Introduction
Ovarian cancer was the fifth-leading cause of
cancer-associated mortality in females in 2015 in the United
States, with 295,414 newly-diagnosed cases globally in 2018 and
184,799 cancer-associated deaths globally in 2018 (1,2).
Statistical analyses indicate that 90% of ovarian cancer cases are
epithelial, with serous carcinoma being the most common
pathological type with a 5-year survival rate of 43% (3). Conventional treatment for epithelial
ovarian cancer involves cytoreductive surgery followed by platinum-
and taxane-based chemotherapy (4).
However, development of resistance to chemotherapy eventually
induces recurrence after treatment (5). An accurate and robust predictive marker
of chemoresistance is urgently required to improve individualized
treatment and enhance the prognosis and survival of patients with
epithelial ovarian cancer. Previous studies have identified a
number of chemoresistance-associated biomarkers, such as reactive
stroma signature, markers of cancer stem cells and miRNAs (6–10), but
they have not been used in clinical practice. Effective predictors
of primary platinum-based chemotherapy resistance would provide
novel strategies for treating patients with epithelial ovarian
cancer.
Dysregulation of genomic expression serves a
critical role in tumorigenesis and chemoresistance in epithelial
ovarian cancer. Previous progress in developing genomics-based and
precision-targeted therapies has provided novel strategies for
treating patients with ovarian cancer (11). However, previous studies have only
focused on gene expression levels rather than investigating how
alternative splicing (AS) can affect transcript architecture
(12,13).
AS is a post-transcriptional modification process
that produces a variable mature mRNA transcript from a single gene
by removing different intronic or exonic regions from the precursor
mRNA and subsequently combining the spliced exons (14,15). AS
generates mRNAs with different stabilities or coding potentials,
enabling quantitative control of protein production and achieving
distinct protein functions (16). AS
serves crucial roles in specialized muscle functions (17), angiogenesis (18) and pathological processes, including
hearing loss (19), Huntington's
disease (20) and cancer (21). Emerging evidence suggests that AS is
associated with tumorigenic processes, such as tumor proliferation,
invasion, metastasis and apoptosis (22). Splicing factors perform splicing by
binding to pre-mRNAs, influencing exon selection and selecting the
splicing site (23). Splicing
factors are expressed differentially between normal and cancerous
tissues (24,25). Therefore, identifying AS signature
profiles and exploring splicing factors may reveal useful cancer
biomarkers.
An analysis of AS in cancer has become possible with
the advent of deep-sequencing techniques that allow the discovery
of previously unknown prognostic and therapeutic biomarkers for
patients with cancer. Prognostic predictors based on AS events have
been identified in patients with various types of cancer, including
ovarian cancer (26–28). However, to the best of our knowledge,
no systematic analyses of chemoresistance-associated AS in ovarian
cancer have been performed, even though these are urgently required
due to the major role of chemoresistance in disease recurrence. In
the present study, The Cancer Genome Atlas (TCGA) RNA-sequencing
(RNA-seq) data was used to investigate whether AS events could
serve as predictors of primary platinum-based chemotherapy
resistance in serous ovarian carcinoma.
Materials and methods
Data acquisition
AS profiles were analyzed using the TCGA SpliceSeq
tool version 1 provided by the MD Anderson Cancer Center
(https://bioinformatics.mdanderson.org/TCGASpliceSeq/)
(29). Seven types of AS events were
quantified using the percent spliced-in (PSI) value: Exon skip
(ES), alternate promoter (AP), alternate terminator (AT),
alternative acceptor site (AA), alternate donor site (AD), retained
intron (RI) and mutually exclusive exons (ME). The PSI values for
the seven types of AS in ovarian serous cystadenoma (OV) were
downloaded from TCGA SpliceSeq. AS events with a standard deviation
>0.05 and a PSI value >75% were included. Clinical
information for the TCGA-OV cohort was obtained from the TCGA
database (https://portal.gdc.cancer.gov/projects/TCGA-OV)
(30). Individuals who met the
following criteria were included in the present study: i) Patients
diagnosed with serous ovarian cancer; ii) patients who received
platinum-based chemotherapy; and iii) patients with well-defined
responses to chemotherapy. Patients without AS information were
excluded from the present study. A total of 63 splicing factors and
their information were obtained from SpliceAid 2 (31). Level three mRNA expression data of
splicing factors were also acquired from the TCGA database.
Statistical analysis
Univariate logistic regression analyses were
performed to assess the predictive value of AS events for primary
platinum-based chemotherapy resistance. Subsequently, the top 30
most significant AS events from the univariate analyses were
included in multivariate logistic regression analyses to build
prediction models for each type of AS event individually and for
all types of AS events combined. The Akaike information criterion
was applied to select the most appropriate risk model (32). The prediction accuracy of the risk
models was evaluated by receiver operating characteristic (ROC)
analysis. Patients were classified into high- and low-risk groups,
with the median score as the cut-off value. Kaplan-Meier analysis
and a log-rank test were performed to estimate the difference in
overall survival (OS) time between the high- and low-risk
groups.
Resistance-associated splicing factor genes were
identified using univariate logistic regression analysis. Pearson's
correlation test was used to determine whether expression of the
splicing factor genes was significantly associated with the PSI
values of resistance-associated AS events. The regulatory network
map was built based on the significantly correlated splicing
factors and AS events.
All analyses were performed using R (version 3.5.2;
www.r-project.org). P<0.05 was considered to
indicate a statistically significant difference, unless otherwise
specified. Differences in clinicopathologic parameters between
chemosensitive and chemoresistant groups, including age, grade,
FIGO (International Federation of Gynecology and Obstetrics) stage
and debulking status (33), were
tested by unpaired t-test or the χ2 test.
Procedures
R was used to perform the univariate and
multivariate logistic analyses and build chemoresistance prediction
models. UpSet plots were generated using UpSetR (version 1.4.0;
http://cran.r-project.org/web/packages/UpSetR/index.html).
The pROC package (version 1.13.0; http://cran.r-project.org/src/contrib/Archive/pROC/)
was used to create ROC curves and to calculate the area under the
curve (AUC). The Functional Annotation Result Summary tool version
6.8 (https://david.ncifcrf.gov/summary.jsp) from the
Database for Annotation, Visualization, and Integrated Discovery
(version 6.8) was used for Gene Ontology (GO) (http://geneontology.org) analysis of the corresponding
genes (34). The gene interaction
network and correlation network were visualized using Cytoscape
(version 3.7.1; http://cytoscape.org).
Results
Comprehensive analysis of AS events in
the OV data
The overall process of the present study is
described in Fig. 1A. Integrated AS
event signatures for 320 patients with OV were curated from the
TCGA database (Table I). Seven types
of AS events were identified, as shown in Fig. 1B. A total of 22,036 AS events were
detected in 7,404 genes, suggesting that one gene might have had
more than one AS event. The following numbers of AS events were
detected for each type: 8,280 ES events in 3,835 genes; 1,535 RI
events in 1,073 genes; 4,841 AP events in 2,196 genes; 3,806 AT
events in 1,801 genes; 1,735 AD events in 1,291 genes; 1,741 AA
events in 1,357 genes; and 98 ME events in 96 genes (Fig. 1C). The most common type of AS events
was ES, followed by AP and AT events.
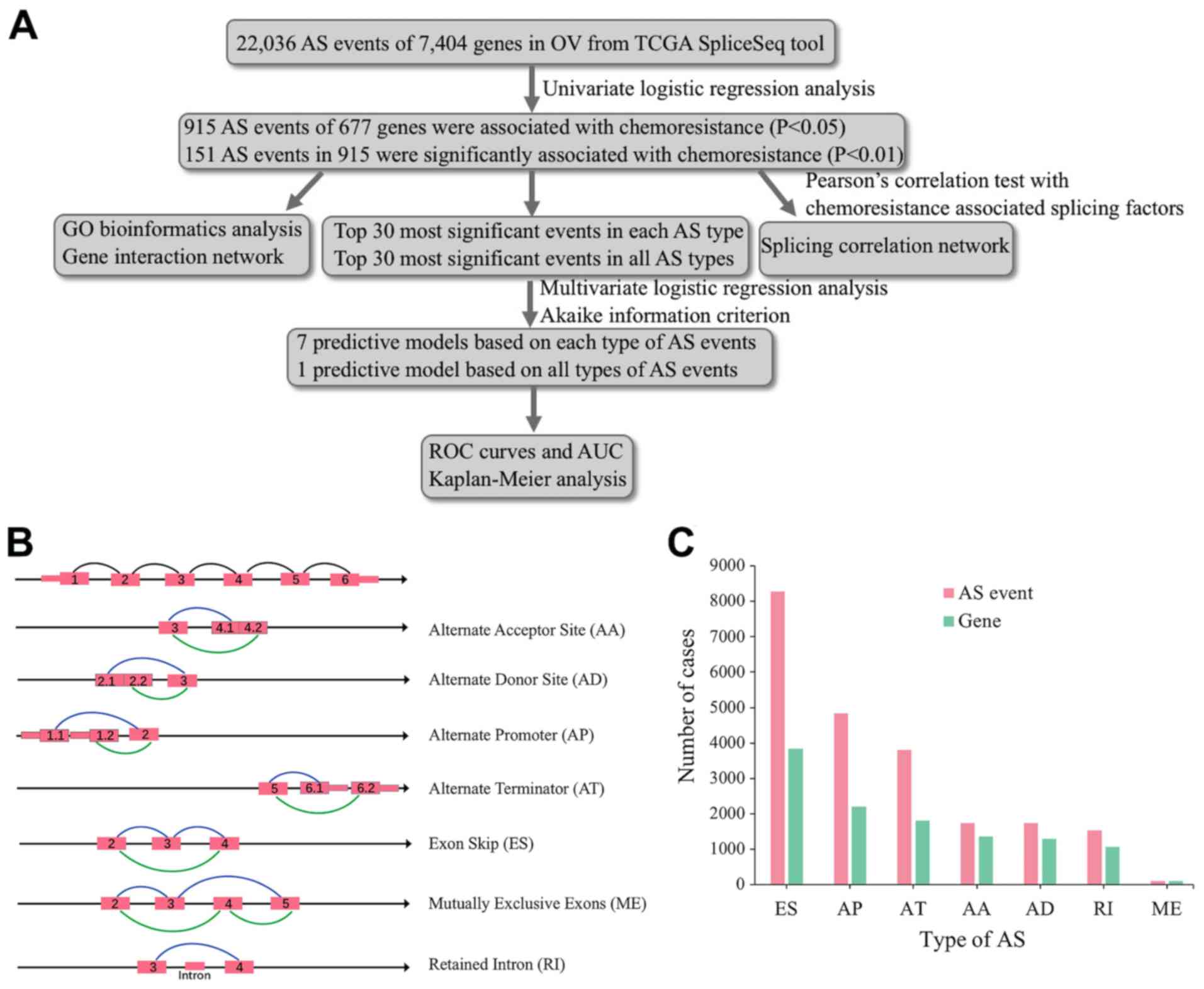 | Figure 1.Overview of the seven types of AS.
(A) Flowchart of the present study. (B) Illustrations of the seven
types of AS events, including AA, AD, AP, AT, ES, ME and RI. (C)
Number of AS events and involved genes from 320 patients with OV.
AS, alternative splicing; OV, ovarian serous cystadenocarcinoma;
AA, alternate acceptor site; AD, alternate donor site; AP,
alternate promoter; AT, alternate terminator; ES, exon skip; ME,
mutually exclusive exons; RI, retained intron; TCGA, The Cancer
Genome Atlas; GO, Gene Ontology; ROC, receiver operating
characteristic; AUC, area under the curve. |
 | Table I.Demographic and clinical
characteristics of ovarian serous cystadenocarcinoma cases in The
Cancer Genome Atlas datasets involved in developing alternative
splicing signatures to predict primary platinum-based
chemoresistance. |
Table I.
Demographic and clinical
characteristics of ovarian serous cystadenocarcinoma cases in The
Cancer Genome Atlas datasets involved in developing alternative
splicing signatures to predict primary platinum-based
chemoresistance.
|
Characteristics | Resistance cases,
n | Sensitive cases,
n | P-value |
|---|
| Sample number | 95 | 225 |
|
| Age, years |
|
| 0.734 |
|
<60 | 56 | 128 |
|
|
≥60 | 39 | 97 |
|
| Stage |
|
| 0.027 |
| FIGO
I/II | 1 | 16 |
|
| FIGO
III/IV | 94 | 209 |
|
| Grade |
|
| 0.788 |
|
Low | 12 | 26 |
|
|
High | 81 | 194 |
|
|
Unknown | 2 | 5 |
|
| Debulking
status |
|
| <0.001 |
|
Optimal | 53 | 164 |
|
|
Suboptimal | 36 | 36 |
|
|
Unknown | 6 | 25 |
|
Chemoresistance-associated AS events
in the OV data
The univariate logistic regression analyses of OV
data from the TCGA database identified 915 AS events associated
with chemotherapy resistance in patients with OV (P<0.05;
Table SI). Among these, 151 AS
events were significantly associated with chemotherapy resistance
(P<0.01; Table SII), 407 AS
events were risk factors for chemotherapy resistance [odds ratio
(OR)>1], and 508 were protective factors for chemotherapy
resistance (OR<1). The distribution of 677 genes involved in 915
AS events was visualized in the UpSet plot (Fig. 2A). A total of 640 genes had only one
type of AS event associated with chemoresistance, whereas 37 genes
had more than one type of AS event associated with them. For
example, ES, AA and AD events in GPR56 were all significantly
associated with chemoresistance (Table
SI).
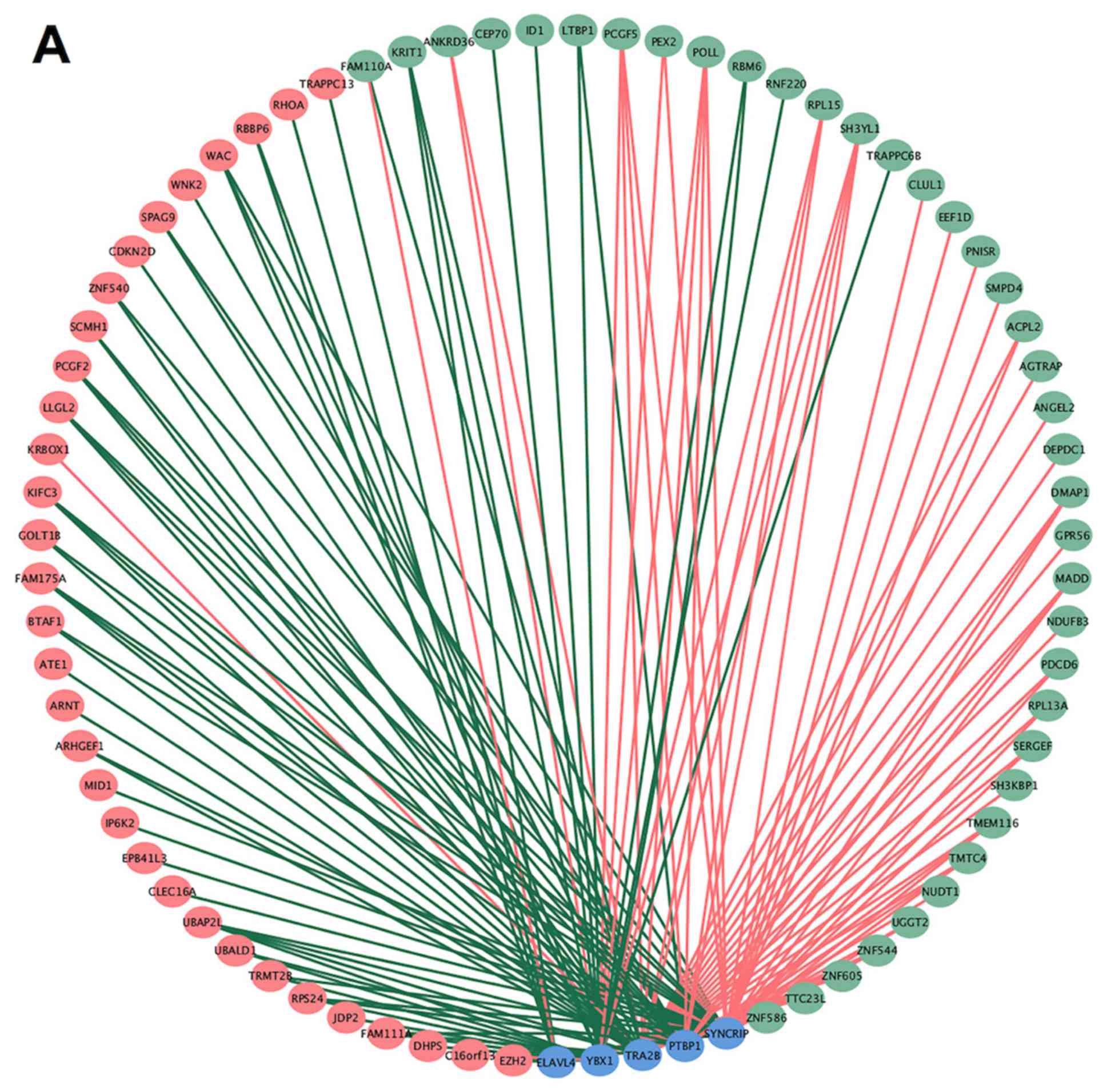
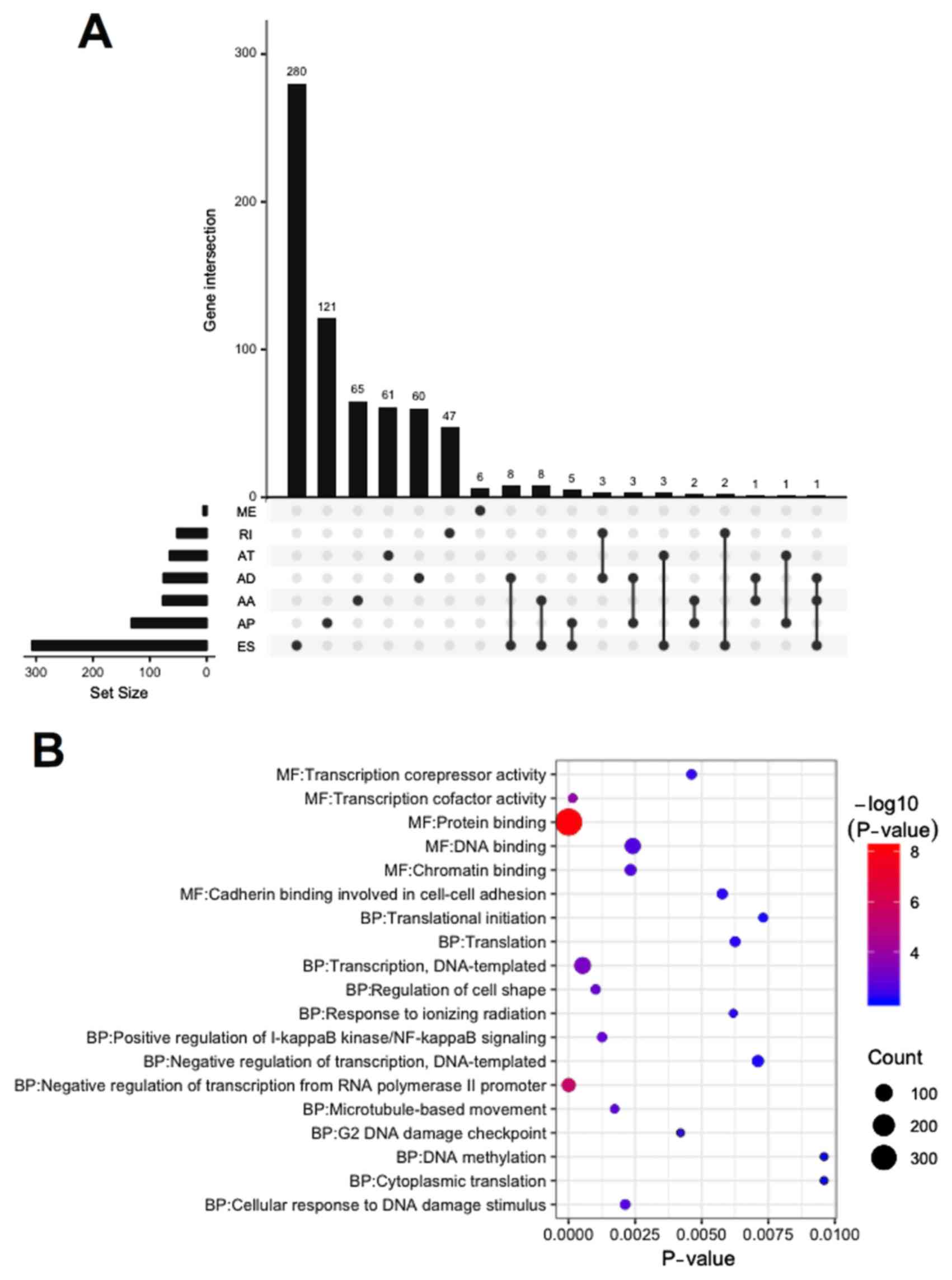 | Figure 2.UpSet plot, GO analysis and gene
network of chemoresistance-associated AS in OV. (A) UpSet plot of
interactions between the seven types of chemoresistance-associated
AS events in OV. (B) GO analysis of chemoresistance-associated AS
events in OV. (C) Gene network of chemoresistance-associated AS in
OV generated by Cytoscape. AS, alternative splicing; OV, ovarian
serous cystadenocarcinoma; AA, alternate acceptor site; AD,
alternate donor site; AP, alternate promoter; AT, alternate
terminator; ES, exon skip; ME, mutually exclusive exons; RI,
retained intron; GO, Gene Ontology; BP, biological process; MF,
molecular function. |
GO bioinformatics analysis was performed on 677
genes with AS events. A total of 13 biological processes and 6
molecular functions were identified in the GO analysis (P<0.01;
Fig. 2B). These genes were found to
be significantly associated with ‘protein binding’ and ‘negative
regulation of transcription from RNA polymerase II promoter’. The
gene interaction network analysis for these 677 genes revealed a
hub that included RHOA, POLR2G, RPS9, DYNLL1 and RPL13A (the top 5
genes with higher degree of connectivity) (Fig. 2C).
Chemoresistance predictors for
patients with OV
The top 30 most significant events for each AS type
(except for ME, which had only 6 events) and for all types of AS
events were selected as candidates to identify the independent
predictive model for chemoresistance in OV (Table SIII). Multivariate logistic
regression analysis was performed for the 30 candidate events for
each AS type and for all AS types combined, and the Akaike
information criterion was used to select the most appropriate risk
model (32). The predictive models
are presented in Table II. The
median score was used as the cut-off value, the patients were
divided into high- and low-risk groups, and the OR for each model
was calculated. ROC curves were generated and the AUCs were
determined to evaluate the effectiveness of the chemoresistance
predictive models. The seven predictors that were built using the
seven types of AS events displayed considerable power in
distinguishing the chemotherapy response of patients with OV. The
model based on ES events was the most effective predictor among the
models based on each type of AS event, with an AUC of 0.894
(Fig. 3). The model based on all
types of AS events exhibited the best efficiency with an AUC of
0.931. The information for AS event candidates involved in this
model is presented in Table III.
This model was utilized in univariate and multivariate logistic
analyses of chemotherapy resistance together with common clinical
characteristics. A high-risk score was an independent risk factor
for chemoresistance (Table IV).
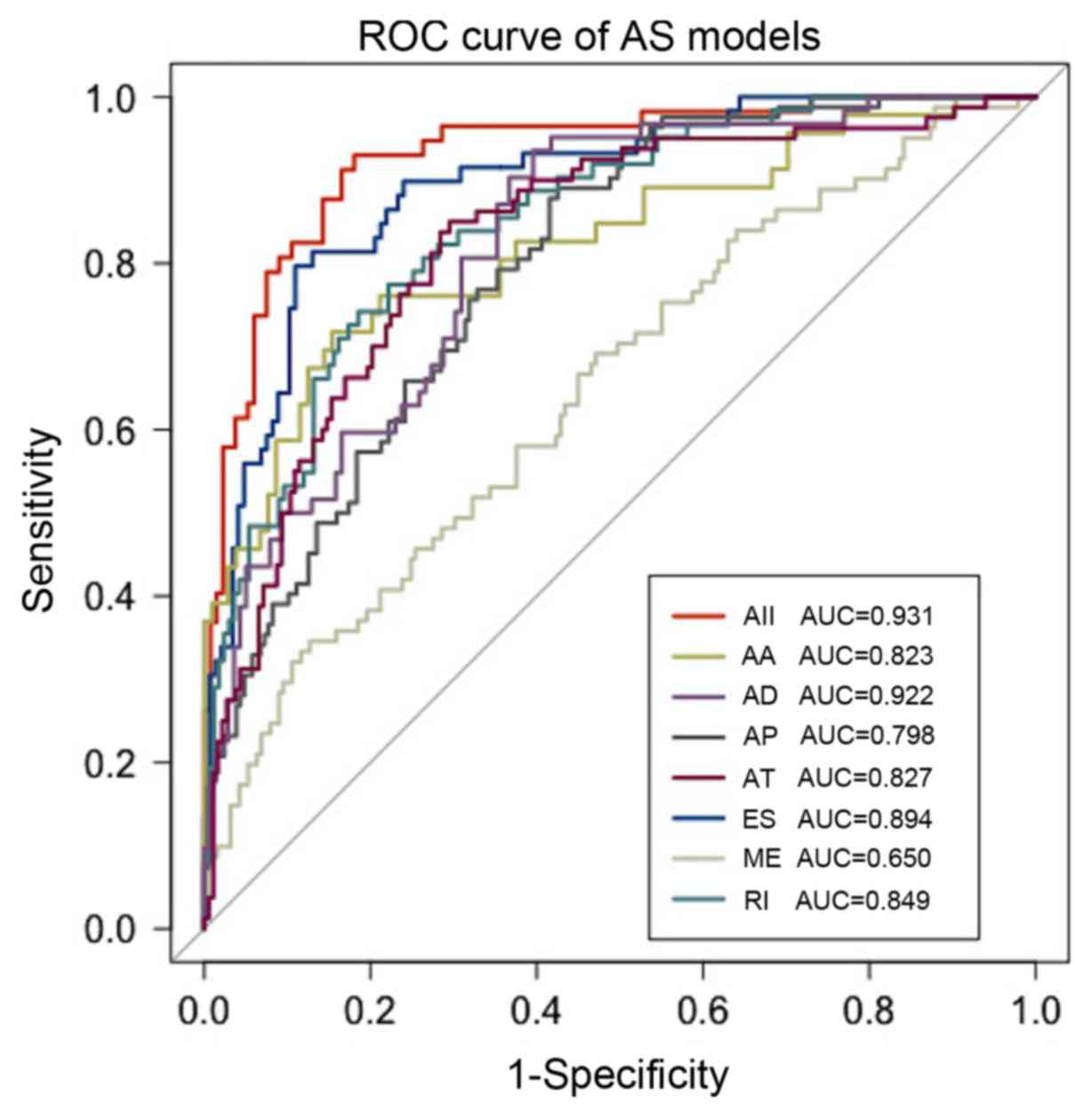 | Figure 3.ROC curves with AUCs of
chemoresistance predictors built by one type or all seven types of
AS events in ovarian serous cystadenocarcinoma. AS, alternative
splicing; ROC, receiver operating characteristic; AUC, area under
the curve; AA, alternate acceptor site; AD, alternate donor site;
AP, alternate promoter; AT, alternate terminator; ES, exon skip;
ME, mutually exclusive exons; RI, retained intron. |
 | Table II.General characteristics of
chemoresistance predictors for ovarian cancer. |
Table II.
General characteristics of
chemoresistance predictors for ovarian cancer.
| Alternative
splicing | Formula | OR (95% CI) (High
vs. low) |
|---|
| AA |
25.108023–4.442929*‘TSACC-8246-AA’+3.315118* | 7.14
(3.65–14.85) |
|
|
‘FAM111A-16027-AA’-5.747130*‘SERPINA1-29121-AA’-7.858303* |
|
|
|
‘TNK1-38931-AA’-10.248344*‘USHBP1-48249-AA’-11.439197* |
|
|
|
‘DNAAF3-52039-AA’-11.843241*‘POLM-79455-AA’ |
|
| AD |
−12.309890–6.338744*‘RNF220-2559-AD’-9.644038*‘GBP3-3711-AD’ | 9.55
(5.18–18.71) |
|
|
+4.0052598*‘CLEC16A-34006-AD’+9.383993*‘GPR56-36585-AD’ |
|
|
|
+14.723866*‘TADA2A-40522-AD’+3.932024*‘YIF1B-49610-AD’ |
|
|
|
+7.967860*‘YBEY-60918-AD’+5.746687*‘ZSCAN25-80706-AD’ |
|
| AP |
8.898032–4.064766*‘ANGEL2-9775-AP’-3.745283* | 6.68
(4.06–11.41) |
|
|
‘KIAA0391-27213-AP’+2.045282*‘PPP1R13L-50435-AP’-9.158386* |
|
|
|
‘FAM110A-58466-AP’-5.969009*‘RBM47-69086-AP’-6.474537* |
|
|
|
‘MYO10-71601-AP’-5.998058*‘NUDT1-78608-AP’+2.415810* |
|
|
|
‘MID1-88461-AP’ |
|
| AT |
1.333149+1.932604*‘FPGT-TNNI3K-3457-AT’+6.408309* | 11.95
(6.78–22.31) |
|
|
‘ADAMTSL4-7486-AT’-6.490989*‘TMEM180-12952-AT’+3.711496* |
|
|
|
‘CSTF3-14883-AT’-5.673945*‘KLC1-29468-AT’+4.2497568* |
|
|
|
‘JUP-40930-AT’-4.365262*‘ZNF544-52425-AT’-5.382822* |
|
|
|
‘RAPH1-57077-AT’+1.901203*‘KRBOX1-64325-AT’-3.998685* |
|
|
|
‘ST3GAL6-65794-AT’ |
|
| ES |
8.884081+12.456874*‘BTAF1-12524-ES’+3.040003*‘SPAG9-42494-ES’ | 22.05
(10.31–54.59) |
|
|
+6.529384*‘GAA-44021-ES’-6.241932*‘RBM6-64950-ES’-5.268264* |
|
|
|
‘SLC10A7-70775-ES’+6.110066*‘TRAPPC13-72245-ES’-6.730836* |
|
|
|
‘PNISR-77056-ES’-4.439848*‘PEX2-84241-ES’-5.337279* |
|
|
|
‘EEF1D-98099-ES’-12.321102*‘COL1A2-1412008-ES’ |
|
| ME |
−2.554826+2.383275*‘ATE1-91855-ME’+2.300138*‘GOLT1B-92984-ME’ | 2.13
(1.36–3.35) |
|
|
+5.824629*‘RAB28-265743-ME’ |
|
| RI |
−14.038586+3.701273*‘POLR2G-16420-RI’+11.008279*‘GLG1-37565-RI’ | 10.23
(5.52–20.39) |
|
|
−5.099955*‘C17orf58-43119-RI’+3.088192*‘CDKN2D-47553-RI’-4.112494* |
|
|
|
‘UNC50-54643-RI’-10.031071*‘ID1-58896-RI’-7.797206*‘HOPX-69372-RI’ |
|
|
|
−1.959239*‘TTC23L-71732-RI’+4.305603*‘SOD2-78304-RI’+3.626741* |
|
|
|
‘PILRB-80936-RI’+2.983951*‘VPS28-85606-RI’ |
|
| All |
14.333930–8.697005*‘SERPINA1-29121-AA’+5.713988*‘SMIM7-48190-AD’- | 64.88
(22.55–284.86) |
|
|
9.355225*‘TRAPPC6B-27360-ES’+9.130642*‘GAA-44021-ES’-2.458800* |
|
|
|
‘PDE4D-72144-AP’-5.978179*‘RBM6-64950-ES’-4.543145* |
|
|
|
‘SLC10A7-70775-ES’+11.331974*‘TRAPPC13-72245-ES’-5.850540* |
|
|
|
‘FAM49B-85160-ES’-6.577849*‘EEF1D-98099-ES’+5.275202* |
|
|
|
‘CLEC16A-34006-AD’-11.452096*‘COL1A2-1412008-ES’-8.721743* |
|
|
|
‘ZNF544-52425-AT’ |
|
 | Table III.Information for AS event candidates
involved in the model based on all types of AS events. |
Table III.
Information for AS event candidates
involved in the model based on all types of AS events.
| Gene | OR (95% CI) | P-value | Type | Exon |
|---|
| SMIM7 | 41.5291
(8.0671–228.7526) | 0.00024 | AD | 7.2 |
| COL1A2 | 0.0043
(0.0003–0.0549) | 0.00061 | ES |
23:24:25:26:27:28:29:30:31:32:33:34:35:36:37:45:46:47 |
| GAA | 130.2634
(13.7266–1,594.0712) | 0.00075 | ES | 2.2 |
| EEF1D | 0.0247
(0.0035–0.1586) | 0.00133 | ES | 6 |
| ZNF544 | 0.0508
(0.0107–0.2331) | 0.00142 | AT | 10.2 |
| SLC10A7 | 0.0916
(0.0257–0.3090) | 0.00151 | ES | 13 |
| FAM49B | 0.0373
(0.0064–0.2078) | 0.00186 | ES | 5 |
| RBM6 | 0.0705
(0.0163–0.2871) | 0.00233 | ES | 4:05:06 |
| TRAPPC6B | 0.0109
(0.0009–0.1213) | 0.00235 | ES | 4 |
| CLEC16A | 10.9329
(3.0549–40.9032) | 0.00237 | AD | 11.2 |
| PDE4D | 0.3015
(0.1554–0.5813) | 0.00274 | AP | 1 |
| TRAPPC13 | 25.9213
(4.4025–167.8280) | 0.00322 | ES | 9 |
| SERPINA1 | 0.03190
(0.0043–0.2147) | 0.00357 | AA | 2.4 |
 | Table IV.Univariate and multivariate logistic
regression analyses for chemoresistance in The Cancer Genome Atlas
datasets. |
Table IV.
Univariate and multivariate logistic
regression analyses for chemoresistance in The Cancer Genome Atlas
datasets.
|
| Univariate |
| Multivariate |
|
|---|
|
|
|
|
|
|
|---|
|
Characteristics | OR | 95% CI | P-value | OR | 95% CI | P-value |
|---|
| Age, years |
|
|
|
|
|
|
|
<60 | 1 (reference) |
|
| 1 (reference) |
|
|
|
≥60 | 0.90 | 0.52–1.53 | 0.741 | 1.72 | 0.77–3.91 | 0.270 |
| Stage |
|
|
|
|
|
|
| FIGO
I/II | 1 (reference) |
|
| 1 (reference) |
|
|
| FIGO
III/IV | 4.10 | 0.94–41.14 | 0.186 | inf | 0-inf | 0.989 |
| Grade |
|
|
|
|
|
|
|
Low | 1 (reference) |
|
| 1 (reference) |
|
|
|
High | 0.99 | 0.43–2.42 | 0.978 | 0.37 | 0.06–1.61 | 0.290 |
| Debulking
status |
|
|
|
|
|
|
|
Optimal | 1 (reference) |
|
| 1 (reference) |
|
|
|
Suboptimal | 2.53 | 1.38–4.63 | 0.012 | 5.13 | 2.01–15.21 | 0.007 |
| Risk score |
|
|
|
|
|
|
|
Low | 1 (reference) |
|
| 1 (reference) |
|
|
|
High | 64.88 | 22.55–284.86 | <0.001 | 192.07 | 41.20–2087.85 | <0.001 |
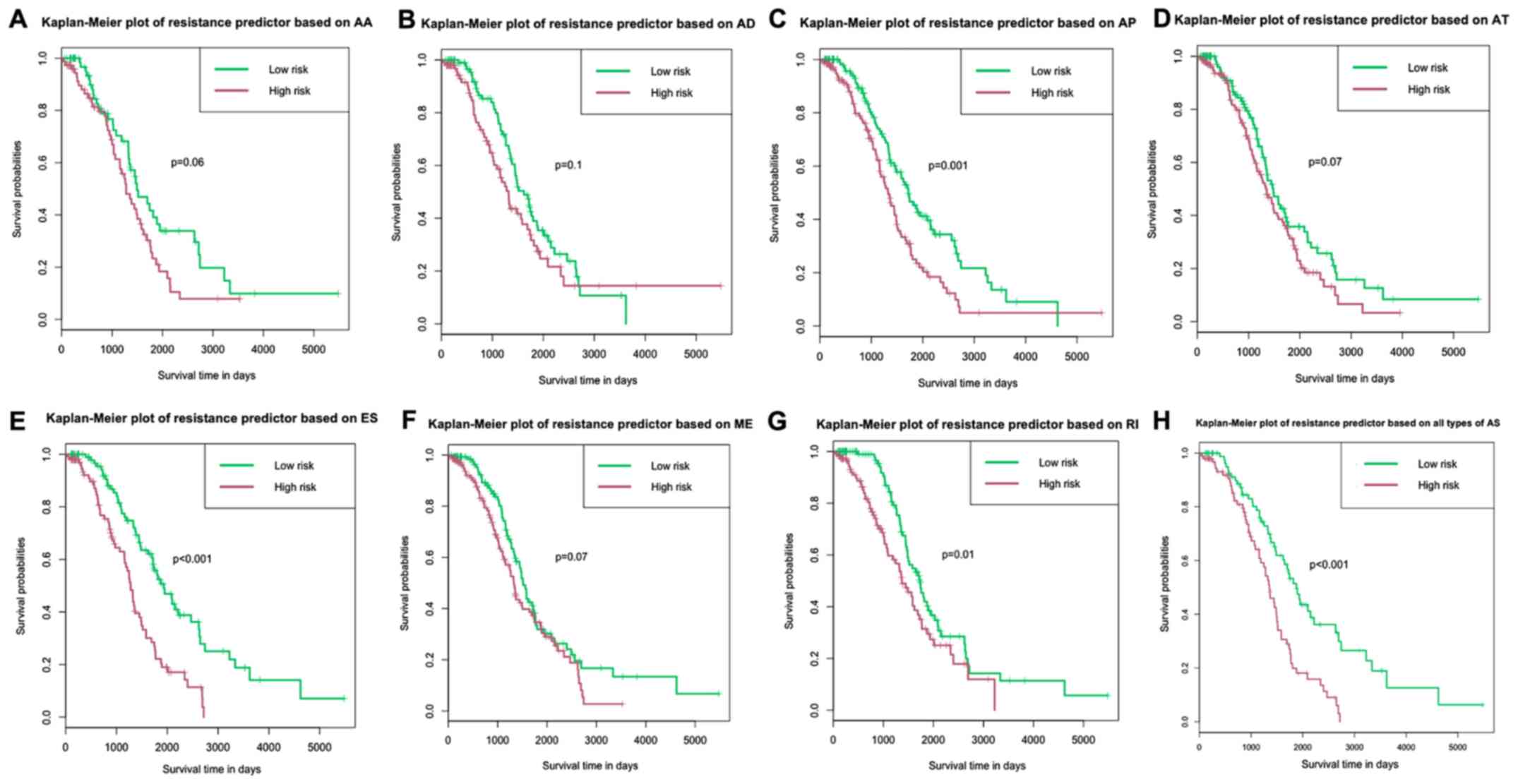
To verify the prognostic value of these predictive
models, Kaplan-Meier analysis and log-rank tests were performed for
each model. The results indicated that the patients in the
high-risk groups in risk models based on AP, ES, RI and all types
of AS events had shorter survival time compared with patients in
the low-risk groups (Fig. 4). In the
risk model based on all types of AS events, the median OS time for
the high- and low-risk groups were 1,341 and 1,875 days,
respectively (Fig. 4H).
Potential correlation network of AS
splicing factors
AS is regulated primarily by splicing factors.
Therefore, it is crucial to determine whether key splicing factors
regulate chemoresistance-associated AS events in OV. Univariate
logistic analyses revealed that the mRNA expression levels of five
splicing factors were associated with chemoresistance. Information
of these splicing factors was obtained from SpliceAid2 and was
shown in Table V. Subsequently,
correlation analyses of the expression levels of the five splicing
factors and the PSI values of 151 AS events were performed
(P<0.01 in univariate analyses). A splicing correlation network
was generated from the significant correlations (P<0.05;
Fig. 5A) between 70
chemoresistance-associated AS events, including 38 protective and
32 adverse AS events, and the 5 splicing factors. Most of the
protective AS events were positively correlated with the expression
of splicing factors, such as AP PSI value of SH3YL1 with expression
of PTBP1, AD PSI value of RPL15 with expression of YBX1, AP PSI
value of CLUL1 with expression of SYNCRIP. Most of the adverse AS
events were negatively correlated with the expression of splicing
factors, such as AT PSI value of UBAP2L with expression of TRA2B,
ES PSI value of RPS24 with expression of SYNCRIP, ES PSI value of
RHOA with expression of ELAVL4. Representative correlations between
AS events and splicing factors are shown in the dot plots (Fig. 5B-G).
 | Table V.Information for splicing factors in
the correlation network from SpliceAid 2. |
Table V.
Information for splicing factors in
the correlation network from SpliceAid 2.
| Splicing
factors | Gene names | Descriptions | Expression in
normal ovary tissue | Expression in
ovarian cancer |
|---|
| hnRNP I (PTB) | PTBP1 | Polypyrimidine
tract binding protein 1. In the context of CALCA gene, PTB enhances
exon 4 inclusion (PMID:9858533). nPTB functionally compensates for
PTB and is upregulated when PTB is removed (PMID:17679092). | P(3)_M(4) | H(1)_M(4)_L(5) |
| HTra2beta1 | TRA2B | Splicing factor
arginine/serine-rich 10. | P(3)_L(4) | H(1)_M(4)_L(5) |
| HuD | ELAVL4 | Embryonic lethal
abnormal vision Drosophila-like 4 (Hu antigen D). | A(4) | A(1)_L(4)_A(5) |
| YB-1 | YBX1 | Y box binding
protein 1. | P(3)_M(4) | H(1)_M(4)_M(5) |
| hnRNP Q | SYNCRIP | Synaptotagmin
binding cytoplasmic RNA interacting protein. | P(3)_M(4) | M(1)_M(4)_L(5) |
Discussion
Previous studies have focused on the function of
single AS events associated with ovarian cancer. Elevated
expression of glutathione-specific γ-glutamylcyclotransferase
splicing variants has been related to poor outcomes in ovarian
cancer (35). Researchers have also
found that an increased level of the mesenchymal spliced variant
CD44s and reduced expression of the epithelial variant CD44v
promotes epithelial-mesenchymal transition and invasion of ovarian
cancer cells (36). A splice variant
of the tetraspanin KAI1 mitigates its tumor-suppressive function,
inducing cell migration and resulting in poor prognosis (37). Chemotherapy sensitivity is the main
factor influencing survival in serous ovarian cancer (38). However, to the best of our knowledge,
only a few studies have investigated the potential role of AS
events in chemotherapy resistance of ovarian cancer (39,40). AS
events of the multidrug resistance-associated protein 1 gene in
ovarian tumors have been reported to confer resistance to
doxorubicin therapy (39).
Overexpression of the VIII-deficient excision repair
cross-complementing group 1 (ERCC1) exon is able to enhance
cisplatin sensitivity in ovarian cancer cell lines by reducing the
protein expression levels of ERCC1 (40). The present study demonstrated that
the ES event of the ERCC1 gene was a protective factor for
chemotherapy resistance, with an OR of 0.069 and a 95% CI of
0.008–0.638 (Table SI), indicating
that these results are consistent with the aforementioned study.
Hence, these studies demonstrated the potential role of AS in
chemotherapy resistance of OV, and further systematic studies of AS
signatures in OV may help to identify potential biomarkers and
targets for chemoresistance.
The present study systemically analyzed the role of
AS signatures in chemotherapy resistance using data from 320
patients with OV from the TCGA database, and then built powerful
resistance predictors. A total of 22,036 AS events were detected in
7,404 genes. Approximately 38% of the AS events were ES, and the
risk model based on ES events exhibited high efficiency. ES events
can be validated by PCR. Thus, future research should investigate
associations between ES events and chemotherapy resistance in more
detail. The predictive model based on all types of AS had the best
efficiency, with the AUC of the ROC curve reaching 0.931. This was
much higher than the AUC for models based on a single type of AS
and was more efficient than previous predictors based on single
mRNA expression (AUC, 0.8056) (41),
the lncRNA signature (AUC, 0.83) (42) or the clinical serum CA125/ascites
leptin (AUC, 0.846) (43). These
combined results suggest that this model could provide accurate
predictions of chemotherapy resistance in patients with OV.
Additionally, the present study investigated the
potential role of splicing factors in chemotherapy resistance. Five
splicing factors were associated with chemotherapy resistance, and
their possible targets were identified. These results suggested
that splicing factors were involved in chemotherapy resistance in
patients with serous ovarian cancer. Further work is required to
determine whether regulation of these specific splicing factors
could increase the sensitivity to chemotherapy and prevent disease
recurrence.
The present study presented some limitations. The
present study was based on RNA-seq data from the TCGA database.
Validation using other databases or larger cohorts is required in
future studies. Numerous splicing events and splicing factors that
may be associated with the biological behavior of OV were
identified and should be further evaluated in future experimental
studies.
In summary, the present study determined that AS
events provided valuable predictors for chemotherapy resistance.
The model used provided efficient risk stratification for
predicting chemotherapy resistance in patients with OV. A splicing
correlation network was generated to explore the potential
relationship between splicing factors and AS. A number of valuable
targets were identified for future validation. The present study
elucidated the role of AS events in primary platinum-based
chemoresistance in patients with serous ovarian cancer and provided
potential targets to overcome chemoresistance.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by The National
Natural Science Foundation of China (grant no. 81872125) and The
Research Fund for the Science and Welfare Career of Liaoning
Province (grant no. 20170017).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
TS and QY designed the study. TS performed the
statistical analyses and wrote the manuscript. QY revised and
edited the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
In the original article of the datasets, the trials
were approved by the local institutional review boards of all
participating centers, and informed consent was obtained from all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torre LA, Trabert B, DeSantis CE, Miller
KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A and Siegel RL:
Ovarian cancer statistics, 2018. CA Cancer J Clin. 68:284–296.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jessmon P, Boulanger T, Zhou W and
Patwardhan P: Epidemiology and treatment patterns of epithelial
ovarian cancer. Expert Rev Anticancer Ther. 17:427–437. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bowtell DD, Böhm S, Ahmed AA, Aspuria PJ,
Bast RC, Beral V, Berek JS, Birrer MJ, Blagden S, Bookman MA, et
al: Rethinking ovarian cancer II: Reducing mortality from
high-grade serous ovarian cancer. Nat Rev Cancer. 15:668–679. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ryner L, Guan Y, Firestein R, Xiao Y, Choi
Y, Rabe C, Lu S, Fuentes E, Huw LY, Lackner MR, et al: Upregulation
of Periostin and reactive Stroma is associated with primary
chemoresistance and predicts clinical outcomes in epithelial
ovarian cancer. Clin Cancer Res. 21:2941–2951. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Muñoz-Galván S, Felipe-Abrio B,
García-Carrasco M, Domínguez-Piñol J, Suarez-Martinez E,
Verdugo-Sivianes EM, Espinosa-Sánchez A, Navas LE, Otero-Albiol D,
Marin JJ, et al: New markers for human ovarian cancer that link
platinum resistance to the cancer stem cell phenotype and define
new therapeutic combinations and diagnostic tools. J Exp Clin
Cancer Res. 38:2342019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
van Zyl B, Tang D and Bowden NA:
Biomarkers of platinum resistance in ovarian cancer: What can we
use to improve treatment. Endocr Relat Cancer. 25:R303–R318. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shu T, Li Y, Wu X, Li B and Liu Z:
Down-regulation of HECTD3 by HER2 inhibition makes serous ovarian
cancer cells sensitive to platinum treatment. Cancer Lett.
411:65–73. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bai L, Wang A, Zhang Y, Xu X and Zhang X:
Knockdown of MALAT1 enhances chemosensitivity of ovarian cancer
cells to cisplatin through inhibiting the Notch1 signaling pathway.
Exp Cell Res. 366:161–171. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bai H, Cao D, Yang J, Li M, Zhang Z and
Shen K: Genetic and epigenetic heterogeneity of epithelial ovarian
cancer and the clinical implications for molecular targeted
therapy. J Cell Mol Med. 20:581–593. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Konecny GE, Winterhoff B and Wang C:
Gene-expression signatures in ovarian cancer: Promise and
challenges for patient stratification. Gynecol Oncol. 141:379–385.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chon HS and Lancaster JM: Microarray-based
gene expression studies in ovarian cancer. Cancer Control. 18:8–15.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Salton M and Misteli T: Small molecule
modulators of Pre-mRNA splicing in cancer therapy. Trends Mol Med.
22:28–37. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Narayanan SP, Singh S and Shukla S: A saga
of cancer epigenetics: Linking epigenetics to alternative splicing.
Biochem J. 474:885–896. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nilsen TW and Graveley BR: Expansion of
the eukaryotic proteome by alternative splicing. Nature.
463:457–463. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nakka K, Ghigna C, Gabellini D and
Dilworth FJ: Diversification of the muscle proteome through
alternative splicing. Skelet Muscle. 8:82018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chang SH, Elemento O, Zhang J, Zhuang ZW,
Simons M and Hla T: ELAVL1 regulates alternative splicing of eIF4E
transporter to promote postnatal angiogenesis. Proc Natl Acad Sci
USA. 111:18309–18314. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Y, Liu Y, Nie H, Ma X and Xu Z:
Alternative splicing of inner-ear-expressed genes. Front Med.
10:250–257. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin L, Park JW, Ramachandran S, Zhang Y,
Tseng YT, Shen S, Waldvogel HJ, Curtis MA, Faull RL, Troncoso JC,
et al: Transcriptome sequencing reveals aberrant alternative
splicing in Huntington's disease. Hum Mol Genet. 25:3454–3466.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Urbanski LM, Leclair N and Anczuków O:
Alternative-splicing defects in cancer: Splicing regulators and
their downstream targets, guiding the way to novel cancer
therapeutics. Wiley Interdiscip Rev RNA. 9:e14762018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Martinez-Montiel N, Rosas-Murrieta NH,
Anaya Ruiz M, Monjaraz-Guzman E and Martinez-Contreras R:
Alternative splicing as a target for cancer treatment. Int J Mol
Sci. 19(pii): E5452018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dvinge H, Kim E, Abdel-Wahab O and Bradley
RK: RNA splicing factors as oncoproteins and tumour suppressors.
Nat Rev Cancer. 16:413–430. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sveen A, Kilpinen S, Ruusulehto A, Lothe
RA and Skotheim RI: Aberrant RNA splicing in cancer; Expression
changes and driver mutations of splicing factor genes. Oncogene.
35:2413–2427. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen S, Wang Y, Wang C, Wu YN and Xing Y:
SURVIV for survival analysis of mRNA isoform variation. Nat Commun.
7:115482016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Song J, Liu YD, Su J, Yuan D, Sun F and
Zhu J: Systematic analysis of alternative splicing signature
unveils prognostic predictor for kidney renal clear cell carcinoma.
J Cell Physiol. 234:22753–22764. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhu GQ, Zhou YJ, Qiu LX, Wang B, Yang Y,
Liao WT, Luo YH, Shi YH, Zhou J, Fan J and Dai Z: Prognostic
alternative mRNA splicing signature in hepatocellular carcinoma: A
study based on large-scale sequencing data. Carcinogenesis. May
17–2019.doi: 10.1093/carcin/bgz073 (Epub ahead of print).
View Article : Google Scholar
|
|
28
|
Zhu J, Chen Z and Yong L: Systematic
profiling of alternative splicing signature reveals prognostic
predictor for ovarian cancer. Gynecol Oncol. 148:368–374. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ryan M, Wong WC, Brown R, Akbani R, Su X,
Broom B, Melott J and Weinstein J: TCGASpliceSeq a compendium of
alternative mRNA splicing in cancer. Nucleic Acids Res.
44:D1018–D1022. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Berger AC, Korkut A, Kanchi RS, Hegde AM,
Lenoir W, Liu W, Liu Y, Fan H, Shen H, Ravikumar V, et al: A
Comprehensive Pan-cancer molecular study of gynecologic and breast
cancers. Cancer Cell. 33:690–705.e9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Piva F, Giulietti M, Burini AB and
Principato G: SpliceAid 2: A database of human splicing factors
expression data and RNA target motifs. Hum Mutat. 33:81–85. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Akaike H: Information theory and an
extension of the maximum likelihood principle. 2nd Int. Sympo. on
Information Theor, 1972. https://doi.org/10.1007/978-1-4612-0919-5_38
|
|
33
|
Jayson GC, Kohn EC, Kitchener HC and
Ledermann JA: Ovarian cancer. Lancet. 384:1376–1388. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Goebel G, Berger R, Strasak AM, Egle D,
Müller-Holzner E, Schmidt S, Rainer J, Presul E, Parson W, Lang S,
et al: Elevated mRNA expression of CHAC1 splicing variants is
associated with poor outcome for breast and ovarian cancer
patients. Br J Cancer. 106:189–198. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bhattacharya R, Mitra T, Ray Chaudhuri S
and Roy SS: Mesenchymal splice isoform of CD44 (CD44s) promotes
EMT/invasion and imparts stem-like properties to ovarian cancer
cells. J Cell Biochem. 119:3373–3383. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Upheber S, Karle A, Miller J, Schlaugk S,
Gross E and Reuning U: Alternative splicing of KAI1 abrogates its
tumor-suppressive effects on integrin αvβ3-mediated ovarian cancer
biology. Cell Signal. 27:652–662. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Markman M: Antineoplastic agents in the
management of ovarian cancer: Current status and emerging
therapeutic strategies. Trends Pharmacol Sci. 29:515–519. 2018.
View Article : Google Scholar
|
|
39
|
He X, Ee PL, Coon JS and Beck WT:
Alternative splicing of the multidrug resistance protein 1/ATP
binding cassette transporter subfamily gene in ovarian cancer
creates functional splice variants and is associated with increased
expression of the splicing factors PTB and SRp20. Clin Cancer Res.
10:4652–4660. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sun Y, Li T, Ma K, Tian Z, Zhu Y, Chen F
and Hu G: The impacts of ERCC1 gene exon VIII alternative splicing
on cisplatin-resistance in ovarian cancer cells. Cancer Invest.
27:891–897. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhao H, Sun Q, Li L, Zhou J, Zhang C, Hu
T, Zhou X, Zhang L, Wang B, Li B, et al: High expression levels of
AGGF1 and MFAP4 predict primary platinum-based chemoresistance and
are associated with adverse prognosis in patients with serous
ovarian cancer. J Cancer. 10:397–407. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu R, Zeng Y, Zhou CF, Wang Y, Li X, Liu
ZQ, Chen XP, Zhang W and Zhou HH: Long noncoding RNA expression
signature to predict platinum-based chemotherapeutic sensitivity of
ovarian cancer patients. Sci Rep. 7:182017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Matte I, Garde-Granger P, Bessette P and
Piché A: Serum CA125 and ascites leptin level ratio predicts
baseline clinical resistance to first-line platinum-based treatment
and poor prognosis in patients with high grade serous ovarian
cancer. Am J Cancer Res. 9:160–170. 2019.PubMed/NCBI
|