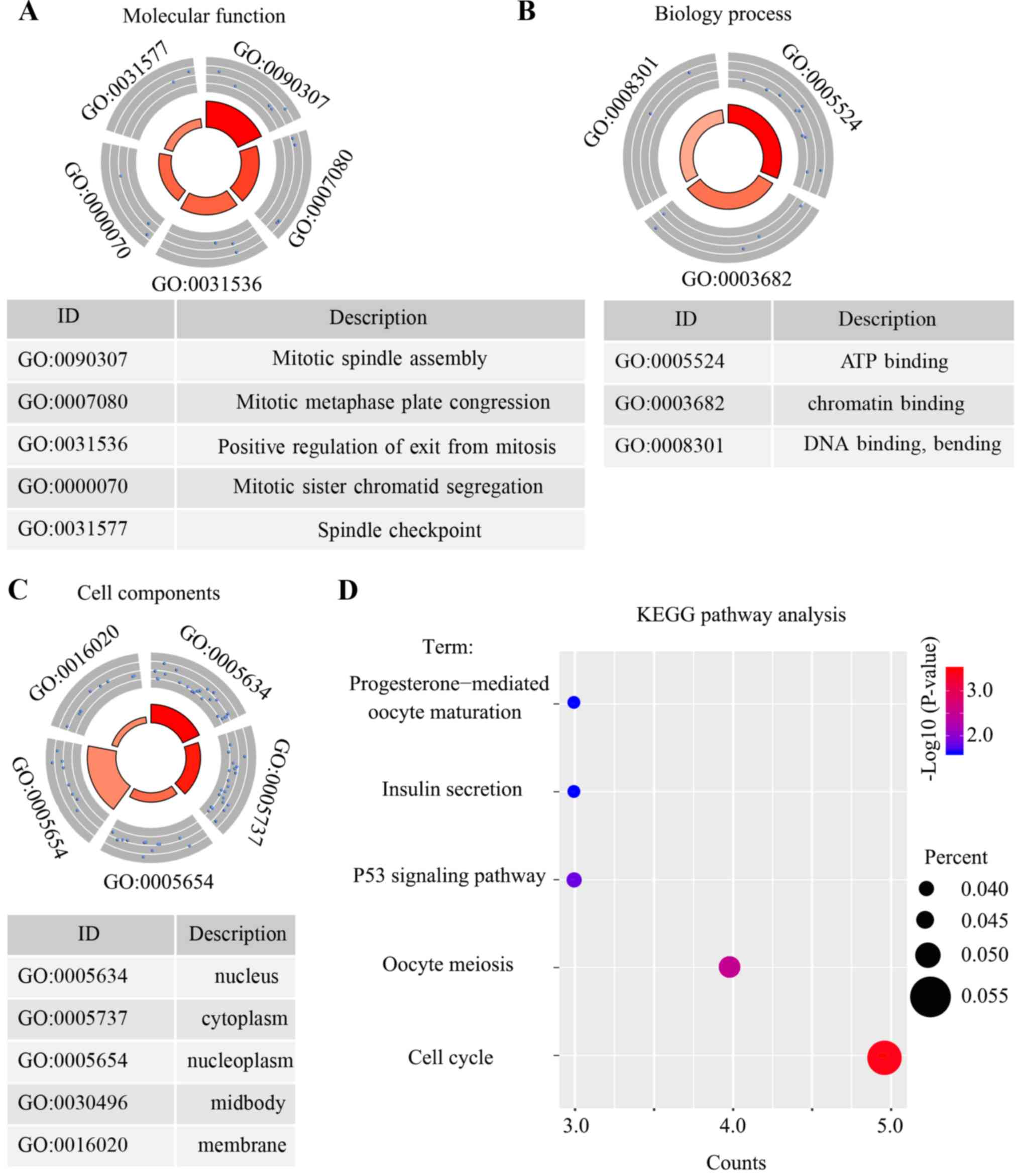
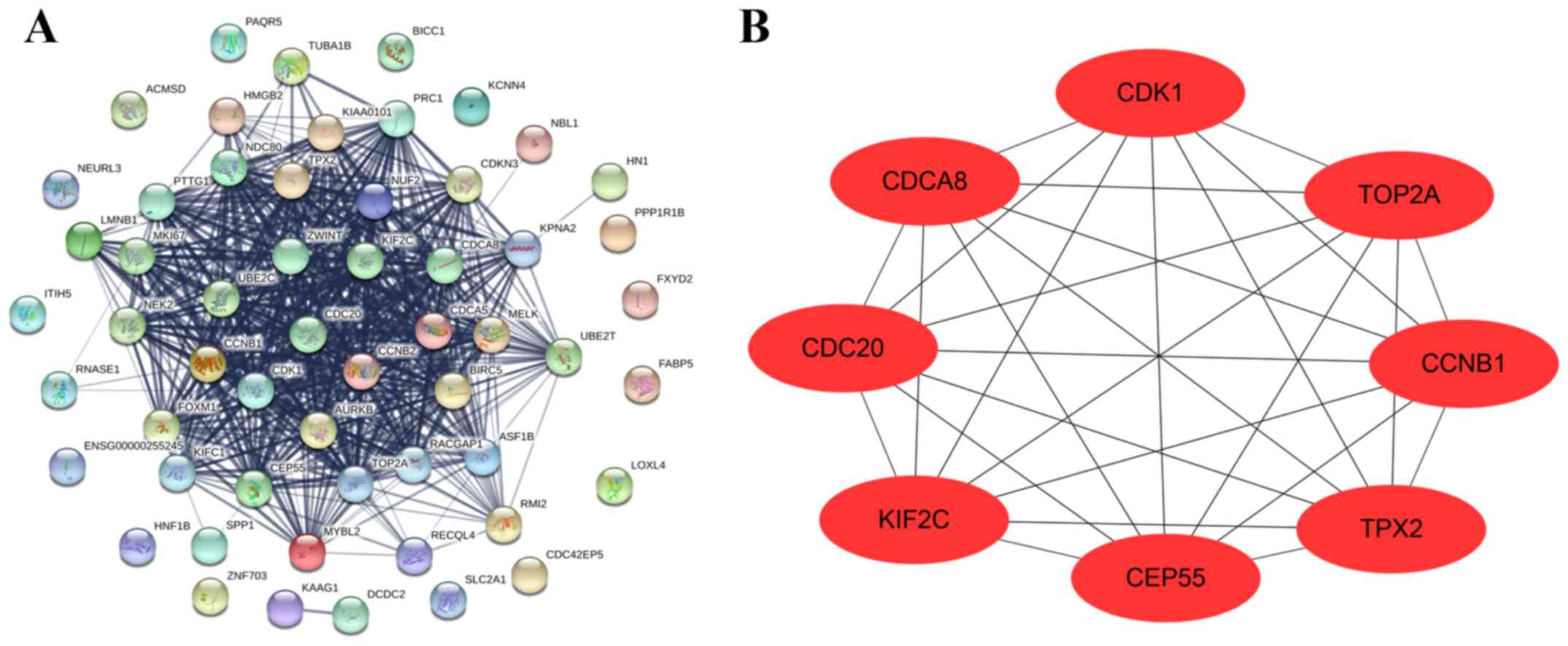
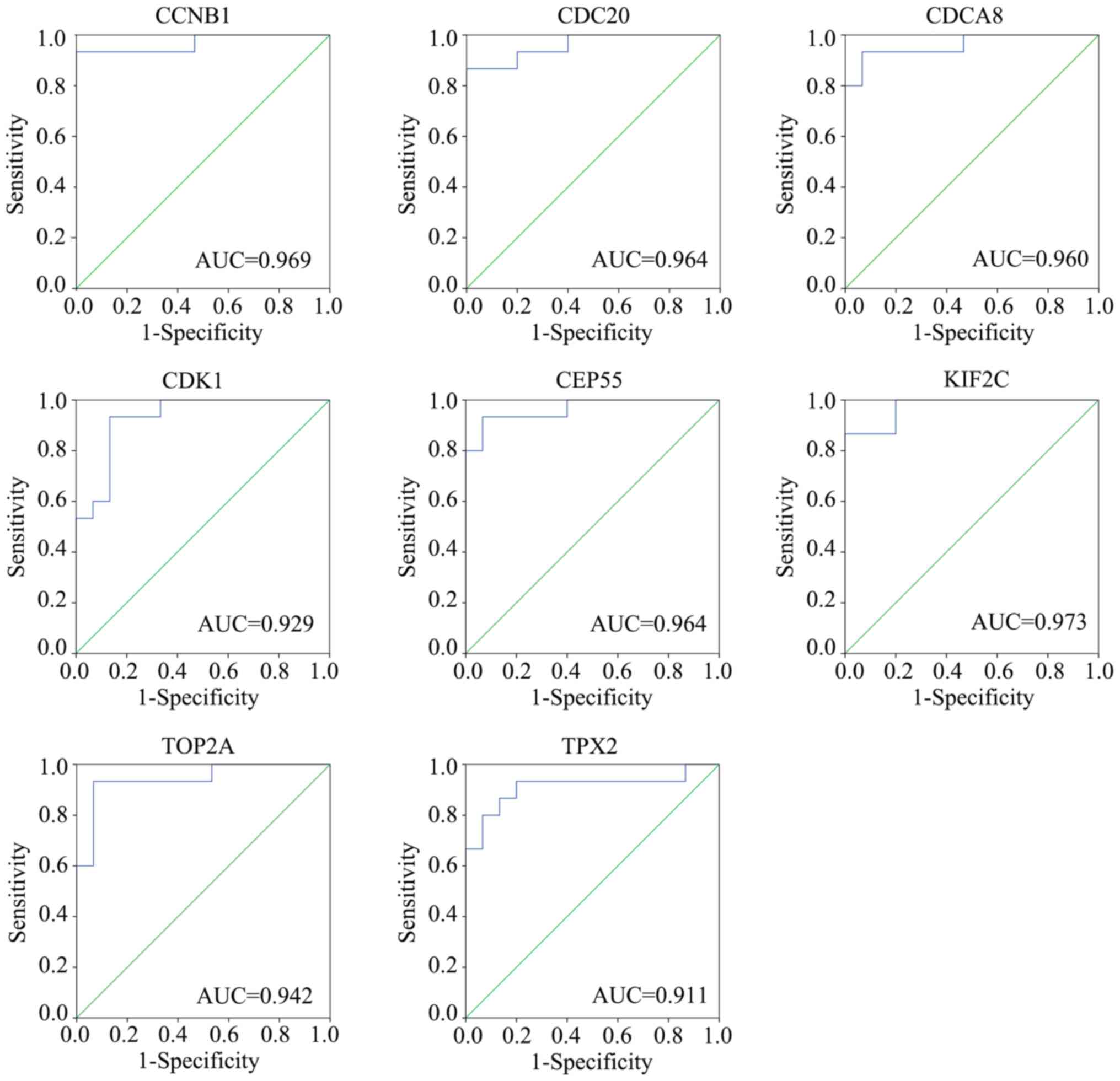
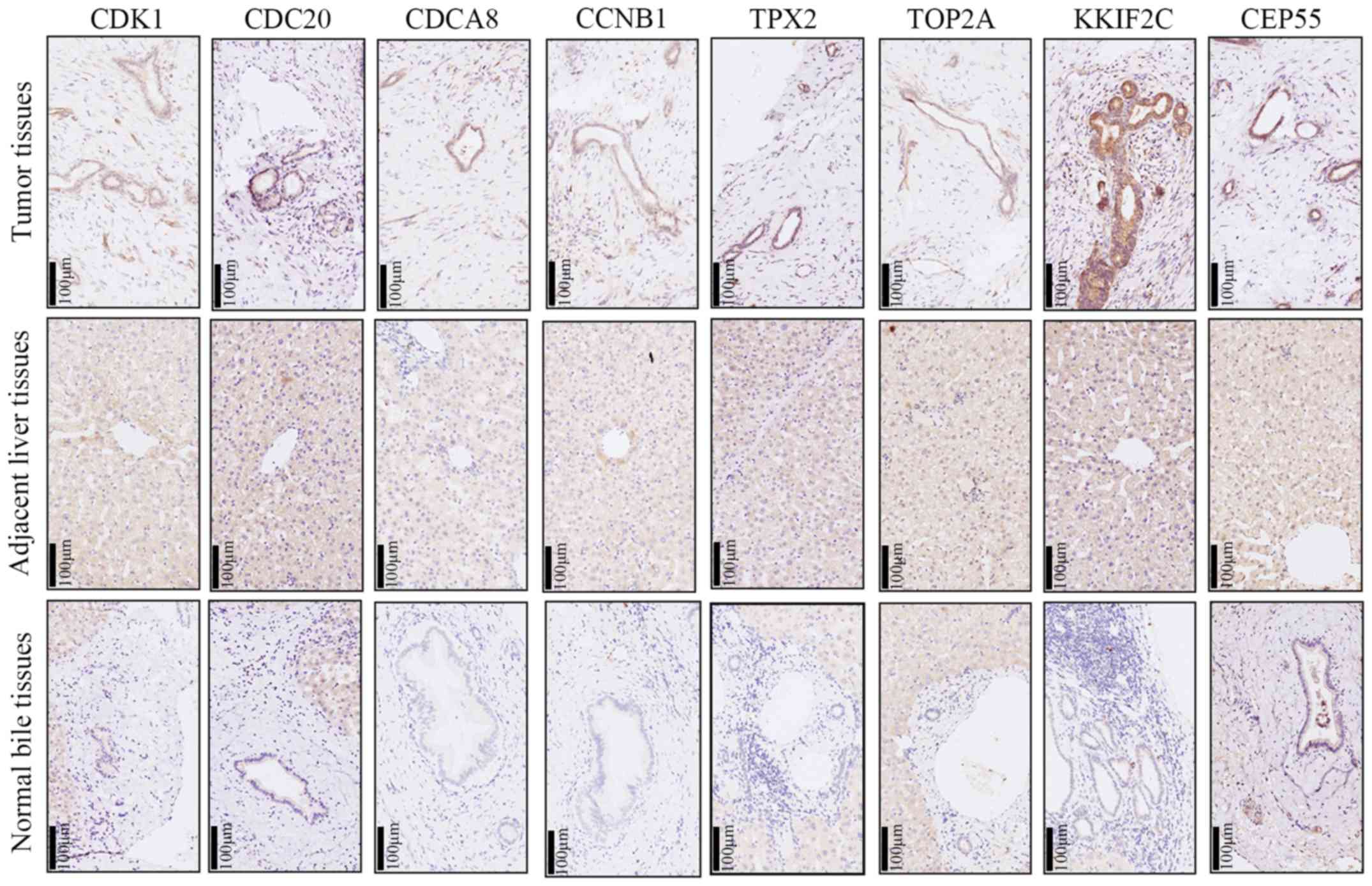
|
1
|
Chun YS and Javle M: Systemic and adjuvant
therapies for intrahepatic cholangiocarcinoma. Cancer Control.
24:10732748177292412017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nakano M, Ariizumi SI and Yamamoto M:
Intrahepatic cholangiocarcinoma. Semin Diagn Pathol. 34:160–166.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhu Q, Sun Y, Zhou Q, He Q and Qian H:
Identification of key genes and pathways by bioinformatics analysis
with TCGA RNA sequencing data in hepatocellular carcinoma. Mol Clin
Oncol. 9:597–606. 2018.PubMed/NCBI
|
|
4
|
Tang D, Zhao X, Zhang L, Wang Z and Wang
C: Identification of hub genes to regulate breast cancer metastasis
to brain by bioinformatics analyses. J Cell Biochem. 120:9522–9531.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ma X, Tao R, Li L, Chen H, Liu Z, Bai J,
Shuai X, Wu C and Tao K: Identification of a 5microRNA signature
and hub miRNAmRNA interactions associated with pancreatic cancer.
Oncol Rep. 41:292–300. 2019.PubMed/NCBI
|
|
6
|
Xiao H, Xu D, Chen P, Zeng G, Wang X and
Zhang X: Identification of five genes as a potential biomarker for
predicting progress and prognosis in adrenocortical carcinoma. J
Cancer. 9:4484–4495. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang Y, Huang Y, Zhang L, Chang A, Zhao
P, Chai X and Wang J: Identification of crucial genes and
prediction of small molecules for multidrug resistance of Hodgkin's
lymphomas. Cancer Biomark. 23:495–503. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Edgar R, Domrachev M and Lash AE: Gene
Expression Omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang K, Yu J, Yu X, Han Z, Cheng Z, Liu F
and Liang P: Clinical and survival outcomes of percutaneous
microwave ablation for intrahepatic cholangiocarcinoma. Int J
Hyperthermia. 34:292–297. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Meng ZW, Pan W, Hong HJ, Chen JZ and Chen
YL: Modified staging classification for intrahepatic
cholangiocarcinoma based on the sixth and seventh editions of the
AJCC/UICC TNM staging systems. Medicine (Baltimore). 96:e78912017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic acids Res. 47:D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu L, Tsilimigras DI, Farooq A, Hyer JM,
Merath K, Paredes AZ, Mehta R, Sahara K, Shen F and Pawlik TM:
Potential survival benefit of radiofrequency ablation for small
solitary intrahepatic cholangiocarcinoma in nonsurgically managed
patients: A population-based analysis. J Surg Oncol. 120:1358–1364.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Laurent S, Verhelst X, Geerts A, Geboes K,
De Man M, Troisi R, Vanlander A, Rogiers X, Berrevoet F and Van
Vlierberghe H: Update on liver transplantation for
cholangiocarcinoma: A review of the recent literature. Acta
Gastroenterol Belg. 82:417–420. 2019.PubMed/NCBI
|
|
17
|
Wang Z, Fan M, Candas D, Zhang TQ, Qin L,
Eldridge A, Wachsmann-Hogiu S, Ahmed KM, Chromy BA, Nantajit D, et
al: Cyclin B1/Cdk1 coordinates mitochondrial respiration for
cell-cycle G2/M progression. Dev Cell. 29:217–232. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xie B, Wang S, Jiang N and Li JJ: Cyclin
B1/CDK1-regulated mitochondrial bioenergetics in cell cycle
progression and tumor resistance. Cancer Lett. 443:56–66. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huskey NE, Guo T, Evason KJ, Momcilovic O,
Pardo D, Creasman KJ, Judson RL, Blelloch R, Oakes SA, Hebrok M and
Goga A: CDK1 inhibition targets the p53-NOXA-MCL1 axis, selectively
kills embryonic stem cells, and prevents teratoma formation. Stem
Cell Rep. 4:374–389. 2015. View Article : Google Scholar
|
|
20
|
Lu M, Breyssens H, Salter V, Zhong S, Hu
Y, Baer C, Ratnayaka I, Sullivan A, Brown NR, Endicott J, et al:
Restoring p53 function in human melanoma cells by inhibiting MDM2
and Cyclin B1/CDK1-phosphorylated nuclear iASPP. Cancer Cell.
30:822–823. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kapanidou M, Curtis NL and Bolanos-Garcia
VM: Cdc20: At the crossroads between chromosome segregation and
mitotic exit. Trends Biochem Sci. 42:193–205. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang L, Zhang J, Wan L, Zhou X, Wang Z and
Wei W: Targeting Cdc20 as a novel cancer therapeutic strategy.
Pharmacol Ther. 151:141–151. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu F, Lin Y, Cui P, Li H, Zhang L, Sun Z,
Huang S, Li S, Huang S, Zhao Q and Liu Q: Cdc20/p55 mediates the
resistance to docetaxel in castration-resistant prostate cancer in
a Bim-dependent manner. Cancer Chemother Pharmacol. 81:999–1006.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Paul D, Ghorai S, Dinesh US, Shetty P,
Chattopadhyay S and Santra MK: Cdc20 directs proteasome-mediated
degradation of the tumor suppressor SMAR1 in higher grades of
cancer through the anaphase promoting complex. Cell Death Dis.
8:e28822017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Y, Xue YB, Li H, Qiu D, Wang ZW and
Tan SS: Inhibition of cell survival by curcumin is associated with
downregulation of cell division cycle 20 (Cdc20) in pancreatic
cancer cells. Nutrients. 9:E1092017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hindriksen S, Meppelink A and Lens SM:
Functionality of the chromosomal passenger complex in cancer.
Biochem Soc Trans. 43:23–32. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ci C, Tang B, Lyu D, Liu W, Qiang D, Ji X,
Qiu X, Chen L and Ding W: Overexpression of CDCA8 promotes the
malignant progression of cutaneous melanoma and leads to poor
prognosis. Int J Mol Med. 43:404–412. 2019.PubMed/NCBI
|
|
28
|
Bi Y, Chen S, Jiang J, Yao J, Wang G, Zhou
Q and Li S: CDCA8 expression and its clinical relevance in patients
with bladder cancer. Medicine (Baltimore). 97:e118992018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Phan NN, Wang CY, Li KL, Chen CF, Chiao
CC, Yu HG, Huang PL and Lin YC: Distinct expression of CDCA3,
CDCA5, and CDCA8 leads to shorter relapse free survival in breast
cancer patient. Oncotarget. 9:6977–6992. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bu Y, Shi L, Yu D, Liang Z and Li W: CDCA8
is a key mediator of estrogen-stimulated cell proliferation in
breast cancer cells. Gene. 703:1–6. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jeffery J, Sinha D, Srihari S, Kalimutho M
and Khanna KK: Beyond cytokinesis: The emerging roles of CEP55 in
tumorigenesis. Oncogene. 35:683–690. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang Y, Jin T, Dai X and Xu J:
Lentivirus-mediated knockdown of CEP55 suppresses cell
proliferation of breast cancer cells. Biosci Trends. 10:67–73.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu S, Wu D, Pan Y, Liu H, Shao Z and Wang
M: Correlation between EZH2 and CEP55 and lung adenocarcinoma
prognosis. Pathol Res Pract. 215:292–301. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ritter A, Kreis NN, Louwen F, Wordeman L
and Yuan J: Molecular insight into the regulation and function of
MCAK. Crit Rev Biochem Mol Biol. 51:228–245. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bie L, Zhao G, Wang YP and Zhang B:
Kinesin family member 2C (KIF2C/MCAK) is a novel marker for
prognosis in human gliomas. Clin Neurol Neurosurg. 114:356–360.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dai X, Hua T and Hong T: Integrated
diagnostic network construction reveals a 4-gene panel and 5 cancer
hallmarks driving breast cancer heterogeneity. Sci Rep. 7:68272017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shimo A, Tanikawa C, Nishidate T, Lin ML,
Matsuda K, Park JH, Ueki T, Ohta T, Hirata K, Fukuda M, et al:
Involvement of kinesin family member 2C/mitotic
centromere-associated kinesin overexpression in mammary
carcinogenesis. Cancer Sci. 99:62–70. 2008.PubMed/NCBI
|
|
38
|
Nelson WG, Haffner MC and Yegnasubramanian
S: The structure of the nucleus in normal and neoplastic prostate
cells: Untangling the role of type 2 DNA topoisomerases. Am J Clin
Exp Urol. 6:107–113. 2018.PubMed/NCBI
|
|
39
|
Lan J, Huang HY, Lee SW, Chen TJ, Tai HC,
Hsu HP, Chang KY and Li CF: TOP2A overexpression as a poor
prognostic factor in patients with nasopharyngeal carcinoma. Tumour
Biol. 35:179–187. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
An X, Xu F, Luo R, Zheng Q, Lu J, Yang Y,
Qin T, Yuan Z, Shi Y, Jiang W and Wang S: The prognostic
significance of topoisomerase II alpha protein in early stage
luminal breast cancer. BMC Cancer. 18:3312018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Potkonjak M, Miura JT, Turaga KK, Johnston
FM, Tsai S, Christians KK and Gamblin TC: Intrahepatic
cholangiocarcinoma and gallbladder cancer: Distinguishing molecular
profiles to guide potential therapy. HPB (Oxford). 17:1119–1123.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Neumayer G, Belzil C, Gruss OJ and Nguyen
MD: TPX2: Of spindle assembly, DNA damage response, and cancer.
Cell Mol Life Sci. 71:3027–3047. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hsu CW, Chen YC, Su HH, Huang GJ, Shu CW,
Wu TT and Pan HW: Targeting TPX2 suppresses the tumorigenesis of
hepatocellular carcinoma cells resulting in arrested mitotic phase
progression and increased genomic instability. J Cancer.
8:1378–1394. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen M, Zhang H, Zhang G, Zhong A, Ma Q,
Kai J, Tong Y, Xie S, Wang Y, Zheng H, et al: Targeting TPX2
suppresses proliferation and promotes apoptosis via repression of
the PI3k/AKT/P21 signaling pathway and activation of p53 pathway in
breast cancer. Biochem Biophys Res Commun. 507:74–82. 2018.
View Article : Google Scholar : PubMed/NCBI
|