre LA, Siegel RL, Ward EM and Jemal A:
Global cancer incidence and mortality rates and trends-an update.
Cancer Epidemiol Biomarkers Prev. 25:16–27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zarogoulidis P, Tsakiridis K, Karapantzou
C, Lampaki S, Kioumis I, Pitsiou G, Papaiwannou A,
Hohenforst-Schmidt W, Huang H, Kesisis G, et al: Use of proteins as
biomarkers and their role in carcinogenesis. J Cancer. 6:9–18.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He L, Zhang Y, Sun H, Jiang F, Yang H, Wu
H, Zhou T, Hu S, Kathera CS, Wang X, et al: Targeting DNA flap
endonuclease 1 to impede breast cancer progression. EBioMedicine.
14:32–43. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Thomas C, Ji Y, Lodhi N, Kotova E, Pinnola
AD, Golovine K, Makhov P, Pechenkina K, Kolenko V and Tulin AV:
Non-NAD-Like poly(ADP-Ribose) polymerase-1 inhibitors effectively
eliminate cancer in vivo. EBioMedicine. 13:90–98. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Papaemmanuil E, Hosking FJ, Vijayakrishnan
J, Price A, Olver B, Sheridan E, Kinsey SE, Lightfoot T, Roman E,
Irving JA, et al: Loci on 7p12.2, 10q21.2 and 14q11.2 are
associated with risk of childhood acute lymphoblastic leukemia. Nat
Genet. 41:1006–1010. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wei CC, Nie FQ, Jiang LL, Chen QN, Chen
ZY, Chen X, Pan X, Liu ZL, Lu BB and Wang ZX: The pseudogene
DUXAP10 promotes an aggressive phenotype through binding with LSD1
and repressing LATS2 and RRAD in non small cell lung cancer.
Oncotarget. 8:5233–5246. 2017.PubMed/NCBI
|
|
18
|
Panagopoulos I, Brunetti M, Stoltenberg M,
Strandabø RAU, Staurseth J, Andersen K, Kostolomov I, Hveem TS,
Lorenz S, Nystad TA, et al: Novel GTF2I-PDGFRB and IKZF1-TYW1
fusions in pediatric leukemia with normal karyotype. Exp Hematol
Oncol. 8:122019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Studd JB, Yang M, Li Z, Vijayakrishnan J,
Lu Y, Yeoh AE, Paulsson K and Houlston RS: Genetic predisposition
to B-cell acute lymphoblastic leukemia at 14q11.2 is mediated by a
CEBPE promoter polymorphism. Leukemia. 33:1–14. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou Y, Wang L, Ban X, Zeng T, Zhu Y, Li
M, Guan XY and Li Y: DHRS2 inhibits cell growth and motility in
esophageal squamous cell carcinoma. Oncogene. 37:1086–1094. 2018.
View Article : Google Scholar : PubMed/NCBI
|