Introduction
Laryngeal squamous cell carcinoma (LSCC), a common
malignant tumor, is closely associated with tobacco usage (1). In 2018, there were an estimated 13,150
new cases of LSCC in the USA alone (2). The symptoms of early-stage LSCC
including, mild hoarseness, dysphonia, a cough and a hypopharyngeal
foreign body sensation, are atypical and easy to ignore. Thus, in
clinical practice, LSCC is most typically diagnosed via a
pathological biopsy using a laryngoscope or imaging techniques
(3). The survival rate of patients
with early-stage LSCC is ~80-90%, which is significantly higher
compared with patients with advanced LSCC (4,5);
however, at the time of diagnosis, the majority of patients are
already in the advanced stages of LSCC or exhibit neck lymph node
metastasis, demonstrating a survival rate of 50% (4). Unfortunately, to date, there are no
specific diagnostic markers or prognostic/predictive markers for
early-stage LSCC. Thus, the requirement to identify effective
prognostic biomarkers for patients with LSCC remains crucial.
The pathogenesis of LSCC is highly complex and
involves not only environmental factors but also genetic
dysregulation (3,6). Long non-coding RNAs (lncRNAs) are a
class of RNAs of >200 nucleotides in length that have no protein
coding ability (7). In fact, the
Genome Project reported that >98% of genes in the entire genome
are non-protein coding genes that are transcribed into non-coding
RNA (ncRNA), of which 76% of ncRNAs are transcribed into lncRNAs
(8,9).
lncRNAs have attracted increasing attention in the
previous few years in the field of cancer research and have been
proven to serve a core regulatory role in protein synthesis
(10). lncRNAs also influence
genomic loci imprinting, chromosomal conformational shaping,
allosterically regulating enzyme activity and numerous other
important biological activities, including coordinating the cell
status and differentiation (7,11,12).
Notably, the upregulation, deletion and mutation of lncRNA genes
has been implicated in numerous types of human disease, especially
cancer (13).
In our previous study, the expression levels of
AC023794.4-201 were analyzed in LSCC tissues, adjacent
non-cancerous tissues and lymphatic metastatic tissues;
AC023794.4-201 expression levels were decreased in the carcinoma
tissues of patients with LSCC with low differentiation, high T
staging and cervical lymph node metastasis compared with other
tissue samples (14). In addition,
low AC023794.4-201 expression levels were discovered to be a poor
prognostic factor for LSCC (14). In
the present study, cell function assays were performed and a
xenograft nude mouse model was constructed to further investigate
the functions of AC023794.4-201 in LSCC.
Materials and methods
Cell culture
Two human LSCC cell lines, TU-212 and AMC-HN8, were
obtained from Jennio Biotech Co., Ltd. and the BeNa Culture
Collection, respectively. The cell lines were authenticated by
short tandem repeat profiling and tested for mycoplasma. TU-212
cells were cultured in RPMI-1640 medium (HyClone; GE Healthcare
Life Sciences), supplemented with 10% FCS (PAN-Biotech GmbH), and
AMC-HN8 cells were cultured in high-glucose DMEM (HyClone; GE
Healthcare Life Sciences), supplemented with 10% FCS (PAN Biotech
UK, Ltd.). Both cell lines were cultured in an incubator at 37°C
with 5% CO2.
Lentiviral transfection
To increase the expression levels of AC023794.4-201
in LSCC cells, AC023794.4-201 overexpression lentiviruses and empty
lentiviruses vector (LV5-EF1a-GFP/Puro) were purchased from
Shanghai GenePharma Co., Ltd. to transfect LSCC cells. Vectors of
lentiviruses are plasmids. According to the GenePharma lentivirus
operation manual, the cells (3×105 per hole plate) were
transfected with the lentiviruses (5×108 TU/ml) in a
6-well plate in their respective medium, supplemented with 10% FCS.
Following 24 h of incubation, the original medium containing the
lentiviruses was removed and replaced with Opti-MEM I reduced serum
medium (Gibco; Thermo Fisher Scientific, Inc.). LSCC cells
overexpressing AC023794.4-201 were named
LV5-AC023794.4-201-transfected cells and empty
lentivirus-transfected LSCC cells carrying no target gene were
named LV5-negative control (NC)-transfected cells. A group of cells
not transfected with lentivirus was set as the blank group.
Quantitative real-time reverse
transcription-polymerase chain reaction (qRT-PCR)
Total RNA was extracted from experimental and
control cells of two cell lines (AMC-HN8 and TU-212) and xenograft
tumor samples using TRIzol® reagent (Thermo Fisher
Scientific Inc.). The concentration and purity of the total RNA was
measured using a SmartSpec Plus spectrophotometer (Bio-Rad,
Laboratories); when the A260/A280 value was between 1.8 and 2.0,
the purity of the total extracted RNA was considered acceptable.
Total RNA from each sample was reverse transcribed into cDNA using
the GoScript™ Reverse Transcription system kit (Promega
Corporation) according to the manufacturer's instructions. qPCR was
subsequently performed using a Mx3005P fluorescence quantitative
PCR instrument (Stratagene; Agilent Technologies, Inc.), according
to the manufacturer's protocol, and a GoTaq® qPCR Master
mix kit (Promega Corporation). The primers used for the PCR were
synthesized by Sangon Biotech Co., Ltd. and the sequences are
presented in Table I. The following
thermocycling conditions were used for the qPCR: Initial
denaturation at 95°C for 5 min; and 40 cycles of denaturation at
94°C for 15 sec, annealing at 55°C for 30 sec and elongation at
72°C for 30 sec. Expression levels were determined using the
2−ΔΔcq method (15) and
normalized to GAPDH, the internal reference gene. All samples were
tested in triplicate and the results were averaged.
 | Table I.Primer sequences of AC023794.4-201
and GAPDH used for reverse transcription-quantitative PCR. |
Table I.
Primer sequences of AC023794.4-201
and GAPDH used for reverse transcription-quantitative PCR.
| Gene | Primer sequence
(5′→3′) |
|---|
| AC023794.4-201 | F:
AGAAGAGGGGGAAAAAAG |
|
| GATGAAG |
|
| R:
CGATGGTTAGGGGTGGGAAAGTC |
| GAPDH | F:
ACCCACTCCTCCACCTTTGAC |
|
| R:
TGTTGCTGTAGCCAAATTCGTT |
Cell proliferation assay
The proliferation of LSCC cells was analyzed using a
real-time cell analyzer (RTCA) assay and the cell index (CI) was
regarded as an indicator of cell proliferation. Briefly, 24 h after
transfection, 5×105 cells/ml cultured in normal media
were plated into an E-plate 96, which was subsequently placed in an
RTCA (Agilent Technologies, Inc.) to monitor cell proliferation.
Proliferation was monitored every 15 min for a total of 96 h.
Colony formation assay
A colony formation assay was also performed to
investigate cell proliferation. Following treatment for 24 h, the
cells were dissociated with EDTA containing 0.25% trypsin and 500
cells/well were plated into a 6-well plate. The 6-well plate was
incubated in a cell culture incubator at 37°C for 2 weeks.
Following the incubation at room temperature the colonies were
fixed with 4% paraformaldehyde for 15 min and stained with a 0.1%
crystal violet for 20 min staining solution. The stained colonies
were manually counted by eye. Each clone contained >50 cells,
ranging in size from 0.3–1.0 mm.
Flow cytometric analysis of the cell
cycle
Cells were serum-starved for 24 h to synchronize the
cells in the G0 phase. The cells were then transfected
and collected 24 h following transfection. Subsequently, cells were
rinsed with PBS and fixed in 70% ethanol at −20°C for 24 h. After
fixation, pre-cooled (4°C) PBS was added to rehydrate the cells for
15 min. According to the manufacturer's protocol, 500 µl propidium
iodide (PI)/RNase cell cycle staining kit reagent [Hangzhou Multi
Sciences (Lianke) Biotech Co., Ltd.] was added and incubated at
37°C for 30 min in the dark. The cell cycle distribution of the
stained cells was determined using a BD FACSCalibur™ flow cytometer
(BD Biosciences).
Flow cytometric analysis of
apoptosis
Software FlowJo® v.10.5.2 (FlowJo LLC)
was used to analyze results. Transfected cells were dissociated
with 0.25% trypsin (EDTA-free) and washed with PBS. According to
the manufacturer's instructions, the resuspended
cells(1×106/ml) were stained with Annexin V-FITC and PI
using the Annexin V-FITC/PI cell apoptosis staining kit (BD
Biosciences). Following 15 min of incubation at room temperature,
apoptotic cells were analyzed using a BD FACSCalibur™ flow
cytometer (BD Biosciences).
Transwell assay
Cells (4×105/ml) were plated in the upper
chambers of Transwell plates in serum-free culture medium specific
for each cell line. Cell culture medium for each cell line,
supplemented with 10% FCS, was plated in the lower chambers.
Following 24 h of incubation at 37°C, the non-migratory cells
remaining on the membrane were removed. The migratory cells in the
lower chamber were stained with 4% methanol and 0.1% crystal violet
(Sigma-Aldrich; Merck KGaA) at room temperature for 10 min. Stained
cells were visualized and imaged using a binocular inverted
microscope (Olympus, Japan; Magnification, ×40).
Xenograft nude mouse model
experiment
All animal studies were approved by and strictly
followed the guidelines of the Animal Ethics and Welfare Committee
of Ningbo University (approval no. AWEC-2015-10). A total of 32
BALB/c nude mice (age, 4 weeks; sex, male; weight, 14–16 g) were
provided by Shanghai SIPPR-Bk Lab Animal Co., Ltd. All nude mice
were raised and tested at the Ningbo University Laboratory Animal
Center at room temperature, 25°C and humidity 70%. The mice had
sterile water and food sterilized by radiation and their cage liner
was replaced every 2 days. A cell suspension with a cell
concentration of 5×107/ml was made by mixing the
AMC-HN-8 cells and the matrigel (BD Biosciences). Each nude mouse
armpit was subcutaneously inoculated with 0.2 ml AMC-HN-8 cell
suspension that is mentioned previously. Ten days after
inoculation, the mice were stochastically assigned to 4 groups (8
mice/group): i) Blank control group (no treatment); ii) LV5-NC
group, which were injected in xenograft with 50 µl LV5-NC
twice/week for 28 days; iii) low-dose group, which were injected in
xenograft with 5×107 TU/ml LV5-AC023794.4-201 twice/week
for 28 days; and iv) high-dose group, which were injected in
xenograft with 5×108 TU/ml LV5-AC023794.4-201 twice/week
for 28 days. The weight, maximum length and maximum width of the
tumors were measured twice/week. The maximum length(a) and maximum
width(b) of the tumors were measured with a vernier caliper. Tumor
volume (mm3) is equal to 1/2 ab2. The maximum
diameter and volume of a single tumor was 18.20 mm and 1,114.76
mm3, respectively. On day 29, the first day of drug
withdrawal, the mice were sacrificed with an intraperitoneal
injection of 200 mg/kg pentobarbital and the tumors were harvested.
Tumor specimens were fixed for histopathological examination and
transmission electron microscopy (TEM).
Hematoxylin and eosin (H&E)
staining
Tumor tissues were fixed in 10% neutral formalin for
3 days at 25°C. The fixed tumor tissues were dehydrated with an
ascending concentration of alcohol (Sinopharm Chemical Reagent Co.,
Ltd) (75, 85 and 100%) and washed with xylene. Sections were
embedded in paraffin and cut into 5-µm-thick sections that were
incubated at 60°C in an electric thermostatic blast oven for 3 h.
The following experiments are performed at room temperature. The
tissue sections were subsequently deparaffinized in 20 ml xylene,
rehydrated in a descending alcohol series (5 ml; 100, 85 and 75%)
and soaked with distilled water for 5 min. Sections were stained
with 0.3% hematoxylin for 10 min and subsequently differentiated in
0.5% alcohol hydrochloride and 0.25% ammonia water for 5 sec. Then,
sections were stained with 0.5% eosin solution for 1 min and
dehydrated with an ascending alcohol series (75, 85 and 100%) for 5
min. Finally, sections were cleared with xylene, mounted with
neutral gum and visualized using an inverted fluorescence
microscope (Nikon, Corporation; magnification, ×100).
TEM
Tumors were cut into 1 mm3-thick tissue
blocks and fixed with 3% glutaraldehyde at 4°C for 2 h. Tissues
were rinsed with 0.1 M phosphate rinsing solution 3 times for 15
min, then fixed with 1% osmium tetroxide for 2 h and rinsed with
0.1 M phosphate rinsing solution 3 times for 15 min at 4°C. Rinsed
tissue blocks were dehydrated in an ascending ethanol series (50,
70 and 90%), (90% ethanol and 90% acetone at a 1:1 equivalent
ratio), 90 and 100% acetone for 15 min at 4°C and subsequently
dehydrated with 100% acetone at room temperature for 15 min. The
tissues were impregnated with 618 epoxy resin (Shanghai Resin
Factory Co., Ltd). The embedded samples were cut into 70 nm-thick
sections with an ultrathin slicer. The sections were stained with
2%uranyl acetate for 10 min and 2%lead citrate for 5 min at room
temperature. Finally, the apoptotic morphological changes of cells
were visualized using a Hitachi H-7650 transmission electron
microscope (Hitachi, Ltd.; magnification, ×15,000).
Statistical analysis
Cell function experiments including RTCA, cell
cloning, flow cytometry, migration experiment and PCR were repeated
3 times. Statistical analysis was performed using SPSS 18.0
software (SPSS Inc.) and all quantitative data are presented as the
mean ± SD. Unpaired Student's t-tests were used to analyze the
statistical differences between the groups in the cell functional
assays, whereas one-way ANOVA was used to determine the statistical
differences between >2 groups, followed by a Tukey's test for
multiple comparisons. P<0.05 was considered to indicate a
statistically significant difference.
Results
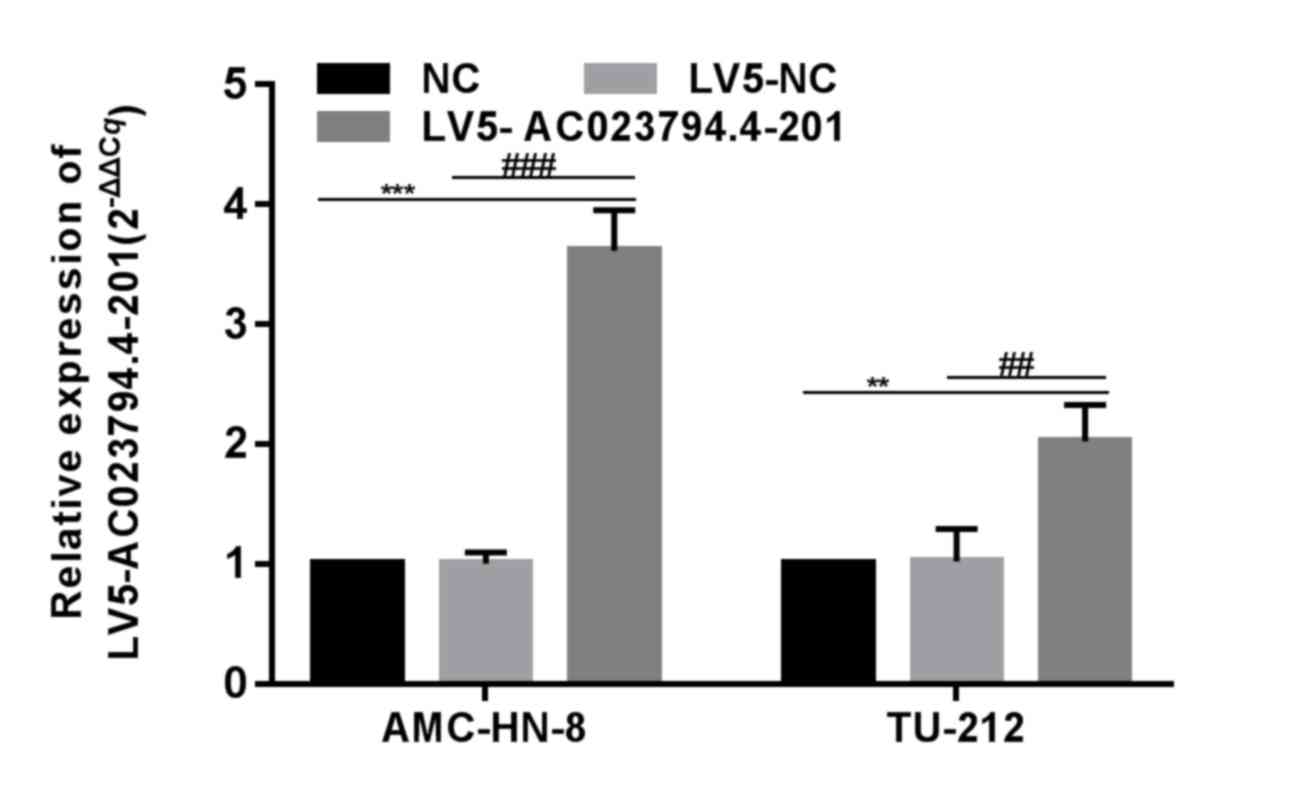
Overexpression of AC023794.4-201 in
LSCC cell lines
AC023794.4-201 was previously discovered to be
expressed at low levels in the cancerous tissues of patients with
LSCC with low differentiation, a high T stage and cervical lymph
node metastasis (14). Thus, to
assess the role of AC023794.4-201 in tumor progression,
AC023794.4-201 was overexpressed in two LSCC cell lines using
AC023794.4-201 overexpression lentiviruses. AC023794.4-201
expression levels in LV5-AC023794.4-201-transfected cells were
significantly increased compared with the LV5-NC-transfected cells,
whereas there was no significant difference observed between the NC
group and LV5-NC group (Fig. 1).
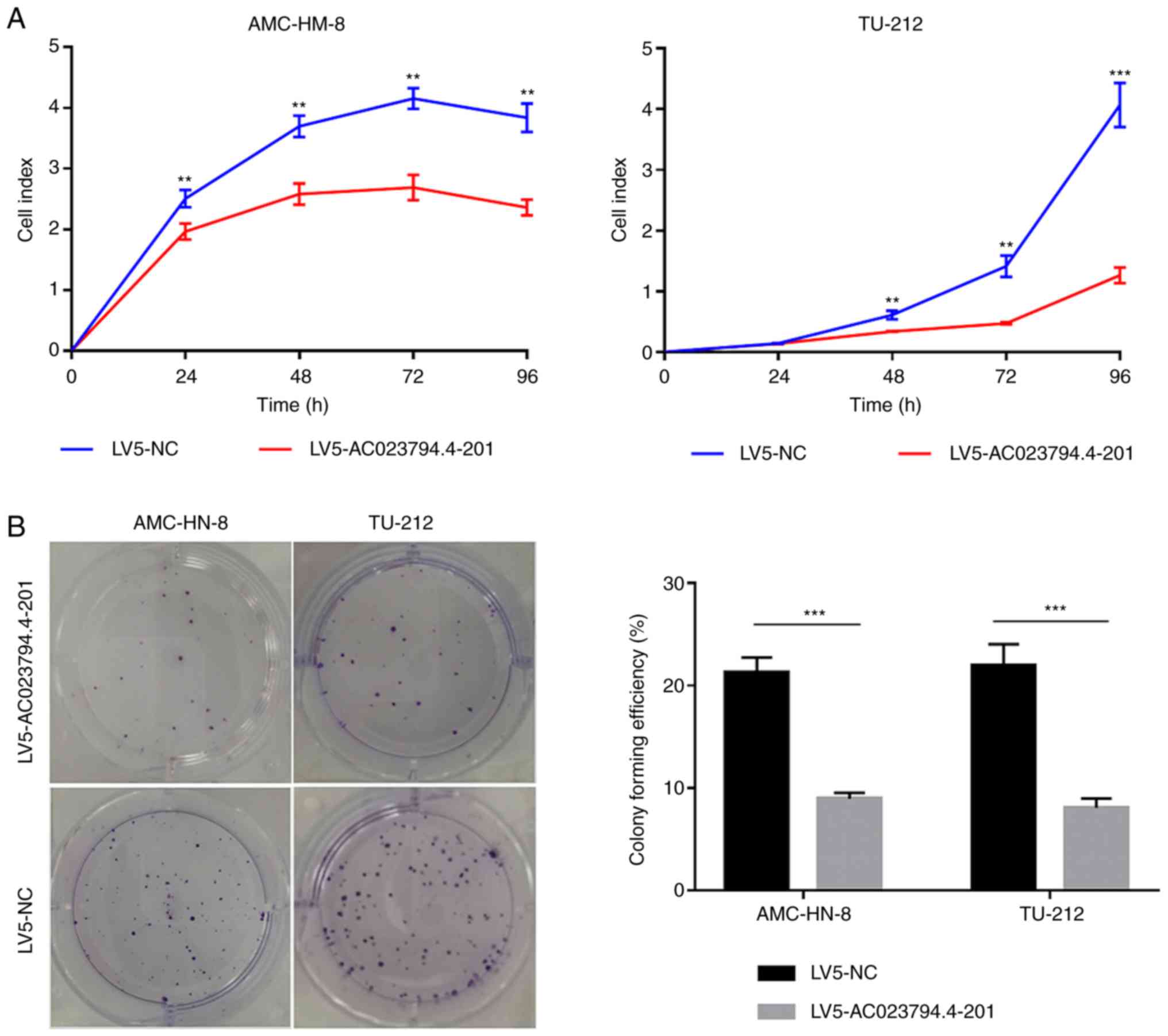
AC023794.4-201 inhibits LSCC cell
proliferation and colony formation
Cell proliferation following AC023794.4-201
lentiviral transfection was analyzed using a RTCA assay. The cell
index of LV5-AC023794.4-201-transfected cells in both LSCC cell
lines was significantly decreased compared with the LV5-NC groups
(Fig. 2A). Moreover, the colony
forming ability of LV5-AC023794.4-201-transfected cells was
significantly decreased in both cell lines compared with the
LV5-NC-transfected cells (Fig.
2B).
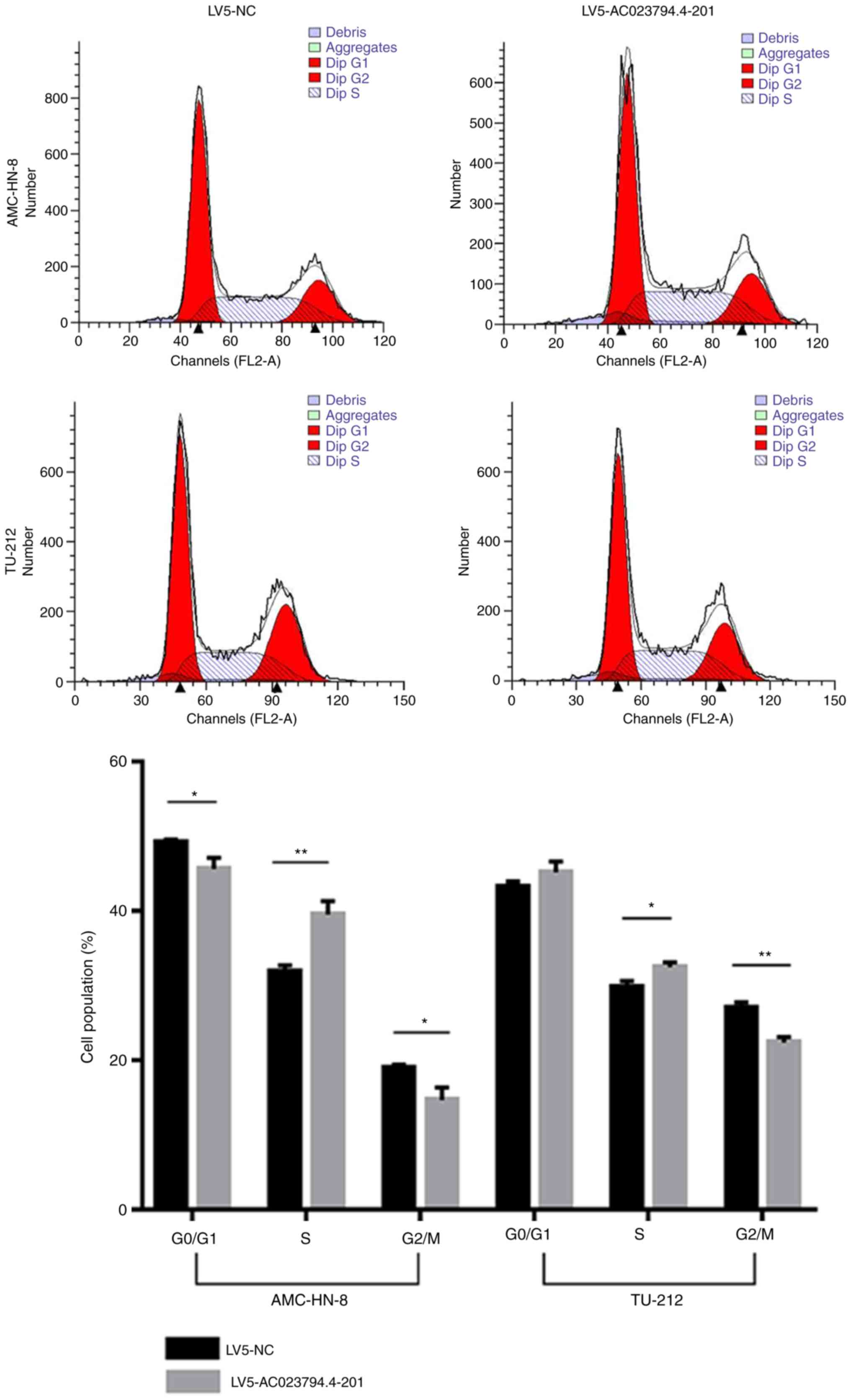
Ac026166.2-001 blocks cell cycle
progression from the S phase to G2/M phase and promotes
cell apoptosis
Cells in the G0 and G1 phases
of the cell cycle were analyzed together as the DNA of both was the
same and the 2 phases could not be distinguished from one another.
Flow cytometry was used to analyze the cell cycle distribution and
rate of apoptosis, and to further investigate the mechanism by
which AC023794.4-201 may inhibit cell proliferation. The proportion
of LV5-AC023794.4-201-transfeced cells in the S phase was
significantly increased compared with the LV5-NC-transfected cells
in both cell lines (Fig. 3).
Conversely, the proportion of cells in the G2/M phase
was significantly decreased in the LV5-AC023794.4-201 group
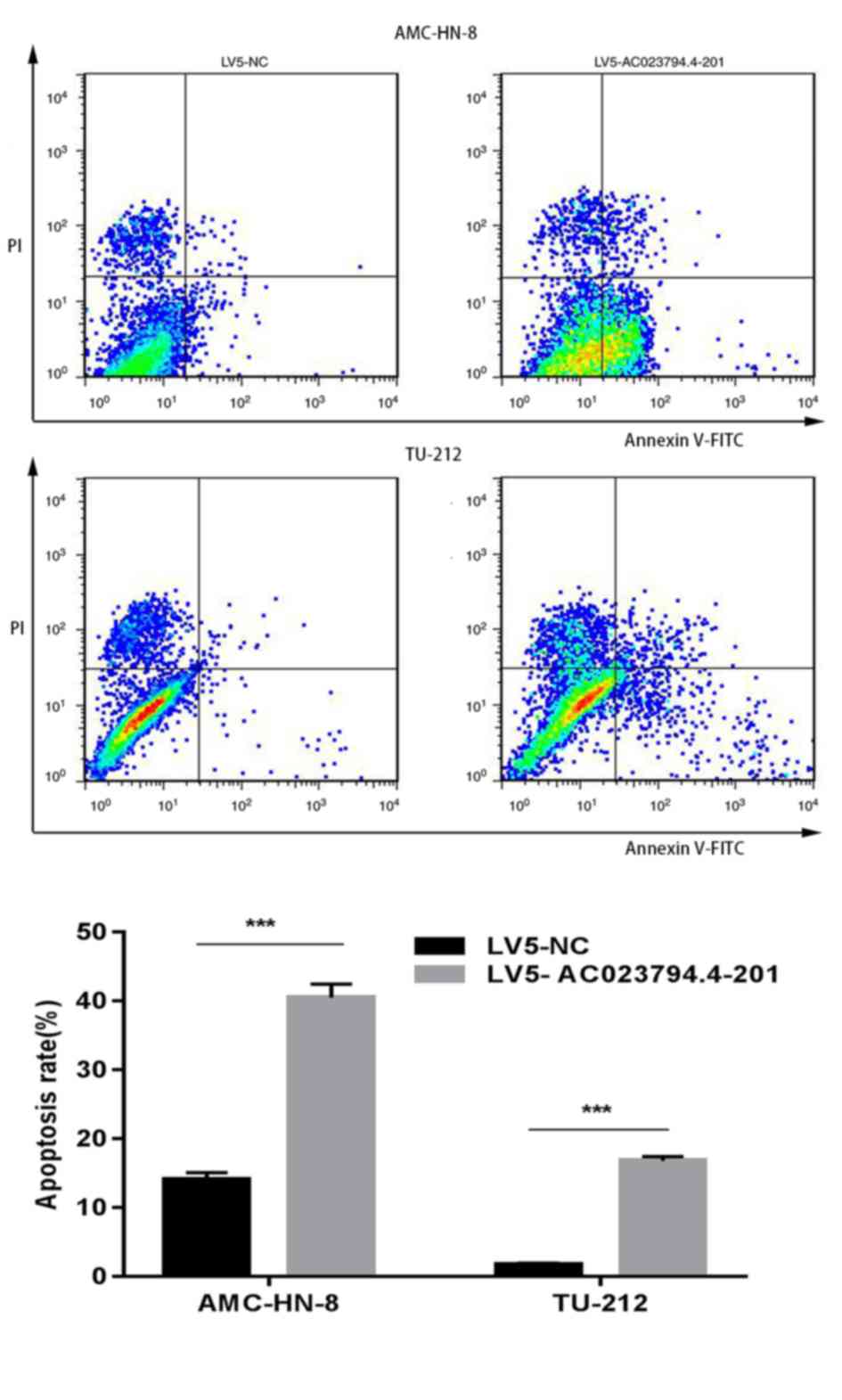
compared with the LV5-C group in both cell lines (Fig. 3). Furthermore, the apoptosis assay
indicated that the percentage of LV5-AC023794.4-201-transfected
apoptotic cells was significantly increased compared with the
LV5-NC-transfected apoptotic cells in both LSCC cell lines
(Fig. 4). Thus, these finding
suggest that the overexpression of AC023794.4-201 may arrest the
cell cycle and promote apoptosis, thereby inhibiting cell
proliferation.
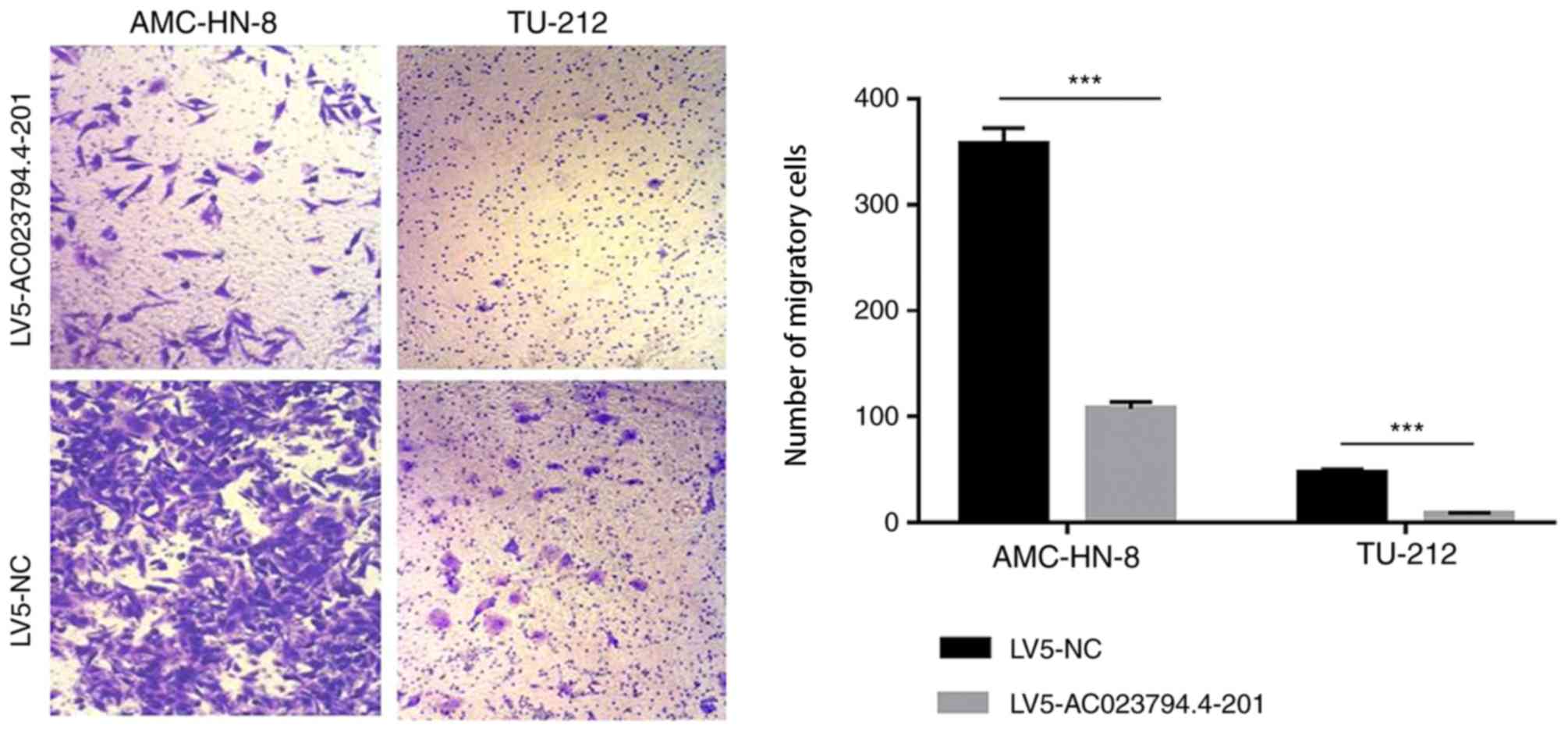
AC023794.4-201 inhibits LSCC cell
migration
Transwell assays were used to assess the rate of
cell migration in the cell lines. The results indicated that
AC023794.4-201 overexpression significantly decreased the migratory
ability of both LSCC cell lines compared with the LV5-NC group
(Fig. 5). These findings suggest
that the overexpression of AC023794.4-201 may inhibit LSCC cell
proliferation, colony formation and migration.
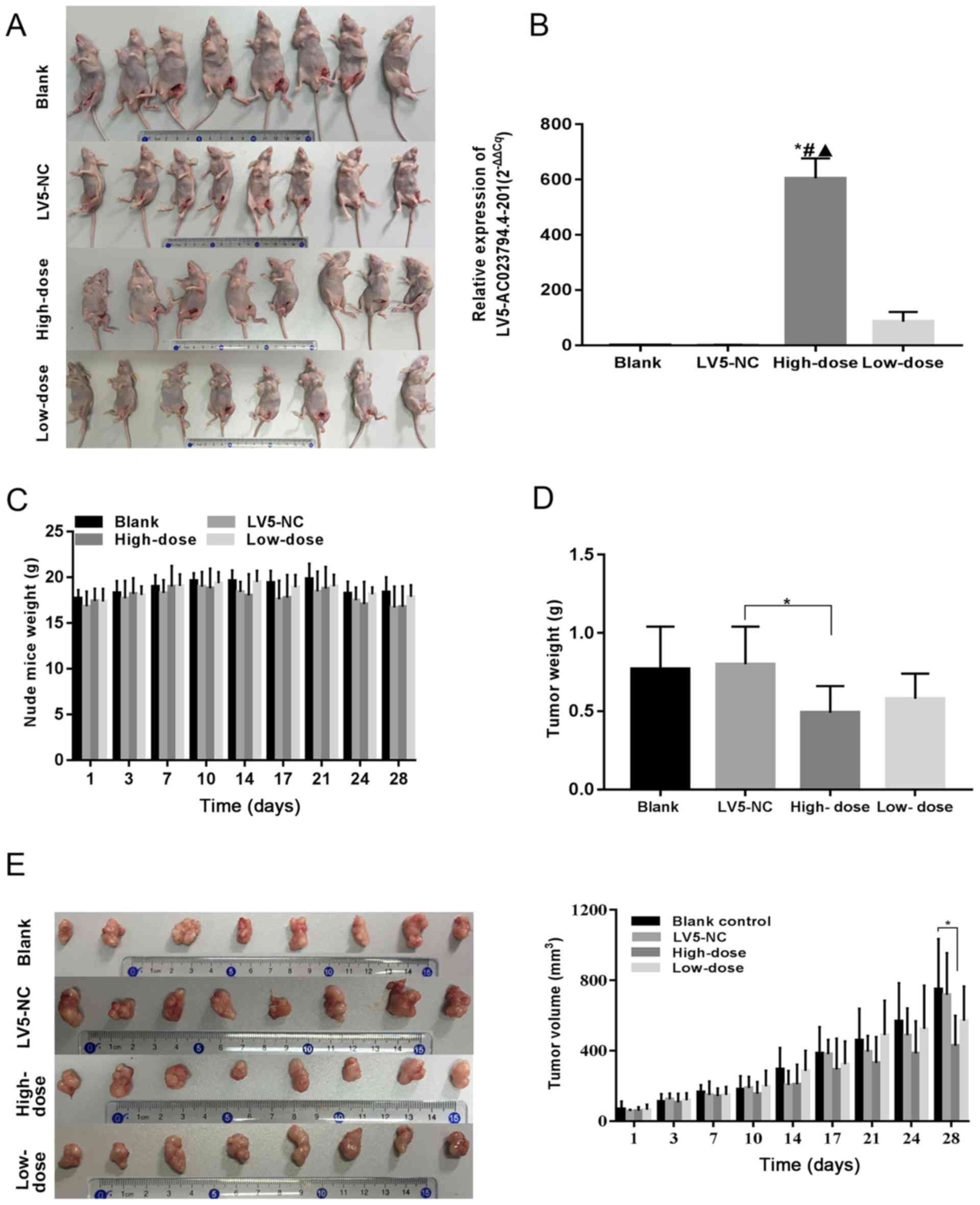
AC023794.4-201 inhibits LSCC growth in
vivo
A xenograft mouse model was constructed to further
investigate whether AC023794.4-201 exerts an antitumor effect in
vivo (Fig. 6A); all nude mice
subcutaneously inoculated with the AMC-HN-8 cell suspension
demonstrated tumor formation. Following injection of the tumors
with the corresponding lentiviruses, significant differences in the
expression levels of AC023794.4-201 were observed in the xenograft
tumors between 4 groups (P<0.001; Table II); the expression levels of
AC023794.4-201 in the high-dose group were significantly increased
compared with the other three groups (*P<0.05 vs. blank group;
#P<0.05 vs. LV5-NC group; ▲P<0.05 vs.
low-dose group; Fig. 6B). There was
no expression difference between the low-dose group and the control
group, as low concentration of LV5-AC023794.4-201 lentiviral
solution has a poor transfection effect. There was no significant
difference observed in the weight of mice in each group (Fig. 6C). Significant differences in the
weight of xenograft tumors among the 4 groups (P<0.05; Table III) and the weight of tumors
injected with a high-dose of the AC023794.4-201 overexpression
lentivirus were significantly decreased compared with the LV5-NC
group (P<0.05; Fig. 6D). In
addition, no significant differences were observed in the tumor
volume of nude mice between groups on days 1, 3, 7, 10, 14, 17, 21
and 24, whereas on day 28, significant differences were observed in
the tumor volume between the 4 groups (P<0.05; Table IV); the tumor volumes of nude mice
in the high-dose AC023794.4-201 group were significantly decreased
compared with the blank group (P<0.05; Fig. 6E). The tumor volumes of nude mice in
the high-dose AC023794.4-201 group were not significantly decreased
compared with the LV5-NC group.
 | Table II.Relative expression of AC023794.4-201
in xenograft. |
Table II.
Relative expression of AC023794.4-201
in xenograft.
| Group | Number of
samples | Mean | Sth. Deviation |
|---|
| Blank | 3 | 1.1472 | 0.5811 |
| LV5-NC | 3 | 1.2413 | 0.1609 |
| High-dose | 3 | 604.4310 | 59.3730 |
| Low-dose | 3 | 85.5558 | 28.8316 |
| Total | 12 | 173.0938 | 264.8346 |
| F | 154.7410 |
|
|
| P-value | 0.0000 |
|
|
 | Table III.Tumor weight of 4 experimental groups
of nude mice. |
Table III.
Tumor weight of 4 experimental groups
of nude mice.
| Group | Number of
samples | Mean | Sth. Deviation |
|---|
| Blank | 8 | 0.7738 | 0.2689 |
| LV5-NC | 8 | 0.7975 | 0.2366 |
| High-dose | 8 | 0.4875 | 0.1666 |
| Low-dose | 8 | 0.5838 | 0.1585 |
| Total | 24 | 0.6606 | 0.2414 |
| F | 3.9700 |
|
|
| P-value | 0.0180 |
|
|
 | Table IV.Volumes of xenograft tumors in 4
groups on day 28 of experiment. |
Table IV.
Volumes of xenograft tumors in 4
groups on day 28 of experiment.
| Group | Number of
samples | Mean | Sth. Deviation |
|---|
| Blank | 8 | 750.5288 | 284.7804 |
| LV5-NC | 8 | 717.8588 | 237.3654 |
| High-dose | 8 | 428.5950 | 171.0821 |
| Low-dose | 8 | 570.4538 | 195.0623 |
| Total | 24 | 616.8591 | 251.3606 |
| F | 3.4210 |
|
|
| P-value | 0.0310 |
|
|
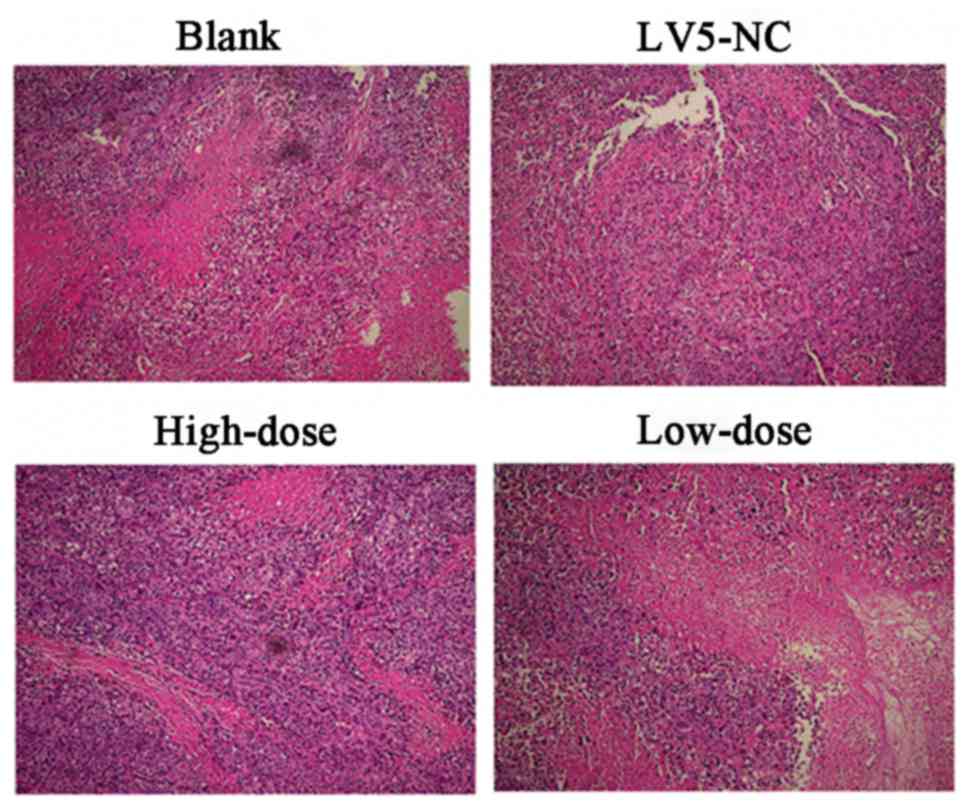
In addition, pathological examinations of all tumor
specimens were performed, alongside TEM. Following HE staining of
the 4 groups of xenograft tissues, a large number of tumor cell
clusters of various sizes and shapes with scattered small pieces or
focal necrosis were observed using light microscopy which indicated
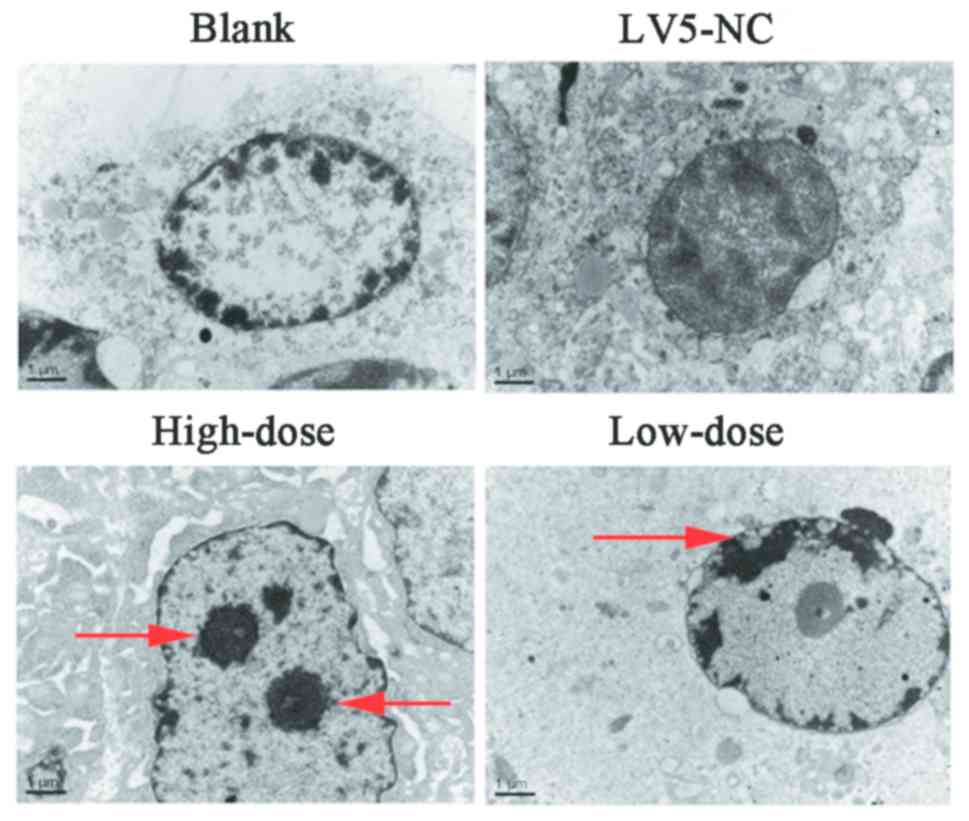
that the xenograft model was successfully constructed (Fig. 7). Using transmission electron
microscopy, chromatin clustered to one side of the nuclei in the
high and low dose groups, which looked like a crescent or a round
body indicating that the tumor cells were undergoing apoptosis.
However, the tumor cells of the control groups did not exhibit
these characteristics by TEM (Fig.
8). Thus, the xenograft nude mouse model revealed that
AC023794.4-201 may inhibit LSCC growth in vivo.
Discussion
The development of gene sequencing and microarray
techniques has facilitated the increasing number of transcript
studies. Previous studies have reported that lncRNAs are closely
associated with the proliferation, apoptosis, migration and
invasion of LSCC cells; for example, AC026166.2-001 inhibited the
proliferation and migration of LSCC cells via modulating the
microRNA (miRNA/miR)-24-3p/p27 axis in cell experiments and nude
mice xenograft models (16) and
LOC554202 was revealed to be highly expressed in LSCC, and promoted
the growth and invasion of LSCC via downregulating miR-31 in cell
experiments (17). In addition, the
roles of lncRNAs have been widely reported in other areas of cancer
research; for instance, HOX transcript antisense RNA (HOTAIR)
expression levels were discovered to be increased in oral squamous
cell carcinoma (OSCC), which inhibited the expression of E-cadherin
by initiating the binding of enhancer of zeste homolog 2 (EZH2) and
H3K27me3 with the E-cadherin promoter, to subsequently promote
tumor cell invasion and metastasis. HOTAIR may be one of critical
targets in progression and metastasis in OSCC (18). Silencing FOXD2 adjacent opposite
strand RNA 1 (FOXD2-AS1) was also revealed to inhibit the growth of
nasopharyngeal carcinoma (NPC) in vitro, whereas the
overexpression of FOXD2-AS1 increased the growth of NPC (19). Long intergenic non-protein coding RNA
regulator of reprogramming was found to be upregulated in papillary
thyroid carcinoma and served an oncogenic role during thyroid
cancer progression (20). HOTAIR
promoted the growth of cervical cancer cells by modulating Bcl-2
via the targeting of miR-143-3p (21). Finally, maternally expressed 3 (MEG3)
was revealed to be downregulated in tongue squamous cell carcinoma
(TSCC), and the overexpression of MEG3 inhibited TSCC cell
proliferation and promoted apoptosis (22). Thus, these findings suggested that
aberrantly expressed lncRNAs may serve as tumor suppressors or
oncogenes and have potential significance in the diagnosis and
treatment of tumors.
AC023794.4-201, which is located on the reverse
strand of chromosome 12 (54,085,132-54,125,992), is one of the
transcripts of the AC023794.4 (ENSG00000250432) gene (23). This lncRNA is 499 nucleotides in
length and using a lncRNA expression microarray profile of LSCC,
our previous study discovered that AC023794.4-201 expression levels
were dysregulated, demonstrating increased expression levels in 86%
(6/7) of the tumor tissues examined (24). Subsequently, 92 pairs of LSCC and
corresponding paracancerous tissues were used to verify the
dysregulation of AC023794.4-201 (14); however, the results revealed that
AC023794.4-201 expression levels were decreased in 79% (73/92) of
the LSCC tissue samples examined. The expression levels of
AC023794.4-201 in the tumor tissues of patients with Tumor (T)3 and
T4 LSCC were decreased compared with patients with T1 and T2 LSCC,
and the expression levels of AC023794.4-201 in the tumor tissues of
patients with lymphatic metastasis tissues (LMT) were decreased
compared with patients with LSCC without LMT. Furthermore,
Kaplan-Meier survival curve analysis revealed that the
postoperative survival rate was markedly decreased in the group
expressing low levels of AC023794.4-201 compared with the group
expressing high levels of AC023794.4-201 (14). These findings indicated that the
results of microarray expression profiling of a small sample set
can be used only as a reference, and conclusive results need to be
based on large-scale PCR analysis, cell functional experiments,
animal models and even further studies on the mechanisms and
pathways involved.
To further understand the role of AC023794.4-201 in
LSCC, in the present study, AC023794.4-201 overexpression
lentiviruses were used to overexpress AC023794.4-201 in LSCC cells.
Cell proliferation was subsequently analyzed using a RTCA assay;
AC023794.4-201 overexpression inhibited cell proliferation, with a
similar result obtained from the colony formation assay, in which
AC023794.4-201 upregulation inhibited colony formation. Flow
cytometric analysis of the cell cycle and apoptosis was used to
further investigate the role of AC023794.4-201 in LSCC. The results
demonstrated that AC023794.4-201 inhibited the progression of the
cell cycle from the S phase to the G2/M phase and
promoted apoptosis. To investigate the effect of AC023794.4-201 on
the migration of LSCC cells, a Transwell assay was performed and it
was discovered that AC023794.4-201 overexpression inhibits the
migration of LSCC cells. This result is consistent with that of our
previous analysis of clinicopathological factors; that is,
AC023794.4-201 levels in the tumor tissues of patients with LMT
were downregulated (14). In
addition, the results of the xenograft mouse model experiment
further confirmed that AC023794.4-201 may serve a tumor-suppressive
role in LSCC. The volume and weight of the transplanted tumors
injected with the AC023794.4-201 overexpression lentivirus were
significantly decreased compared with the control groups. As
demonstrated using PCR, the AC023794.40291 expression in the
high-dose group was higher compared with the other 3 groups, which
had a greater impact on the volume and weight of the transplanted
tumor. The typical features of cell apoptosis (chromatin clustered
to one side of the nuclei, appearance as a crescent or a round
body) were observed. Following comprehensive analysis of the
clinical data, the results of the cell functional assays and the
xenograft mouse model experiment, it was concluded that
AC023794.4-201 may have a tumor-suppressive function in LSCC.
However, in our animal experiments, the apoptosis of tumor tissues
was limited to observation by TEM, and no experiments were
performed to investigate the expression levels of pathological
markers of proliferation and apoptosis. There are numerous
pathological markers of proliferation and apoptosis that have been
applied to LSCC studies, such as Ki-67 and cyclin D1, which are
associated with proliferation, and caspase-3 and p27 protein, which
are associated with apoptosis (25–28).
Thus, related immunohistochemistry experiments to assess the
expression levels of these markers in vivo will be used in a
follow-up experiment.
Notably, sequence alignments in the Ensembl Genes
database found that Recombinant single strand-selective
monofunctional uracil-DNA glycosylase 1–233 (SMUG1-223) and
AC023794.4-201 had partially overlapping gene loci, and that both
SMUG1-223 and AC023794.4-201 are transcribed in the same direction.
SMUG1-223 is one of the transcripts of SMUG1 (ENSG00000123415),
which is located on the reverse strand of chromosome 12:
54,121,277-54,173,229 (23).
Previous studies have reported that SMUG1 encodes a protein that
participates in base excision repair by removing uracil and certain
oxidized pyrimidines from DNA, repairing oxidative damage to DNA
and controlling ribosomal (r) RNA quality (29–31).
AC023794.4-201 and SMUG1-223 occupy the same site (23), indicating that these two genes may
interact or have similar functions. Based on these findings, it was
hypothesized that AC023794.4-201 may also repair oxidative DNA
damage and control rRNA quality to exert its tumor-suppressive
function; however, the mechanism of action, targets, regulatory
pathways and other factors involved in this hypothesis require
clarification through further studies.
Numerous previous studies have discovered that
lncRNAs can be used as potential diagnostic markers; Gong et
al (32) performed a comparative
study of serum samples from 151 patients with colorectal carcinoma
and 160 healthy individuals, and found that serum hypoxia-inducible
factor 1α-antisense RNA 1(HIF1A-AS1) can serve as a latent
biomarker for the diagnosis and prognosis of colorectal carcinoma.
Chen et al (33) performed
RT-qPCR on bladder cancer associated transcript 1 (BLACAT1)
expression levels and obtained data from The Cancer Genome Atlas
database, and reported that BLACAT1 expression levels may be used
as a non-specific cancer diagnostic biomarker. The aforementioned
study included 12 different types of cancer including:
hepatocellular carcinoma, lung cancer, breast cancer, ovarian
cancer, endometrial cancer, cervical cancer, prostate cancer,
gastric cancer, esophagus cancer, thyroid cancer, bladder cancer
and nasopharynx cancer. In addition, serum actin filament
associated protein 1-antisense RNA 1 was identified as a potential
diagnostic biomarker for non-small cell lung cancer (34). In fact, previous reports have
demonstrated that combining multiple genetic biomarkers increased
the diagnostic value of these biomarkers in cancer (35); for example, Madhavan et al
(36) reported that the concomitant
assessment of pancreatic cancer-initiating cells and miRNA
serum-exosome marker groups significantly improved the sensitivity
and specificity of pancreatic cancer diagnoses (35); and the combination of serum
prostate-specific antigen (PSA) and plasma expression levels of
let-7c, miR-30c, miR-141 and miR-375 were discovered to be
non-invasive diagnostic biomarkers for prostate cancer screening,
which were all demonstrated to be more accurate compared with PSA
detection alone (37). Therefore,
the identification of suitable biomarkers for the diagnosis of LSCC
in combination with AC023794.4-201 represents a promising future
research direction.
lncRNA AC023794.4-201 inhibits laryngeal cancer cell
proliferation and promotes apoptosis in vitro and in
vivo. lncRNA AC023794.4-201 plays the role of a tumor
suppressor gene in the occurrence and development of LSCC. However,
the specific mechanism and regulatory pathways by which lncRNA
AC023794.4-201 acts needs further investigation. lncRNA
AC023794.4-201 can be used as a potential diagnostic and prognostic
biomarker for LSCC.
Acknowledgements
Not applicable.
Funding
This work was supported by grants from the Zhejiang
Provincial Natural Science Foundation of China (grant no.
LY14H160003), the Ningbo Health Branding Subject Fund (grant no.
PPXK2018-02) and the Medical and Health Research Project of
Zhejiang Province (grant no. 2018RC063).
Availability of data and materials
Previously reported data and the microarray profile
used to support this study are available on the NCBI GEO database
(accession no. GSE59652). The datasets used and/or analyzed in the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
ZSS collected the data and analyzed the feasibility
of the experiment. JY designed the experiments and the data
collection strategy. QLT and WJH purchased the experimental
materials and conducted the experiments. HXD and QL conducted the
statistical analysis and confirmed all statistical outcomes. CCZ,
YH and JX critically analyzed the data and wrote the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All animal studies were approved by and strictly
followed the guidelines of the Animal Ethics and Welfare Committee
of Ningbo University (approval no. AWEC-2015-10).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pantel M and Guntinas-Lichius O: Laryngeal
carcinoma: Epidemiology, risk factors and survival. HNO. 60:32–40.
2012.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Steuer CE, El-Deiry M, Parks JR, Higgins
KA and Saba NF: An update on larynx cancer. CA Cancer J Clin.
67:31–50. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Forastiere AA, Ismaila N, Lewin JS, Nathan
CA, Adelstein DJ, Eisbruch A, Fass G, Fisher SG, Laurie SA, Le QT,
et al: Use of larynx-preservation strategies in the treatment of
laryngeal cancer: American society of clinical oncology clinical
practice guideline update. J Clin Oncol. 36:1143–1169. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Berrino F, De Angelis R, Sant M, Rosso S,
Bielska-Lasota M, Coebergh JW and Santaquilani M; EUROCARE Working
group, : Survival for eight major cancers and all cancers combined
for European adults diagnosed in 1995-99: Results of the EUROCARE-4
study. Lancet Oncol. 8:773–783. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Forrest ME and Khalil AM: Review:
Regulation of the cancer epigenome by long non-coding RNAs. Cancer
Lett. 407:106–112. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stein LD: Human genome: End of the
beginning. Nature. 431:915–916. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
ENCODE Project Consortium, . An integrated
encyclopedia of DNA elements in the human genome. Nature.
489:57–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang X, Zhou X, Hu Q, Sun B, Deng M, Qi X
and Lü M: Advances in esophageal cancer: A new perspective on
pathogenesis associated with long non-coding RNAs. Cancer Lett.
413:94–101. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–616. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Flynn RA and Chang HY: Long noncoding RNAs
in cell-fate programming and reprogramming. Cell Stem Cell.
14:752–761. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gugnoni M and Ciarrocchi A: Long
non-coding RNA and epithelial-mesenchymal transition in cance. Int
J Mol Sci. 20:19242019. View Article : Google Scholar
|
|
14
|
Tong Q, Shen Z, Li Q, Hao W and Zhou C:
Expression and clinical significance of long non-coding RNA
ac023794.4-201 in laryngeal squamous cell carcinoma. Chin Arch
Otolaryngol Head Neck Surg. 10:564–566. 2018.(In Chinaese).
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shen Z, Hao W, Zhou C, Deng H, Ye D, Li Q,
Lin L, Cao B and Guo J: Long non-coding RNA AC026166.2-001 inhibits
cell proliferation and migration in laryngeal squamous cell
carcinoma by regulating the miR-24-3p/p27 axis. Sci Rep.
8:33752018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang S, Wang J, Ge W and Jiang Y: Long
non-coding RNA LOC554202 promotes laryngeal squamous cell carcinoma
progression through regulating miR-31. J Cell Biochem.
119:6953–6960. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu Y, Zhang L, Zhang L, Wang Y, Li H, Ren
X, Wei F, Yu W, Liu T, Wang X, et al: Long non-coding RNA HOTAIR
promotes tumor cell invasion and metastasis by recruiting EZH2 and
repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol.
46:2586–2594. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen G, Sun W, Hua X, Zeng W and Yang L:
Long non-coding RNA FOXD2-AS1 aggravates nasopharyngeal carcinoma
carcinogenesis by modulating miR-363-5p/S100A1 pathway. Gene.
645:76–84. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang R, Hardin H, Huang W, Buehler D and
Lloyd RV: Long non-coding RNA Linc-ror is upregulated in papillary
thyroid carcinoma. Endocr Pathol. 29:1–8. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu M, Jia J, Wang X, Liu Y, Wang C and
Fan R: Long non-coding RNA HOTAIR promotes cervical cancer
progression through regulating BCL2 via targeting miR-143-3p.
Cancer Biol Ther. 19:391–399. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jia LF, Wei SB, Gan YH, Guo Y, Gong K,
Mitchelson K, Cheng J and Yu GY: Expression, regulation and roles
of miR-26a and MEG3 in tongue squamous cell carcinoma. Int J
Cancer. 135:2282–2293. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ensembl Genes. (DB/OL), . https://www.ensembl.org/Homo_sapiens/Gene/Summary?db=core;g=ENSG00000250432;r=12:5
4085132-54125992June 20–2019
|
|
24
|
Shen Z, Li Q, Deng H, Lu D, Song H and Guo
J: Long non-coding RNA profiling in laryngeal squamous cell
carcinoma and its clinical significance: Potential biomarkers for
LSCC. PLoS One. 9:e1082372014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gioacchini FM, Alicandri-Ciufelli M,
Magliulo G, Rubini C, Presutti L and Re M: The clinical relevance
of Ki-67 expression in laryngeal squamous cell carcinoma. Eur Arch
Otorhinolaryngol. 272:1569–1576. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jovanovic IP, Radosavljevic GD,
Simovic-Markovic BJ, Stojanovic SP, Stefanovic SM, Pejnovic NN and
Arsenijevic NN: Clinical significance of Cyclin D1, FGF3 and p21
protein expression in laryngeal squamous cell carcinoma. J BUON.
19:944–952. 2014.PubMed/NCBI
|
|
27
|
Wang HX and Tang C: Galangin suppresses
human laryngeal carcinoma via modulation of caspase-3 and AKT
signaling pathways. Oncol Rep. 38:703–714. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang JQ, Liang Z, Wu M, Sun YM and Liu HX:
Expression of p27 and PTEN and clinical characteristics in early
laryngeal squamous cell carcinoma and their correlation with
recurrence. Int J Clin Exp Pathol. 8:5715–5720. 2015.PubMed/NCBI
|
|
29
|
Jobert L, Skjeldam HK, Dalhus B,
Galashevskaya A, Vågbø CB, Bjørås M and Nilsen H: The human base
excision repair enzyme SMUG1 directly interacts with DKC1 and
contributes to RNA quality control. Mol Cell. 49:339–345. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Matsubara M, Tanaka T, Terato H and Ide H:
Action mechanism of human SMUG1 uracil-DNA glycosylase. Nucleic
Acids Symp Ser (Oxf). 49:295–296. 2005. View Article : Google Scholar
|
|
31
|
Darwanto A, Theruvathu JA, Sowers JL,
Rogstad DK, Pascal T, Goddard W III and Sowers LC: Mechanisms of
base selection by human single-stranded selective monofunctional
uracil-DNA glycosylase. J Biol Chem. 284:15835–15846. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gong W, Tian M, Qiu H and Yang Z: Elevated
serum level of lncRNA-HIF1A-AS1 as a novel diagnostic predictor for
worse prognosis in colorectal carcinoma. Cancer Biomark.
20:417–424. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen X, Dai M, Zhu H, Li J, Huang Z, Liu
X, Huang Y, Chen J and Dai S: Evaluation on the diagnostic and
prognostic values of long non-coding RNA BLACAT1 in common types of
human cancer. Mol Cancer. 16:1602017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li W, Li N, Kang X and Shi K: Circulating
long non-coding RNA AFAP1-AS1 is a potential diagnostic biomarker
for non-small cell lung cancer. Clin Chim Acta. 475:152–156. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang J, Zhou Y, Lu J, Sun Y, Xiao H, Liu M
and Tian L: Combined detection of serum exosomal miR-21 and HOTAIR
as diagnostic and prognostic biomarkers for laryngeal squamous cell
carcinoma. Med Oncol. 31:1482014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Madhavan B, Yue S, Galli U, Rana S, Gross
W, Müller M, Giese NA, Kalthoff H, Becker T, Büchler MW and Zöller
M: Combined evaluation of a panel of protein and miRNA
serum-exosome biomarkers for pancreatic cancer diagnosis increases
sensitivity and specificity. Int J Cancer. 136:616–627. 2015.
View Article : Google Scholar
|
|
37
|
Kachakova D, Mitkova A, Popov E, Popov I,
Vlahova A, Dikov T, Christova S, Mitev V, Slavov C and Kaneva R:
Combinations of serum prostate-specific antigen and plasma
expression levels of let-7c, miR-30c, miR-141, and miR-375 as
potential better diagnostic biomarkers for prostate cancer. DNA
Cell Biol. 34:189–200. 2015. View Article : Google Scholar : PubMed/NCBI
|