Introduction
Hepatocellular carcinoma (HCC) is the most common
form of liver cancer, the sixth most commonly diagnosed cancer, and
the fourth leading cause of cancer-associated mortality worldwide
(1). In China, HCC is the third
leading cause of cancer-associated mortality (2). Moreover, the mortality rate of
hepatitis B virus (HBV)-associated liver cancer was 16.42 per
100,000 people in 2016 (3). The
annual disability-adjusted life-years of liver cancer caused by HBV
in China have been consistently higher than the global level
(3).
While surgical resection is the most common method
for HCC treatment, radiofrequency ablation (RFA) and microwave
ablation (MWA) have attracted increasing interest due to the
reduced trauma and faster recovery observed with these approaches
(4). Additionally, both RFA and MWA
are accessible to subjects who are not eligible for surgical
resection (5,6). Notably, no difference has been observed
in the overall survival rate and disease-free survival rate in
subjects with early-stage HCC between ablation and surgical
resection (7,8). An alternative tumor-specific target
treatment should generate a tumor-specific immune response in
patients with cancer (9). Previous
clinical studies suggested that RFA induced the expression of
tumor-associated antigen as well as the activation of the immune
response, which not only increased the number of tumor-specific
immune cells, but also the frequency of immune cells specific for
recall antigens (10–12). However, whether the systemic immune
response can be activated after ablation remains unclear.
DNA methylation mediates primary epigenetic
regulation of genome function. Previous studies indicated that DNA
methylation was involved in the development of immune cells
(13), T lymphocyte function,
persistent inflammation, HCC progression (14–16) and
the effector phase of chronic viral infection (17). The inhibition of de novo DNA
methylation programs combined with immune checkpoint blockade can
facilitate the control of chronic viral infection and tumor growth
(18). Our previous study
demonstrated that peripheral blood mononuclear cells (PBMCs) DNA
methylated alterations were enriched in immune-associated canonical
pathways and had immune modulation functions during HCC progression
(19). Thus, it is essential to
investigate the dynamic immune response profile following thermal
ablation by DNA methylation, as well as the potential effective
immune response mechanism.
To the best of the authors knowledge, few studies
have investigated the effects of the systemic immune response and
immune-activated time points following thermal ablation for HCC.
The present study established the PBMC DNA methylation profiles for
HBV-associated HCC in order to describe the epigenetic changes
associated with the immune response following thermal ablation
treatment.
Materials and methods
Sample collection
In the present study, five subjects with HCC (The
nomenclature of the subjects were represented by ‘M + number’,
thereafter referred to as M1, M2, M3, M4 and M5) were recruited at
Beijing Youan Hospital of the Capital Medical University from
January 2018 and March 2018. The diagnosis of HCC was based on the
European Association for the Study of the Liver Clinical Practice
Guidelines (20), and the
classification of HCC stages was based on the Barcelona Clinic
Liver Cancer staging system (21).
The following inclusion criteria were used: i) Liver biopsy or film
degree exam diagnosed as HCC; ii) subjects positive for HBV surface
antigen; iii) liver cirrhosis classified as Child-Pugh class A; iv)
single HCC nodule ≤5 cm; and v) absence of extrahepatic metastasis.
The exclusion criteria were as follows: i) Subjects undergoing
treatment, such as thermal ablation, surgical resection,
chemotherapy, radiotherapy, gene therapy, adoptive cellular
immunotherapy, or any other treatments for HCC; ii) subjects with
coexistent hematological disorders, serious or active infection
before treatment; iii) subjects with other types of cancer; iv)
combinations with other virus infections and autoimmune diseases;
v) no procedure-associated complications occurred; and vi) subjects
undergoing other interventional procedures during hospitalization,
such as biliary drainage or cyst puncture drainage. No subjects
included in the present study had received HCC-associated
treatment. All subjects had undergone abdominal CT or abdominal MRI
before and after thermal ablation (thereafter referred to as
ablation). The present study was approved by The Ethics Committee
of Beijing Youan Hospital, Capital Medical University. Written
informed consent was obtained from all subjects. The present study
was performed according to the guidelines of the Helsinki
Declaration.
A total of 10 ml whole-blood samples were collected
prior to thermal ablation (PR), 1–3 days post-ablation (P1), and
5–7 days post-ablation (P7) following the first ablation treatment
(among them, M3 subject underwent two ablation treatment, and other
subjects underwent one ablation treatment in the hospitalization).
PBMCs were isolated by Ficoll density gradient within 6 h after
peripheral blood collection, then stored in liquid nitrogen until
use.
Interventional treatments
Before transarterial chemo-embolization (TACE;
microguide wire and microcatheter; Asahi Intecc, Co., Ltd.), a
percutaneous liver biopsy was taken. RFA (RF electrode needle;
AngioDynamics, Inc.) or MWA (microwave ablation needle; Nanjing Eco
Microwave System, Co., Ltd.) was performed under local anesthesia 1
week after TACE. The power and duration of RFA or MWA are presented
in Table I. The aforementioned
treatments were performed by an interventional radiologist with
>5 years of experience. Abdominal CT was used to evaluate the
ablation effect by all radiologists in The Interventional Therapy
Center for Oncology.
 | Table I.Demographic and clinical data of five
patients with hepatitis B virus-associated hepatocellular carcinoma
undergoing thermal ablation. |
Table I.
Demographic and clinical data of five
patients with hepatitis B virus-associated hepatocellular carcinoma
undergoing thermal ablation.
| Subjects | Ma1 | Ma2 | Ma3 | Ma4 | Ma5 |
|---|
| Time
point-PRb | 2018/1/8 | 2018/1/24 | 2018/2/23 | 2018/3/16 | 2018/2/23 |
| Time
point-P1c | 2018/1/18 | 2018/1/30 | 2018/3/1 | 2018/3/21 | 2018/3/2 |
| Time
point-P7d | 2018/1/22 | 2018/2/5 | 2018/3/7 | 2018/3/26 | 2018/3/6 |
| Age | 48 | 62 | 46 | 58 | 50 |
| Sex | Male | Female | Male | Male | Male |
| Alcohol | Abstaining for 6
years | No | Consuming for the
past 5 years | No | No |
| Smoking, years | No | 30 | 26 | No | No |
| Cirrhotic
morphology | Yes | Yes | Yes | Yes | Yes |
| Prior
treatment | No | No | No | No | No |
| Tumor nodules,
n | 1 | 1 | 4 | 1 | 1 |
| Maximum diameter,
cm | 4.4 | 2.5 | 4.5,1,1,2 | 2.2 | 2.7 |
| Ablation region
maximum diameter, cm | 5.45 | 7.15 | 6.08, 2.14 | 8.92 | 6.23 |
| Tumor size treated,
cm | 4.4 | 2.5 | 4.5,1 | 2.2 | 2.7 |
| Complete ablation
or note | Yes | Yes | No | No | Yes |
| Total ablation
numberf | 1 | 1 | 2 | 1 | 1 |
| Vascular
invasion | No | Yes | No | Yes | No |
| Distant
metastasis | No | No | No | No | No |
| BCLCg | A | C | B | C | A |
| Surgery duration,
min | 85 | 80 | 100 | 160 | 120 |
| Relapse | No | No | No | Yes | No |
Illumina BeadChip 850K analysis and
methylation assay
DNA was extracted from PBMCs using a QIAamp DNA Mini
kit (Qiagen GmbH) according to the manufacturers protocol. DNA
purity was assessed by measuring the
A260/A280 ratio using a NanoDrop™ instrument
(Thermo Fisher Scientific, Inc.). DNA integrity was evaluated via
agarose gel electrophoresis. An intense band within the
high-molecular weight range (>10 kb) was required to pass the
quality control assessment. Bisulfite conversion was performed
according to the manufacturers recommendations in the EZ DNA
Methylation kit (Zymo Research Corp.), followed by Illumina 850K
BeadChip analysis (Infinium Methylation EPIC BeadChip v.1.0 (8
samples/chip); Illumina, Inc.). DNA methylation levels was detected
using Illumina Infinium HD Methylation Assay (Infinium Methylation
EPIC BeadChip kit v.1.0 (16 samples); Illumina, Inc.) according to
the manufacturers protocol.
Statistical analysis
Illumina arrays were analyzed using Bioconductor
package in R (22–25). Raw data were kept for probes with a
mean detection value of P<0.05. Probes on the X or Y chromosome
were filtered out in order to mitigate sex effects, as well as
probes with single nucleotide polymorphisms and probes that aligned
to multiple locations (24,25). Intra-array normalization adjusting
biased data was performed using preprocess Quantile (24). The present study corrected for batch
effects after normalization and filtering using the sva
Bioconductor package in R (26). The
β-values of the normalized data were used for downstream
statistical analysis. Differentially methylated CG sites (CGs) at
PR, P1 and P7 time points were identified using one-way ANOVA with
the lmFit function in the Bioconductor package Limma (23) and correcting for multiple testing
using Benjamini-Hochberg (adjusted P<0.05), and |log(FC)
|>0.5 was also used to indicate the significant difference,
where FC is the fold-change (23).
Pearson correlation analyses were performed to identify
correlations among the three-time points (r>0.8 or r<-0.8).
Heatmaps and boxplots were generated using the pheatmap (https://CRAN.R-project.org/package=pheatmap) and
ggplot2 (https://CRAN.R-project.org/package=ggplot2) package in
R. After the annotation of differentially methylated CGs on the
promoter, the mean β-value of those CGs was calculated to predict
positive or negative gene expression. Pathway analysis of genes was
determined by Gene Ontology analysis using Ingenuity Pathway
Analysis (IPA; Qiagen GmbH), Fishers exact test was used to
calculate P-values, determining the probability between genes and
the canonical pathways, and any pathway with -log(P)>1 and at
least three genes were included in the analysis. The age of those
patients is expressed as the mean ± standard deviation.
Results
Clinical characteristics
The demographic and clinical characteristics of the
five subjects are presented in Table
I. The mean age was 52.80±6.87 years. Parameters which could
impact the distribution of DNA methylation, such as age, sex,
drinking and smoking, were eliminated by paired analysis with the
samples of each subject before and after ablation treatment.
Distribution of DNA methylation
differs at the PR, P1 and P7 time points
The present study delineated differentially
methylated CGs among the PR, P1 and P7 time points using the
Bioconductor package Limma (23). A
total of 119,440 differentially methylated CGs were identified
(adjusted P<0.05): 115,166 different CGs between the PR and P1
time points, 84,330 CGs between the PR and P7 time points, and
84,673 CGs between the P1 and P7 time points. T-distributed
stochastic neighbor embedding of those 119,440 CGs exhibited a good
separation effect among the three time points (Fig. 1A).
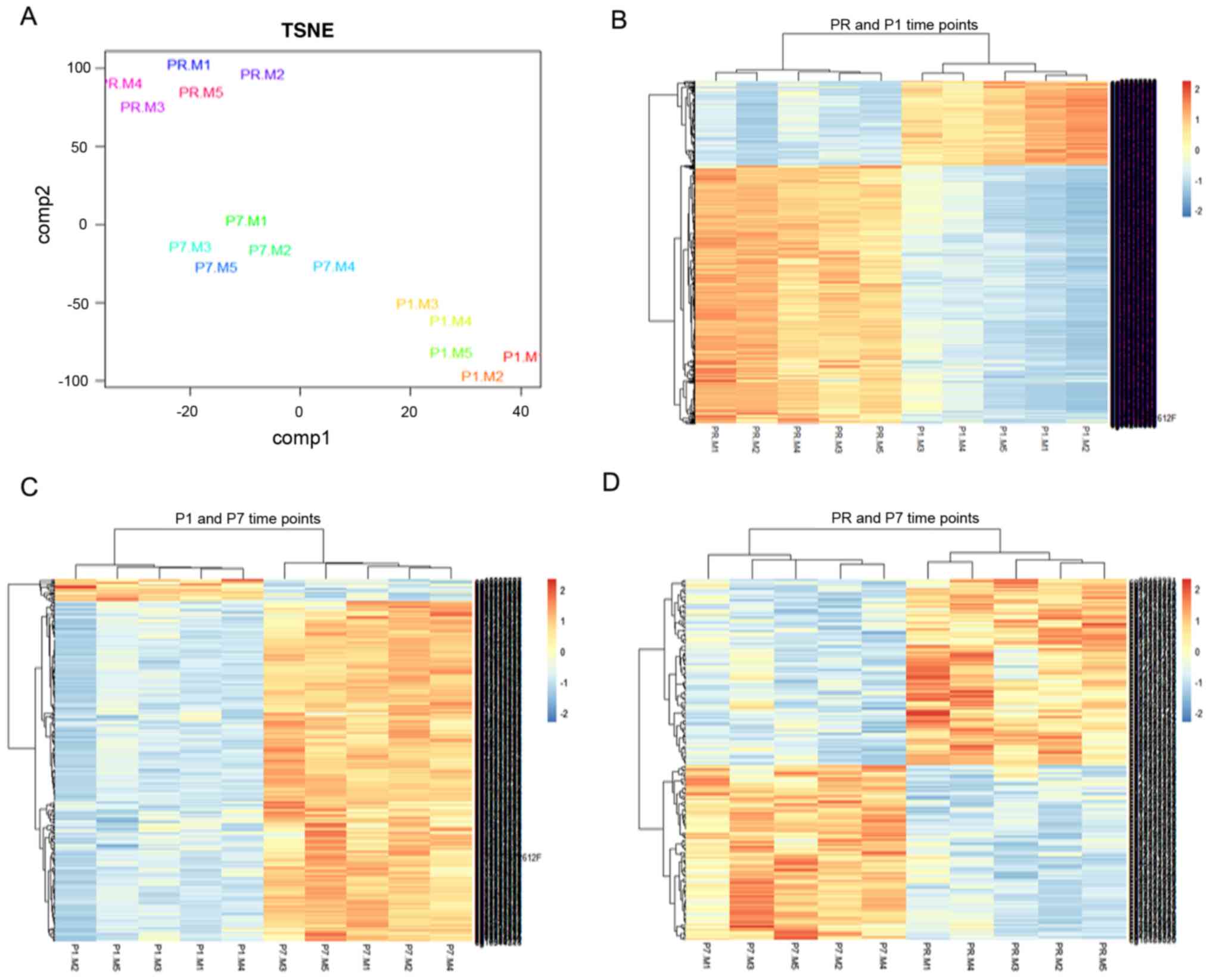 | Figure 1.Differentially methylated CGs at the
PR, P1 and P7 time points. (A) One-way ANOVA was used to compare
differentially methylated CGs among the three time points.
Distributional characteristics of the 119,440 different CGs are
displayed by TSNE. (B) Heatmap of hierarchical clustering by 2,779
differentially methylated CGs between the PR and P1 time points,
the color indicates the CGs methylation level, the deep color
represents greater expression (red denotes hypermethylation; blue
denotes hypomethylation). (C) Heatmap of hierarchical clustering by
374 differentially methylated CGs between the P1 and P7 time
points, the color indicates the CGs methylation level, the deep
color represents greater expression (red denotes hypermethylation;
blue denotes hypomethylation). (D) Heatmap of hierarchical
clustering by 204 differentially methylated CGs between the PR and
P7 time points, the color indicates the CGs methylation level, the
deep color represents greater expression (red denotes
hypermethylation; blue denotes hypomethylation). Adjusted
P<0.05; |log(fold-change)|>0.5. CGs, CG sites; TSNE,
T-distributed stochastic neighbor embedding; PR, prior to thermal
therapy; P1, 1–3 days post-ablation; P7, 5–7 days
post-ablation. |
Paired analysis with Benjamini-Hochberg correction
(adjusted P<0.05; |log(FC)|>0.5) was performed. Finally,
there were 2,779 differentially methylated sites between the PR and
P1 time points (Fig. 1B), 374
differentially methylated sites between the P1 and P7 time points
(Fig. 1C), and 204 differentially
methylated sites between the PR and P7 time points (Fig. 1D). The clustering analysis
demonstrated that all those different CGs could well distinguish
those time points.
Immune activation at the P1 time point
is associated with systemic stress responses
Next, 3,000 differently methylated CGs (adjusted
P<0.05; |log(FC)|>0.5) in all three groups were focused on.
The mean methylation levels of those 3,000 CGs were decreased
between the PR and P1 time points, but increased at the P7 time
point (Fig. 2A). Among the 3,000
CGs, there were 744 (24.8%; 744/3,000) sites with increased
methylation between the PR and P1 time points, which gradually
decreased at the P7 time point (Fig.
2B). At the same time, there were 2,256 (75.2%; 2,256/3,000)
sites with decreased methylation between the PR and P1 time points,
which gradually increased at the P7 time point (Fig. 2C).
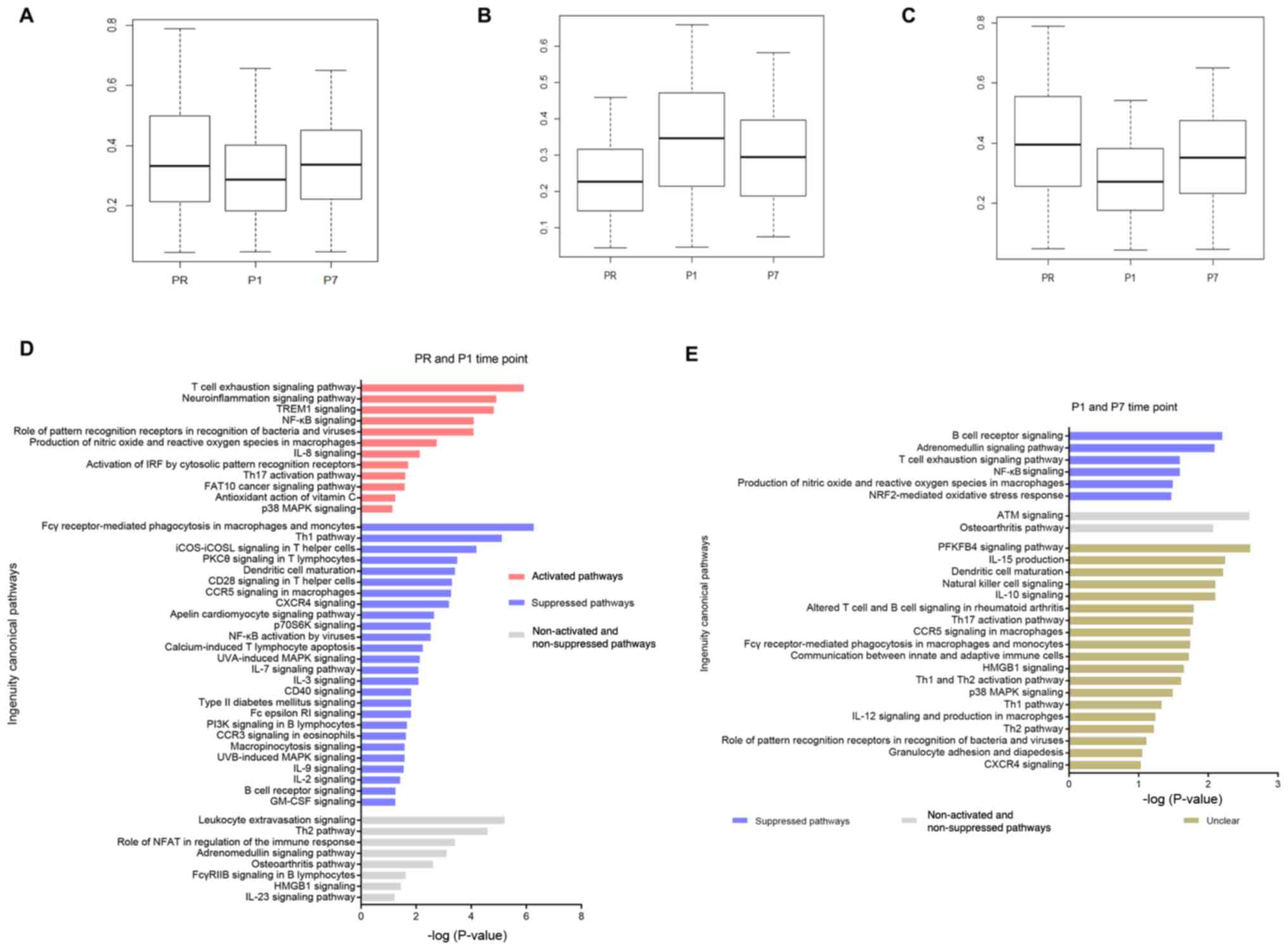 | Figure 2.Change trend of mean methylation
level and pathway analysis at the PR, P1, P7 time points. (A)
Boxplot of DNA methylation mean β-values of 3,000 CGs (adjusted
P<0.05; |log(FC)|>0.5) among the PR, P1 and P7 time points.
(B) Boxplot of DNA methylation mean β-values of 744 (24.8%;
744/3,000) increased CGs (adjusted P<0.05; |log(FC)|>0.5)
between the PR and P1 time points, which were decreased at the P7
time point. (C) Boxplot of DNA methylation mean β-values of 2,256
(75.2%; 2,256/3,000) decreased CGs (adjusted P<0.05;
|log(FC)|>0.5) between the PR and P1 time points, which were
increased at the P7 time point. (D) Pathway analysis between the PR
and P1 time points (-log(P)>1). Unclear pathways are not
presented. (E) Pathway analysis between the P1 and P7 time points
(-log(P)>1). CGs, CG sites; FC, fold-change; PR, prior to
thermal therapy; P1, 1–3 days post-ablation; P7, 5–7 days
post-ablation. |
Gene annotation of differentially methylated sites
located on the promoter indicated that there were 759 different
genes between the PR and P1 time points, and 167 different genes
between the P1 and P7 time points. The signaling pathways
associated with these genes were then identified using IPA. The
results were filtered in order to select immune-associated
pathways, which identified 46 signaling pathways between the PR and
P1 time points, 27 signaling pathways between the P1 and P7 time
points (-log(P)>1, with at least three genes in one
pathway).
For the PR and P1 time points, several systemic
stress response pathways were identified, such as
‘neuroinflammation signaling pathway’ and ‘antioxidant action of
vitamin C’. Innate and adaptive immune pathways were also observed,
such as ‘T cell exhaustion signaling pathway’, ‘production of
nitric oxide and reactive oxygen species in macrophages’ and
‘activation of IRF by cytosolic pattern recognition receptors’.
Moreover, the ‘FAT10 cancer signaling pathway’ was activated.
However, T lymphocyte co-stimulation (‘CD28 signaling in T helper
cells’ and ‘CD40 signaling’), activation (‘PKCθ signaling in T
lymphocytes’), migration (‘CXCR4 signaling’) and the ‘Th1 pathway’
were suppressed (Fig. 2D).
At the P1 and P7 time points, several systemic
stress response pathways were suppressed, such as the
‘adrenomedullin signaling pathway’ and the ‘NRF2-mediated oxidative
stress response’. Several innate and adaptive immune pathways that
were activated at the P1 time point were suppressed at the P7 time
point, such as ‘T cell exhaustion signaling pathway’ and
‘production of nitric oxide and reactive oxygen species in
macrophages’. The change trends of 19 other pathways, including
‘dendritic cell maturation’ and ‘natural killer cell signaling’,
were unclear (Fig. 2E).
Activation of the adaptive immune
response at the P7 time point
Following the gene annotation and signaling pathway
analysis, there were 184 different genes, and 29 signaling pathways
altered between the PR and P7 time points (-log(P)>1, at least
three genes in one pathway). In total, 7 pathways were activated,
including innate immunity (‘dendritic cell maturation’ and
‘production of nitric oxide and reactive oxygen species in
macrophages’). In addition, T cell-related pathways were also
observed, such as ‘CXCR4 signaling’, the ‘Th1 pathway’ and the ‘Th2
pathway’. The ‘osteoarthritis pathway’ was suppressed, and the
change trends for 21 other pathways were unclear (Fig. 3A).
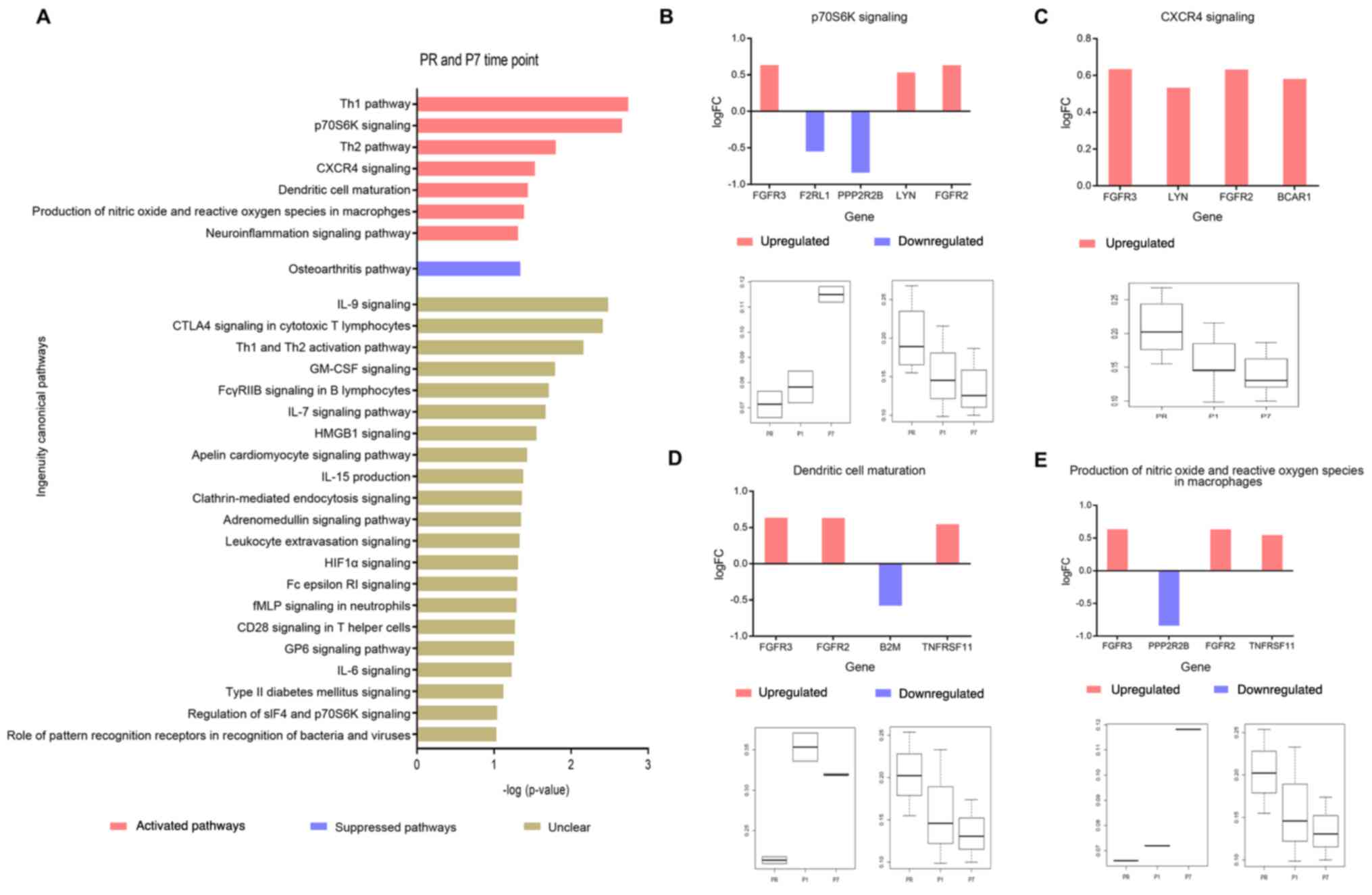 | Figure 3.Pathway analysis between the PR and
P7 time points. (A) Pathway analysis between the PR and P7 time
points (-log(P)>1). (B) Change trend of ‘p30S6K
signaling’-associated genes between PR and P7 time points (upper).
Red indicates activation, blue indicates suppression. Boxplot
(bottom) indicates the mean methylation levels of increased (left)
and decreased (right) CGs in this pathway, whose levels of
methylation were significant between PR and P7 time points
(adjusted P<0.05; |log(FC)|>0.5). (C) Change trend of ‘CXCR4
signaling’-associated genes between PR and P7 time points (upper),
red indicates activation. Boxplot (bottom) shows the mean
methylated levels of those decreased CGs in this pathway, whose
levels of methylation were significant between PR and P7 time
points (adjusted P<0.05; |log(FC)|>0.5). (D) Change trend of
‘dendritic cell maturation’ pathway-associated genes between PR and
P7 time points (upper), red indicates activation, blue indicates
suppression. Boxplot (bottom) indicates the mean methylation levels
of increased (left) and decreased (right) CGs in this pathway,
whose levels of methylation were significant between PR and P7 time
points (adjusted P<0.05; |log(FC)|>0.5), respectively. (E)
Change trend of ‘production of nitric oxide and reactive oxygen
species in macrophages’-associated genes between PR and P7 time
points (upper). Red indicates activation, blue indicates
suppression. Boxplot (bottom) indicates the mean methylation levels
of increased (left) and decreased (right) CGs in this pathway,
whose levels of methylation were significant between PR and P7 time
points (adjusted P<0.05; |log(FC)|>0.5). FC, fold-change;
CGs, CG sites; PR, prior to thermal therapy; P1, 1–3 days
post-ablation; P7, 5–7 days post-ablation. |
Excluding the T helper cell 1 (Th1), T helper cell 2
(Th2) and neuroinflammation pathways, the present study focused on
the remaining four pathways (‘p70S6K signaling’, ‘CXCR4 signaling’,
‘dendritic cell maturation’ and ‘production of nitric oxide and
reactive oxygen species in macrophages’), to further analyze the
increased or decreased expression of the pathway associated CGs
(adjusted P<0.05; |log(FC)|>0.5), and the upregulated or
downregulated expression of the annotated genes (Fig. 3B-E). The majority of CGs had
consistent change trends that there was an increase at the P1 time
point, there was also an increase at the P7 time points; there was
a decrease at the P1 time point, there was also a decrease at the
P7 time points (Fig. 3B-E).
Discussion
An effective tumor-specific immune response and
long-lasting anti-tumor immunity is necessary for the development
of an effective form of immunotherapy (9). The present study focused on the global
genomic DNA methylation profiles of PBMCs to describe the overall
immune change trends following ablation treatment for patients with
HBV-associated HCC. DNA methylation profiles of pathways associated
with the immune system were different at the P1 and P7 time points
following ablation, and affected the systemic stress response at
the P1 time point in particular. The activation of ‘Th1 pathway’,
‘Th2 pathway’ and ‘CXCR4 signaling’, as well as macrophage and
dendritic cell-related pathways indicated that effective immune
responses were observable at the P7 time point. However, DNA
methylation in these pathways were already altered at the P1 time
point, suggesting that changes in DNA methylation occurred early
after ablation. Hence, the timing of combined immunotherapy and
ablation procedures should be chosen with caution.
As a main site of immune tolerance, the liver can
affect pathogen-specific immune responses (27). In addition, the liver can release
HCC-associated circular tumor DNA to the peripheral blood (28,29).
Abnormal liver lesions can also be detected by clinical laboratory
tests, such as alanine transaminase and aspartate aminotransferase.
Our previous study revealed that alterations of DNA methylation in
the immune system were closely associated with the development of
HCC (19). Other previous studies
suggested that ablation could induce tumor antigen-associated T
cell responses (10), as well as
systemic immune responses (12). As
peripheral immune cells can reflect the HCC-associated immune
response to a certain extent, PBMCs were used for analysis in the
present study. Following different methylation CGs analysis and the
estimation of gene expression, the methylation levels of those CGs
and immune-associated pathways were used to observe the methylation
and immune response change trend following ablation. The increased
sites between the PR and P1 time points gradually decreased at the
P7 time point, and the decreased sites between the PR and P1 time
points gradually increased at the P7 time point. Combined with the
pathway analysis, which demonstrated that the activated pathways
between the PR and P1 time points were suppressed at the P1 and P7
time points, and the stress response-associated ‘adrenomedullin
signaling pathway’ and ‘NRF2-mediated oxidative stress response’
were also suppressed at the P1 and P7 time points. Therefore, the
present study suggested that the suppressed effect of those
pathways was caused by a systemic stress response.
Between the PR and P7 time points, 7 signaling
pathways were activated following ablation. Among them, the
neuroinflammation and osteoarthritis pathways (30,31),
Th1, Th2 and p70 ribosomal S6 kinase (p70S6K) signaling were
associated with the change of immune microenvironment. Previous
studies about HCC indicated that Th1 cells mediated the immune
clearance of tumor cells and prevented tumorigenesis (32), and the cytokines that they produced
are strongly associated with good clinical outcome (33), whereas Th2 cells were associated with
tumor growth or metastasis (34). It
is well known that Th1/Th2 balance is broken in the HCC
micro-environment (35). Thus, the
activation of Th1 and Th2 signaling pathways and the significant
activation of Th1 pathway in the present study suggested the
possibility that ablation may reverse the Th1/Th2 immune imbalance
of HCC subjects. p70S6K is a serine/threonine protein kinase, and
the activation of the PI3K/mTOR/p70S6K pathway may serve to
integrate the extracellular signals to regulate the appropriate
differentiation program of naïve CD4+ T cells and the
generation of effector T cells (36). Robust Th1 cell and cytotoxic immune
responses are associated with prolonged survival in patients with
HCC (33). Based on the
aforementioned data, it can be speculated that the activation of
Th1 and p70S6K pathways after ablation were associated with immune
activation.
C-X-C motif chemokine receptor type 4 (CXCR4) is an
α-chemokine receptor that binds stromal-derived factor 1 and can
activate multiple biological processes (37). Following T cell receptor
crosslinking, CXCR4 is recruited and accumulates at the
immunological synapse, resulting in a stronger T
cell/antigen-presenting cell (APC) interaction, high levels of T
cell proliferation and interferon-γ production (38). Meanwhile, the activation of
classically activated macrophages can produce pro-inflammatory
cytokines and reactive oxygen/nitrogen species, which are crucial
for host defense and tumor cell killing, and suppress HCC cell
growth and induce liver tumor regression (39). Phenotypically, they also express high
levels of major histocompatibility complex class II, CD68, CD80 and
CD86 costimulatory molecules (40).
In addition, tumor-specific T cells in pre-malignant liver lesions
encountering tumor antigens on non-professional and/or
non-activated APCs in a non-inflammatory context can lead to
peripheral self-tolerance (14).
Therefore, the activation of CXCR4, dendritic cell maturation and
macrophage signaling can promote the interaction between T cell and
APC, increase host defense function, and are necessary for
effective anti-tumor immune responses.
A correlation analysis among the PR, P1 and P7 time
points demonstrated that the M3 and M4 subjects had a different
change in trend compared with others (data not shown). Considering
the consistency of the inclusion and exclusion criteria, the
potential confounding factors (such as smoking, drinking, sex and
age) can be ignored by paired analysis, so the ablation and
tumor-associated factor may be the main factors accounting for this
difference. Previous studies indicated that incomplete ablation
could activate signaling pathways that may favor pro-tumor
development or metastasis (41–43). In
the present study, two subjects (M3 and M4) received incomplete
ablation at the sample collection time point. According to the
aforementioned analysis, the present study inferred that these two
subjects with incomplete ablation may have a different immune
response trend compared with the complete ablation subjects,
however, this requires further validation.
According to the standard deviation among three time
points (0.016) and the desired power (0.8), the appropriate sample
sizes for microarray experiments were calculated, which suggested
that a small sample size (n=3) was required to detect a δ of 0.1 at
the microarray data set. Thus, the paired sample size of five is
large enough to explain those results. However, there were several
limitations to the present study. First, since there were only five
subjects in the present study, the immune response could not be
compared between incomplete and complete ablation subjects.
Secondly, gene expression is affected not only by DNA methylation,
but also by histone deacetylation (44). Thus, the effects of deacetylation
should also be investigated. Thirdly, a previous study indicated
that a single high dose of radiation could affect the immune
response (45) In the present study,
samples were collected before and after the first ablation
treatment to understand how the immune response is affected by
repeated ablation treatments requires further investigation.
Overall, the results of the present study
demonstrated that DNA methylation could be altered soon after
ablation, while systemic immune responses were induced at the P7
time point. Furthermore, the continuously activated signaling
pathways may not end at the P7 time points. Hence, it is necessary
to investigate the duration of immune activation, and the time
point of immunotherapy, particularly for the immune checkpoint
inhibitor for ablation subjects in a future study.
Acknowledgements
Not applicable.
Funding
The present study was supported by The Beijing
Natural Science Foundation (grant no. 7191004), The Beijing
Municipal Science & Technology Commission (grant no.
Z171100001017078), The Beijing Municipal Administration of
Hospitals (grant nos. DFL20181701 and ZYLX201711), The National S
& T Major Project for Infectious Diseases Control (grant no.
2017ZX10201101001008) and The Biomarkers of Infection-Related
Diseases Beijing Key Laboratory (grant no. BZ0373). The funders had
no role in study design, data collection and analysis, decision to
publish, or preparation of the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
YNZ performed the experiments, data analysis,
writing of the original draft and reviewing the manuscript. KL and
JS performed the experiments and data analysis. JS also wrote and
reviewed the manuscript for important intellectual content. NH and
PZ performed the quality control of the data and algorithms, and
the clinical data evaluation. CZ, XY, CH, JL, HZ and QW collected
the clinical samples and performed the clinical data evaluation.
YHZ and YZ conceptualized and designed the present study, acquired
the funding, performed the project administration, drafted the
original manuscript and reviewed the manuscript for important
intellectual content. All authors have read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethics
Committee of Beijing You'an Hospital, Capital Medical University.
Written informed consent was obtained from all subjects. The
present study was performed according to the guidelines of the
Helsinki Declaration [ethical approval no. JingyouKelunzi (2017)
no. 26].
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HBV
|
hepatitis B virus
|
|
HCC
|
hepatocellular carcinoma
|
|
PBMCs
|
peripheral blood mononuclear cells
|
|
CGs
|
CG sites
|
|
RFA
|
radiofrequency ablation
|
|
MWA
|
microwave ablation
|
|
TACE
|
transarterial chemoembolization
|
|
IPA
|
Ingenuity Pathway Analysis
|
|
FC
|
fold-change
|
|
APC
|
antigen-presenting cell
|
|
Th1
|
T helper cell 1
|
|
Th2
|
T helper cell 2
|
|
p70S6K
|
p70 ribosomal S6 kinase
|
|
CXCR4
|
C-X-C motif chemokine receptor 4
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Feng RM, Zong YN, Cao SM and Xu RH:
Current cancer situation in China: Good or bad news from the 2018
Global cancer statistics? Cancer Commun (Lond). 39:222019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu J, Liang W, Jing W and Liu M:
Countdown to 2030: Eliminating hepatitis B disease. China. Bull
World Health Organ. 97:230–238. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen MS, Li JQ, Zheng Y, Guo RP, Liang HH,
Zhang YQ, Lin XJ and Lau WY: A prospective randomized trial
comparing percutaneous local ablative therapy and partial
hepatectomy for small hepatocellular carcinoma. Ann Surg.
243:321–328. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jiang Y, Zhou S, Shen G, Jiang H and Zhang
J: Microwave ablation combined with transcatheter arterial
chemoembolization is effective for treating unresectable
hepatoblastoma in infants and children. Medicine (Baltimore).
97:e126072018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tang C, Shen J, Feng W, Bao Y, Dong X, Dai
Y, Zheng Y and Zhang J: Combination therapy of radiofrequency
ablation and transarterial chemoembolization for unresectable
hepatocellular carcinoma: A retrospective study. Medicine
(Baltimore). 95:e37542016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang WD, Zhang LH, Ni JY, Jiang XY, Chen
D, Chen YT, Sun HL, Luo JH and Xu LF: Radiofrequency ablation
combined with transcatheter arterial chemoembolization therapy
versus surgical resection for hepatocellular carcinoma within the
milan criteria: A meta-analysis. Korean J Radiol. 19:613–622. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hocquelet A, Balageas P, Laurent C, Blanc
JF, Frulio N, Salut C, Cassinotto C, Saric J, Possenti L, Bernard
PH, et al: Radiofrequency ablation versus surgical resection for
hepatocellular carcinoma within the milan criteria: A study of 281
Western patients. Int J Hyperthermia. 31:749–757. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Greten TF, Manns MP and Korangy F:
Immunotherapy of hepatocellular carcinoma. J Hepatol. 45:868–878.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mizukoshi E, Yamashita T, Arai K,
Sunagozaka H, Ueda T, Arihara F, Kagaya T, Yamashita T, Fushimi K
and Kaneko S: Enhancement of tumor-associated antigen-specific T
cell responses by radiofrequency ablation of hepatocellular
carcinoma. Hepatology. 57:1448–1457. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zerbini A, Pilli M, Penna A, Pelosi G,
Schianchi C, Molinari A, Schivazappa S, Zibera C, Fagnoni FF,
Ferrari C and Missale G: Radiofrequency thermal ablation of
hepatocellular carcinoma liver nodules can activate and enhance
tumor-specific T-cell responses. Cancer Res. 66:1139–1146. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Waitz R and Solomon SB: Can local
radiofrequency ablation of tumors generate systemic immunity
against metastatic disease. Radiology. 251:1–2. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Youngblood B, Hale JS, Kissick HT, Ahn E,
Xu X, Wieland A, Araki K, West EE, Ghoneim HE, Fan Y, et al:
Effector CD8 T cells dedifferentiate into long-lived memory cells.
Nature. 552:404–409. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schietinger A, Philip M, Krisnawan VE,
Chiu EY, Delrow JJ, Basom RS, Lauer P, Brockstedt DG, Knoblaugh SE,
Hämmerling GJ, et al: Tumor-specific T cell dysfunction is a
dynamic antigen-driven differentiation program initiated early
during tumorigenesis. Immunity. 45:389–401. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gao W, Kondo Y, Shen L, Shimizu Y, Sano T,
Yamao K, Natsume A, Goto Y, Ito M, Murakami H, et al: Variable DNA
methylation patterns associated with progression of disease in
hepatocellular carcinomas. Carcinogenesis. 29:1901–1910. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ye C, Tao R, Cao Q, Zhu D, Wang Y, Wang J,
Lu J, Chen E and Li L: Whole-genome DNA methylation and
hydroxymethylation profiling for HBV-related hepatocellular
carcinoma. Int J Oncol. 49:589–602. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ahn E, Youngblood B, Lee J, Lee J, Sarkar
S and Ahmed R: Demethylation of the PD-1 promoter is imprinted
during the effector phase of Cd8 T cell exhaustion. J Virol.
90:8934–8946. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ghoneim HE, Fan Y, Moustaki A, Abdelsamed
HA, Dash P, Dogra P, Carter R, Awad W, Neale G, Thomas PG and
Youngblood B: De novo epigenetic programs inhibit PD-1
blockade-mediated t cell rejuvenation. Cell. 170:142–157 e19. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Y, Petropoulos S, Liu J, Cheishvili
D, Zhou R, Dymov S, Li K, Li N and Szyf M: The signature of liver
cancer in immune cells DNA methylation. Clin Epigenetics. 10:82018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
European Association for the Study of the
Liver. Electronic address; easloffice@easloffice.eu: Corrigendum to
‘EASL clinical practice guidelines: Management of hepatocellular
carcinoma’. J Hepatol 69 (2018) 182–236. J Hepatol.
70:8172019.PubMed/NCBI
|
|
21
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Morris TJ, Butcher LM, Feber A,
Teschendorff AE, Chakravarthy AR, Wojdacz TK and Beck S: ChAMP: 450
k Chip analysis methylation pipeline. Bioinformatics. 30:428–430.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Aryee MJ, Jaffe AE, Corrada-Bravo H,
Ladd-Acosta C, Feinberg AP, Hansen KD and Irizarry RA: Minfi: A
flexible and comprehensive bioconductor package for the analysis of
infinium DNA methylation microarrays. Bioinformatics. 30:1363–1369.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Maksimovic J, Phipson B and Oshlack A: A
cross-package bioconductor workflow for analysing methylation array
data. Version 3. F1000Res. 5:12812016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Leek JT, Johnson WE, Parker HS, Jaffe AE
and Storey JD: The sva package for removing batch effects and other
unwanted variation in high-throughput experiments. Bioinformatics.
28:882–883. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jenne CN and Kubes P: Immune surveillance
by the liver. Nat Immunol. 14:996–1006. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun K, Jiang P, Chan KC, Wong J, Cheng YK,
Liang RH, Chan WK, Ma ES, Chan SL, Cheng SH, et al: Plasma DNA
tissue mapping by genome-wide methylation sequencing for
noninvasive prenatal, cancer, and transplantation assessments. Proc
Natl Acad Sci USA. 112:E5503–E5512. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu RH, Wei W, Krawczyk M, Wang W, Luo H,
Flagg K, Yi S, Shi W, Quan Q, Li K, et al: Circulating tumour DNA
methylation markers for diagnosis and prognosis of hepatocellular
carcinoma. Nat Mater. 16:1155–1161. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Molfino A, Gioia G, Rossi Fanelli F and
Laviano A: Contribution of neuroinflammation to the pathogenesis of
cancer cachexia. Mediators Inflamm. 2015:8016852015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Haywood L, McWilliams DF, Pearson CI, Gill
SE, Ganesan A, Wilson D and Walsh DA: Inflammation and angiogenesis
in osteoarthritis. Arthritis Rheum. 48:2173–2177. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mossanen JC and Tacke F: Role of
lymphocytes in liver cancer. Oncoimmunology. 2:e264682013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fridman WH, Pages F, Sautes-Fridman C and
Galon J: The immune contexture in human tumours: Impact on clinical
outcome. Nat Rev Cancer. 12:298–306. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee HL, Jang JW, Lee SW, Yoo SH, Kwon JH,
Nam SW, Bae SH, Choi JY, Han NI and Yoon SK: Inflammatory cytokines
and change of Th1/Th2 balance as prognostic indicators for
hepatocellular carcinoma in patients treated with transarterial
chemoembolization. Sci Rep. 9:32602019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cao X: Immunology in China: The past
persent and future. Nat Immunol. 9:339–342. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
O'Brien TF and Zhong XP: The role and
regulation of mTOR in T-lymphocyte function. Arch Immunol Ther Exp
(Warsz). 60:173–181. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chatterjee S, Behnam Azad B and Nimmagadda
S: The intricate role of CXCR4 in cancer. Adv Cancer Res.
124:31–82. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Eckert F, Schilbach K, Klumpp L, Bardoscia
L, Sezgin EC, Schwab M, Zips D and Huber SM: Potential role of
CXCR4 targeting in the context of radiotherapy and immunotherapy of
cancer. Front Immunol. 9:30182018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Guerra AD, Yeung OWH, Qi X, Kao WJ and Man
K: The anti-tumor effects of M1 macrophage-loaded poly (ethylene
glycol) and gelatin-based hydrogels on hepatocellular carcinoma.
Theranostics. 7:3732–3744. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Aras S and Zaidi MR: TAMeless traitors:
Macrophages in cancer progression and metastasis. Br J Cancer.
117:1583–1591. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dong S, Kong J, Kong F, Kong J, Gao J, Ke
S, Wang S, Ding X, Sun W and Zheng L: Insufficient radiofrequency
ablation promotes epithelial-mesenchymal transition of
hepatocellular carcinoma cells through Akt and ERK signaling
pathways. J Transl Med. 11:2732013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yuan CW, Wang ZC, Liu K and Liu DJ:
Incomplete radiofrequency ablation promotes the development of
CD133+ cancer stem cells in hepatocellular carcinoma
cell line HepG2 via inducing SOX9 expression. Hepatobiliary
Pancreat Dis Int. 17:416–422. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang N, Ma D, Wang L, Zhu X, Pan Q, Zhao
Y, Zhu W, Zhou J, Wang L, Chai Z, et al: Insufficient
radiofrequency ablation treated hepatocellular carcinoma cells
promote metastasis by up-regulation ITGB3. J Cancer. 8:3742–3754.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Attwood JT, Yung RL and Richardson BC: DNA
methylation and the regulation of gene transcription. Cell Mol Life
Sci. 59:241–257. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zeng H, Zhang W, Gong Y and Xie C:
Radiotherapy activates autophagy to increase CD8+ T cell
infiltration by modulating major histocompatibility complex class-I
expression in non-small cell lung cancer. J Int Med Res.
47:3818–3830. 2019. View Article : Google Scholar : PubMed/NCBI
|

















