Introduction
Lung cancer is the most common cause of
cancer-associated mortalities worldwide, accounting for one in four
mortalities and the incidence rate was 96.8 per 100,000 in 2008 in
the United States (1). It is
estimated that ~2,093,876 new cases are diagnosed annually around
the world and the 5-year survival rate for patients with lung
cancer is 18%, which is the lowest among the major types of cancer,
including breast, colon and liver cancer (1). Non-small cell lung cancer (NSCLC) and
small cell lung cancer (SCLC) account for 85 and 15% of all lung
cancer cases (2), respectively. The
former subgroup is typically comprised of two common subtypes,
adenocarcinoma (~70%) and squamous cell lung cancer (~30%)
(2). The mechanisms underlying lung
carcinogenesis are complex and poorly understood. Numerous cellular
phenomena, including hypoxia, inflammation, the tumor
microenvironment and oxidative stress, coupled with various
molecular events, promote lung cancer initiation and progression
(3–6). The majority of patients with lung
cancer have a poor prognosis due to recurrence and distant
metastasis (7,8). Despite the advancements in
chemotherapy, radiotherapy and surgical techniques for lung cancer
in recent years, the long-time survival rate has failed to improve
(9). The lack of appropriate
molecular biomarkers is one of the main reasons for this failure as
it results in patients being diagnosed at an advanced or distant
metastatic stage, at which point curative treatments are no longer
available (10,11). For patients with lung cancer,
diagnosis and treatment at an early stage of disease can prolong
survival (12). At present, the
clinical diagnosis of lung cancer depends on chest X-ray and low
dose computed tomography scans (13); however, the high false positive rates
(14), side-effects of radiation and
the high costs may influence diagnostic accuracy and limit utility
in lung cancer screening. In addition, approaches such as
bronchoscopy and biopsy can be used, but they are painful, invasive
and laborious (15). As a result, it
is imperative to identify novel, sensitive and reliable biomarkers,
as well as more efficient therapeutic targets to improve the
diagnosis and treatment of patients with lung cancer.
The whole human genome consists of 20,000-25,000
genes; however, only 2% of genome sequences encode proteins
(16) and at least 90% of human
genes are transcribed into non-coding RNAs (ncRNAs) (17). Currently, several ncRNAs have been
discovered, including ribosomal RNAs, transfer RNAs,
piwi-interacting RNAs, microRNAs and long non-coding RNAs
(lncRNAs), which were previously regarded as useless, but they are
now known to have crucial biological functions (18). According to transcript size, the
remaining ncRNAs are classified into small ncRNAs (<200 nt) and
lncRNAs (>200 nt) (13). lncRNAs
can be classified into the following 6 categories: Sense,
antisense, divergent, bidirectional, intronic and intergenic
(19). lncRNAs have attracted the
most attention and an increasing number of studies have
demonstrated that many lncRNAs play crucial regulatory roles in a
wide variety of physiological processes, including development,
differentiation and metabolism at the transcriptional,
post-transcriptional and epigenetic levels (20,21). At
the transcriptional level, lncRNAs regulate gene expression
depending upon the sequence and relative position; for example, by
combining with the promoters of target genes and forming stable
DNA-RNA triplex complexes to inhibit transcription initiation
(22). At the post-transcriptional
level, a number of lncRNAs may competitively bind to the target
mRNA and prevent the binding of transcription factors, resulting in
aberrant mRNA translation, splicing and degradation (23). At the epigenetic level, lncRNAs
promote chromatin reprogramming genome-wide and regulate DNA
methylation (24) and histone
modification (25,26). Genome-scale approaches revealed that
several lncRNAs have a significant secondary structure (24), which is critical to specific binding
and function (27), and shares
certain common sequence features, including paucity of introns, low
GC content, weak start codon and open reading frame contexts
(28). Aberrantly expressed lncRNAs
in various types of cancer are thought to contribute to
tumorigenesis by influencing tumor cell proliferation, resisting
cell death, enabling distant metastasis and replicative
immortality, and inducing angiogenesis (20,29–32).
However, the roles of lncRNAs in lung cancer and their association
with clinicopathological parameters remain largely unknown and
therefore require further study.
The RFPL3 antisense (RFPL3S) gene is located on
chr22:32,359,906-32,382,052 (GRCh38/hg38) and has been suggested to
regulate the sense RFPL genes at the post-transcriptional level
(33). RFPL3S consists of eight
transcripts with a length range between 334 and 1,295 bp (33). The present study further investigated
the expression of RFPL3S in lung cancer based on tissue samples and
databases, with the purpose of investigating the prognostic
implications and expression pattern of RFPL3S, as well as the
associations with clinicopathological characteristics in patients
with lung cancer.
Materials and methods
Tissue specimens
Human lung cancer tissue samples and adjacent
non-cancerous tissues (within 3 cm of cancer tissue) were obtained
from 205 patients who underwent surgical resection of lung cancer
between January 2009 and October 2011 at The First People's
Hospital of Wujiang District (Suzhou, China). Written informed
consent was obtained from all patients and the present study was
approved by the Ethics Committee of the First People's Hospital of
Wujiang District. The collected tissue samples were frozen in
liquid nitrogen until subsequent analysis. None of the patients had
received any therapy, including chemotherapy and radiotherapy,
prior to surgery. The histological grades were classified according
to the World Health Organization guidelines (34) and the tumors were staged using the
7th edition of the tumor-node-metastasis (TNM) staging system
(35).
Reverse transcription-quantitative
(RT-q) PCR analysis
Total RNA was extracted from tissues using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. cDNA was
synthesized with 1 µg total RNA using the Primer Script RT mix
(Takara Biotechnology Co., Ltd.). qPCR analysis was subsequently
performed with the SYBR Green Real-time PCR Master Mix (Takara
Biotechnology Co., Ltd.) in a 20 µl reaction volume. The
thermocycling conditions were as follows: 95°C for 20 sec
(pre-denaturation), followed by 40 cycles of 95°C for 10 sec
(denaturation) and 60°C for 45 sec (extension). The primers were
designed using Primer Premier software (version 5.0; Premier
Biosoft International) and the sequences were as follows: RFPL3S
forward 5′-GTCGTCAGAAATGAGGAGGAAGT-3′ and reverse,
5′-TTGAAGTAGAAGAGAGGCATGGG-3′ and GAPDH forward,
5′-CAAGGTCATCCATGACAACTTTG-3′ and reverse,
5′-GTCCACCACCCTGTTGCTGTAG-3′. The relative gene expression levels
were calculated using the 2−ΔΔCq method (36) and normalized to the internal
reference gene GAPDH. Each sample was run in triplicate and
independently repeated three times.
Immunohistochemistry
Tissue blocks were cut into 5-µm thick sections and
prepared for immunohistochemical staining. The sections were
deparaffinized and rehydrated using a graded series of alcohol
solutions, according to standard protocols. Endogenous peroxidase
activity was blocked by incubating the sections with 3% hydrogen
peroxide in methanol for 30 min at room temperature. For antigen
retrieval, the sections were placed in 10 mM citrate buffer (pH
6.0) for 20 min at 95°C. Sections were blocked with 5% BSA (Dako;
Agilent Technologies, Inc.) for 30 min at room temperature, in
order to inhibit non-specific binding. Subsequently, the sections
were incubated in a humidified chamber for 1 h at room temperature
with primary antibody directed against Ki-67 (1:50; cat. no.
MA5-14520; Thermo Fisher Scientific, Inc.). After washing three
times with ice-cold PBS, the sections were incubated with a
biotinylated goat anti-rabbit antibody secondary antibody (1:1,000;
cat. no. RS0002; Dako; Agilent Technologies, Inc.) for 30 min at
room temperature. The DAB Elite kit (Dako; Agilent Technologies,
Inc.) was employed to visualize peroxidase activity and the
sections were counterstained with Mayer hematoxylin for 1 min at
room temperature, followed by dehydration and mounting. The
expression of Ki-67 was evaluated using the labeling index, which
was determined by counting 500–1,000 tumor cells under a light
microscope (magnification, ×400). Tumor cells with ≥31% Ki-67
staining were defined as high Ki-67, based on the median value of
positive staining. All sections were independently and blindly
evaluated by three pathologists, one from the Department of
Oncology, Suzhou Ninth People's Hospital and two from the
Department of Oncology, The First Affiliated Hospital of Soochow
University (both Suzhou, China). The average of the scores from the
three independent pathologists was the final immunohistochemical
score.
Oncomine analysis
RFPL3S mRNA levels in different types of cancer were
determined using the Oncomine database (www.oncomine.org), which is a publicly accessible
online cancer microarray database that facilitates genome-wide
expression analyses (37). In the
present study, the unpaired student's t-test was used to generate a
P-value for comparison between the tumor and adjacent non-cancerous
tissues. The fold change was defined as 2 and the P-value was set
at 0.01.
Cancer Cell Line Encyclopedia (CCLE)
analysis
RFPL3S mRNA levels in lung cancer were analyzed
using the CCLE database (portals.broadinstitute.org/ccle/home), which is an
online encyclopedia that facilitates the identification of genetic,
lineage and predictors of drug sensitivity (37).
Kaplan-Meier plotter survival
analysis
The prognostic values of lung cancer samples with
high RFPL3S expression were further assessed using the Kaplan-Meier
plotter (kmplot.com/analysis).
Statistical analysis
Statistical analyses were performed using SPSS
software (version 18.0; IBM Corp.) and GraphPad Prism (version 6;
GraphPad Software, Inc.). The prognostic impacts of RFPL3S were
estimated using the Kaplan-Meier method and survival curves were
compared using the log-rank test. The associations between
clinicopathological characteristics and RFPL3S expression were
analyzed by the χ2 test. The data on RFPL3S expression
in paired tumor tissue samples and adjacent non-cancerous tissues
were analyzed via the paired Student's t-test. P<0.05 was
considered to indicate a statistically significant difference and
the data are presented as the mean ± standard deviation.
Results
Association between RFPL3S expression
levels and clinicopathological characteristics
The present study investigated the association
between RFPL3S expression levels and clinicopathological
characteristics, including sex, age, smoking history,
differentiation, histological type, pT factor, pN factor, pTNM
stage and Ki-67 labeling index in patients with lung cancer
(Table I). A total of 42 female and
163 male patients with a median age of 64 years (range, 30–86
years) were subdivided into the following two groups based on their
RFPL3S expression levels, high expression (n=149 patients) and low
expression (n=56 patients). For the lung cancer samples, increased
RFPL3S expression was more frequently observed in well- and
moderately-differentiated tumors than in poorly-differentiated
tumors (P=0.0229). In addition, it was noted that RFPL3S expression
was significantly associated with pN factor (P=0.0267), pTNM stage
(P=0.0117) and Ki-67 labeling index (P<0.0001); however, no
significant associations were demonstrated between RFPL3S
expression and sex (P=0.5672), age (P=0.2603), smoking history
(P=0.3432), histological type (P=0.3995) or pT factor (P=0.0513)
(Table I). These results demonstrate
that increased RFPL3S expression levels tend to be associated with
poorer differentiation, lymph node metastasis, TNM stage and Ki-67
labeling index.
 | Table I.Associations between RFPL3S
expression levels and clinicopathological characteristics in
patients with lung cancer. |
Table I.
Associations between RFPL3S
expression levels and clinicopathological characteristics in
patients with lung cancer.
|
|
| RFPL3S
expression |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | Patient, n | High | Low | P-value |
|---|
| Sex |
|
|
| 0.5672 |
|
Male | 163 | 117 | 46 |
|
|
Female | 42 | 32 | 10 |
|
| Age, years |
|
|
| 0.2603 |
|
<60 | 101 | 77 | 24 |
|
|
≥60 | 104 | 72 | 32 |
|
| Smoking
history |
|
|
| 0.3432 |
|
Smoker | 142 | 106 | 36 |
|
|
Non-smoker | 63 | 43 | 20 |
|
|
Differentiation |
|
|
| 0.0229 |
| Well +
Moderate | 140 | 95 | 45 |
|
|
Poor | 65 | 54 | 11 |
|
| Histological
type |
|
|
| 0.3995 |
|
Squamous | 111 | 78 | 33 |
|
|
Adenocarcinoma | 94 | 71 | 23 |
|
| pT factor |
|
|
| 0.0513 |
|
T1-2 | 148 | 102 | 46 |
|
|
T3-4 | 57 | 47 | 10 |
|
| pN factor |
|
|
| 0.0267 |
| N0 | 95 | 62 | 33 |
|
|
N1-2 | 110 | 87 | 23 |
|
| pTNM stage |
|
|
| 0.0117 |
|
I–II | 137 | 92 | 45 |
|
|
III–IV | 68 | 57 | 11 |
|
| Ki-67 labeling
index |
|
|
| <0.0001 |
|
<31 | 98 | 54 | 44 |
|
|
≥31 | 107 | 95 | 12 |
|
RFPL3S is upregulated in lung
cancer
Based on the Oncomine analysis, no significant
differences were demonstrated in the 56 total unique analysis when
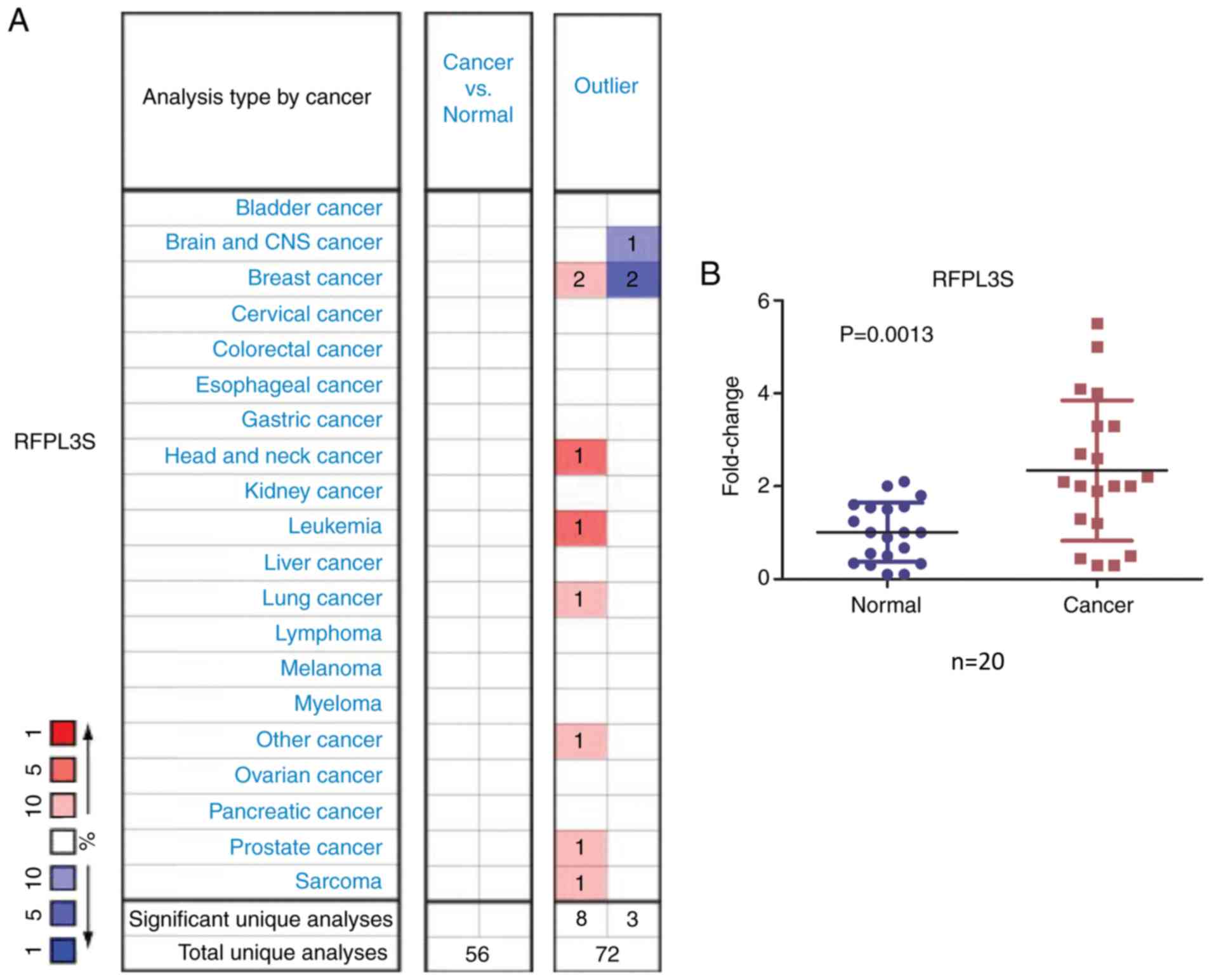
cancer tissues were compared with normal tissues (Fig. 1A). The outlier analysis, used to
determine significant RFPL3S expression in a subset of the patient
samples, suggested that there were eight cases with a significant
increase and three with a significant decrease in RFPL3S
expression. Patients with a significant increase included two
patients with breast cancer and one patient with brain and CNS
cancer (Fig. 1A). In the present
study, RFPL3S was found in several types of human cancers including
sarcoma, leukemia, breast, head and neck, lung and prostate cancer
based on the Oncomine analysis, and significantly increased RFPL3S
levels were detected in leukemia and head and neck cancer.
Furthermore, the present study determined the levels of RFPL3S
expression in 20 pairs of lung cancer tissues and adjacent
non-cancerous tissues and revealed that RFPL3S transcripts were
2.683-fold higher in lung cancer samples compared with normal
tissues (P=0.0013; Fig. 1B).
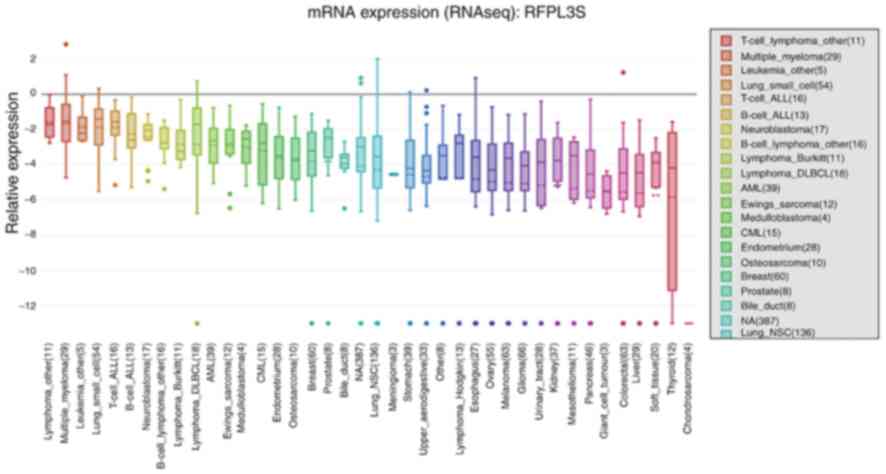
In addition, the mRNA expression of RFPL3S in
different cancer cell lines was obtained via CCLE analysis
(Fig. 2). In line with the results
of the Oncomine analysis, the expression levels of RFPL3S were
demonstrated to be upregulated in lung cancer cell lines compared
with normal cell lines, including SCLC and NSCLC cell lines, and
ranked the fourth highest in SCLC cell lines among the different
types of cancer. Notably, RFPL3S mRNA expression levels ranked the
third highest in leukemia cell lines, behind that of lymphoma and
multiple myeloma, and are also observed in breast and prostate
cancer, which was consistent with the results of the Oncomine
analysis. These results indicate that RFPL3S may play unique roles
in the development of several types of cancer.
Association between RFPL3S expression
and survival in patients with lung cancer
Since RFPL3S was revealed to be highly expressed in
lung cancer, the present study further investigated the potential
prognostic roles of RFPL3S in lung cancer. The association between
patient survival and RFPL3S expression status was analyzed using
KM-plotter, according to the expression of RFPL3S. A total of 1,926
patients with lung cancer were used for survival analysis.
According to the expression of RFPL3S, the samples were divided
into two groups (the low and high expression groups). Kaplan-Meier
analysis demonstrated that high RFPL3S expression was associated
with poorer overall survival (OS) compared with low RFPL3S
expression in all patients with lung cancer [hazard ratio
(HR)=1.38; P=5.1×10−7; Fig.
3A]. In particular, sub-analysis revealed that higher RFPL3S
expression was significantly associated with survival in
adenocarcinoma (HR=1.37; P=7.9 ×10−3 Fig. 3B), but not in squamous cell carcinoma
(HR=1.14; P=0.28; Fig. 3C), and
associated with stage 1 (HR=1.77; P=3.1×10−5 Fig. 3I), but not in stage 2 (HR=1.12;
P=0.54; Fig. 3J). High RFPL3S mRNA
expression was associated with poorer OS in patients with lung
cancer who smoked (HR=1.6; P=9.1×10−6; Fig. 3D), non-smokers (HR=2.32; P=0.0052;
Fig. 3E), male patients (HR=1.22;
P=0.013; Fig. 3H), female patients
(HR=1.71; P=6.7×10−6; Fig.
3G) and patients with tumor-negative surgical margins (HR=2.04;
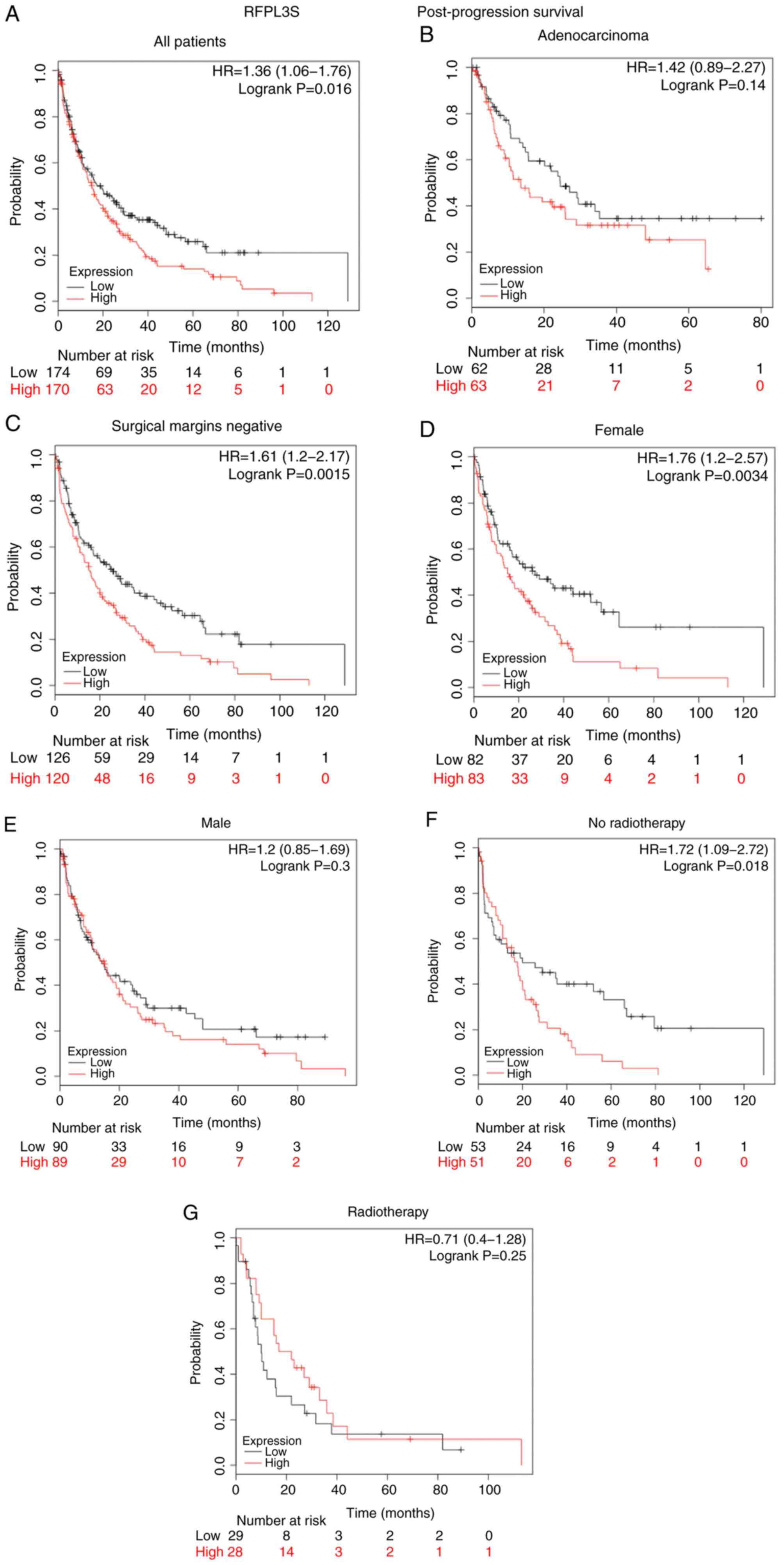
P=2.1×10−9; Fig. 3F).
Subsequently, post-progression survival (PPS) in patients with lung
cancer was analyzed (Fig. 4).
Notably, patients with increased RFPL3S expression exhibited a
shorter PPS than patients with lower RFPL3S expression (HR=1.36;
P=0.016; Fig. 4A), which was
consistent with OS. Furthermore, increased RFPL3S levels were
significantly associated with PPS in patients with tumor-negative
surgical margins (HR=1.61; P=0.0015; Fig. 4C), female patients (HR=1.76;
P=0.0034; Fig. 4D) and patients not
receiving radiotherapy (HR=1.72; P=0.018; Fig. 4F). However, no significant
differences were demonstrated among patients with adenocarcinoma
(HR=1.42; P=0.14; Fig. 4B), male
patients (HR=1.2; P=0.3; Fig. 4E)
and patients receiving radiotherapy (HR=0.71; P=0.25; Fig. 4G). The association between RFPL3S
expression levels and first progression (FP) was analyzed (Fig. 5). The results revealed that FP of all
patients with NSCLC, with increased RFPL3S expression was worse
than patients with lower RFPL3S expression (HR=1.85;
P=4.1×10−10; Fig. 5A),
which was in line with the results of OS and PPS. In addition, the
Kaplan-Meier analysis revealed that FP of patients with high RFPL3S
expression levels was significantly associated with squamous cell
carcinoma (HR=1.62 P=0.035; Fig.
5C), smoking (HR=2.22; P=1.91×10−10; Fig. 5D), not smoking (HR=1.5; P=0.097;
Fig. 5E), male gender (HR=1.73;
P=3.3×10−5; Fig. 5H),
female gender (HR=1.99; P=2.9×10−6; Fig. 5G), tumor-negative surgical margins
(HR=2.11; P=5.7×10−9; Fig.
5F), N0 (HR=1.76; P=5.7×10−4; Fig. 5I), N1 (HR=2.19;
P=8.4×10−4; Fig. 5J) and
T2 stage (HR=1.72; P=3.8×10−4; Fig. 5L). However, no statistically
significant differences were observed for adenocarcinoma (HR=1.23;
P=0.2; Fig. 5B) or T1 stage
(HR=1.46; P=0.14; Fig. 5K). Notably,
the results demonstrated that higher RFPL3S mRNA expression was
significantly associated with shorter survival, including OS, PPS
and FP in patients who had tumor-negative surgical margins.
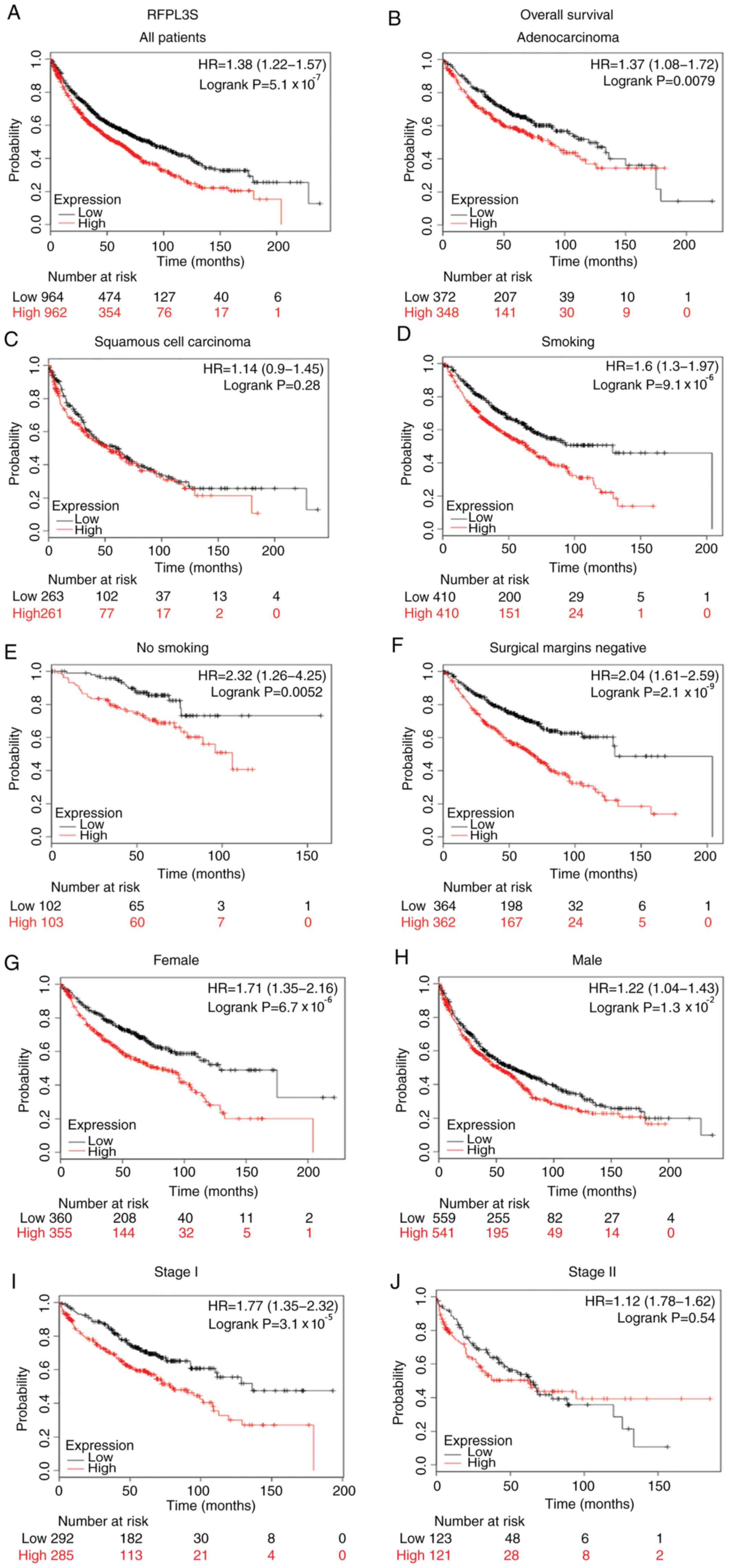 | Figure 3.Kaplan-Meier survival curves of
patients with lung cancer stratified by expression levels of
RPFL3S. Black and red lines indicate patients with low or high
RFPL3S expression, respectively. Overall survival analysis of
RFPL3S expression in lung cancer in (A) all patients, (B)
adenocarcinoma, (C) squamous cell carcinoma, (D) smokers, (E)
non-smokers, (F) tumor-negative surgical margins, (G) females, (H)
males, (I) stage I and (J) stage II. Statistical significance was
calculated using the log-rank test. RFPL3S, RFPL3 antisense. |
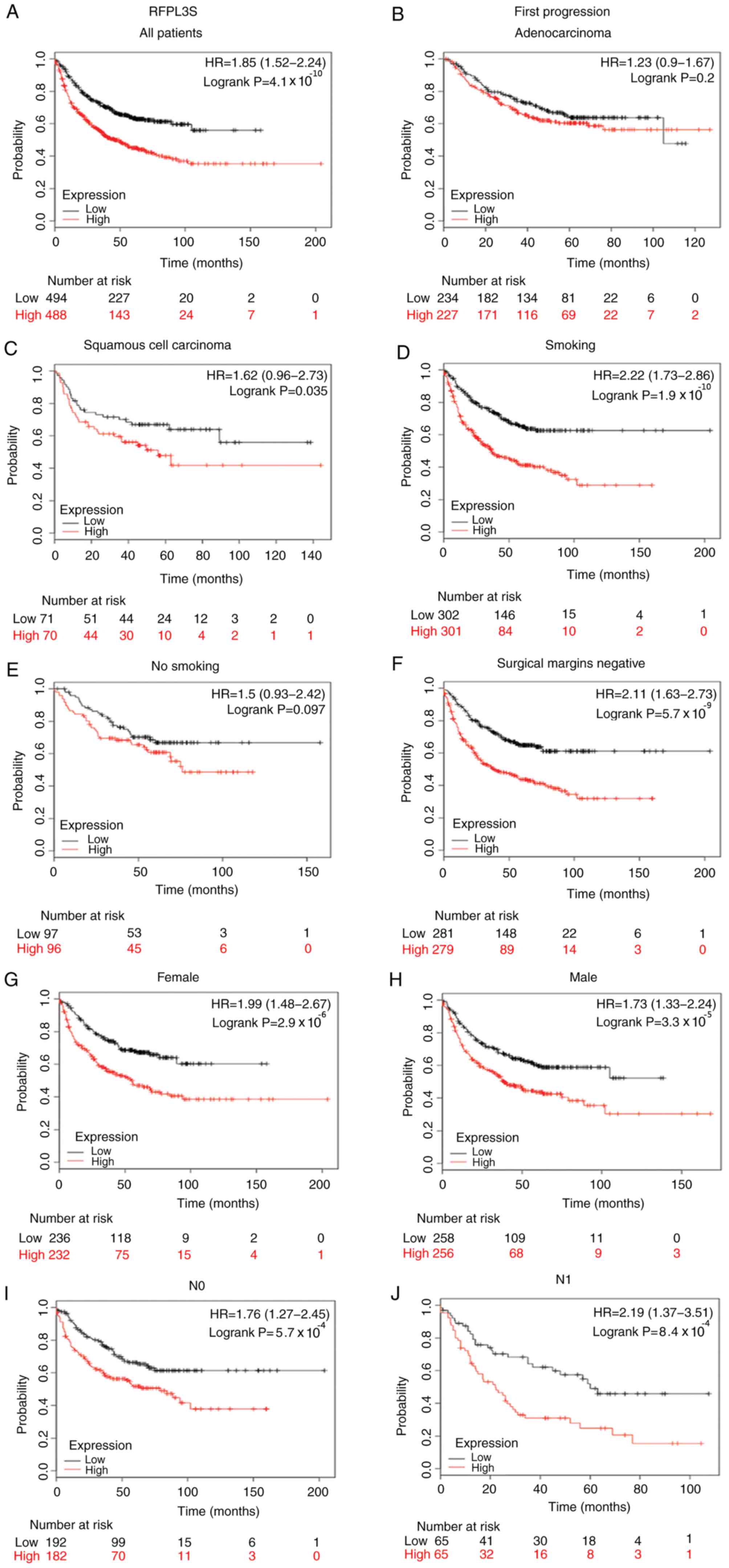 | Figure 5.Kaplan-Meier survival curves of
patients with lung cancer stratified by expression levels of
RPFL3S. Black and red lines indicate patients with low or high
RFPL3S expression, respectively. First-progression survival
analysis of RFPL3S in lung cancer in (A) all patients, (B)
adenocarcinoma, (C) squamous cell carcinoma, (D) smokers, (E)
non-smokers, (F) tumor-negative surgical margins, (G) females, (H)
males, (I) N0 and (J) N1. First-progression survival analysis of
RFPL3S in lung cancer in (K) T1 and (L) T2. Statistical
significance was calculated using the log-rank test. RFPL3S, RFPL3
antisense. |
Discussion
Lung cancer is a heterogeneous disease that consists
of a variety of subtypes with distinct biological and clinical
features, and presents the highest prevalence and mortality among
all malignancies, primarily due to the development of resistance to
targeted therapy and distant metastasis (38–41). It
is challenging but rewarding to illustrate the pathogenesis of lung
cancer, as well as to develop novel biomarkers and discover
effective therapeutic approaches for individual patients.
lncRNAs have been widely recognized as pivotal
regulators in a several types of human cancer, and their aberrant
expression has also been observed in different tumor tissues
(42–44). Furthermore, lncRNAs have been
demonstrated to serve as a novel class of diagnostic biomarkers and
therapeutic targets in cancer (45–47).
Several lncRNAs have been reported to play a critical role in the
development of different human tumors (18,48–52),
thereby providing a logical framework for understanding the
complexities of neoplastic disease (53) and regulated gene expression at
different levels. However, the role of lncRNA expression in lung
cancer remains unclear. To the best of our knowledge, the present
study was the first to investigate the association between RFPL3S
expression and the clinicopathological characteristics and
prognosis of patients with lung cancer. In the present study,
according to the results of the associations between RFPL3S
expression levels and clinicopathological characteristics, RFPL3S
expression levels may be associated with poorer differentiation,
lymph node metastasis, TNM stage and Ki-67 labeling index. A number
of previous studies have indicated that all of the above were
associated with a worse prognosis when compared with other features
(54–56), such as the patients with well
differentiation and absence of lymph node metastasis. Although
statistically significant differences were not observed for the
factors associated with the size of the primary tumor, a tendency
towards a poorer prognosis was observed in patients with increased
RFPL3S expression levels. Oncomine analysis demonstrated increased
RFPL3S expression levels in lung cancer compared with a variety of
other types of cancer; the results were consistent with data
obtained when comparing cancerous and normal tissues. Statistically
significant differences were observed in the survival analyses,
which demonstrated that increased RFPL3S expression was associated
with shorter OS in all patients with lung cancer. Patients with
increased RFPL3S expression levels had shorter survival times,
which is in line with the results regarding PPS and FP. Notably,
patients with a smoking history, which has been thought to be the
predominant cause of multiple types of cancer and increases the
incidence of lung cancer in the general population, demonstrated
the potential of RFPL3S in the poor prognosis of lung cancer
(57–61).
RFPL3S is the antisense transcript of the gene RFPL3
that is comprised of four exons and is formed by transcription in
the opposite direction to the sense RFPL3 transcript, depending on
the structure and position of the splicing sites for RFPL3S
(34). A previous study identified
1.2 kB non-coding antisense mRNAs of RFPL3S genes, which cover
substantial portions of their sense counterparts, suggesting that
RFPL3S is involved in the post-transcriptional regulation of the
sense RFPL genes at different spatial and temporal windows, and no
apparent Open Reading Frame or repetitive elements could be
detected (33). RFPL3, belonging to
the RFPL protein family (including RFPL1, RFPL2 and RFPL3)
(33), can increase telomerase
activity and length (62), and
promote the proliferation of tumor cells by regulating the
expression of the human telomerase reverse transcriptase (hTERT)
gene in NSCLC cells and driving hTERT promoter transcription
(63). Epidermal growth factor
significantly increased the levels of RFPL3 and hTERT proteins in
NSCLC cells via activation of the MEK signaling pathway, resulting
in promoting proliferation (63). In
addition, CREB binding protein has been identified to coordinate
with RFPL3 to regulate hTERT promoter activity through acetylation
to promote lung cancer cell growth (64). However, further research is required
to identify the molecular mechanisms that participate in RFPL3S
upregulation in lung cancer.
Overall, the present study demonstrated that RFPL3S
expression was upregulated and significantly associated with poor
prognosis in lung cancer. Therefore, it may act as a potential
prognostic biomarker in lung cancer. However, further studies are
required to determine the underlying molecular mechanisms that are
associated with RFPL3S expression in lung cancer.
Acknowledgements
Not applicable.
Funding
The present study was funded by National Natural
Science Foundation of China (grant nos. 81402176, 81402093 and
81472296), the Natural Science Foundation of Jiangsu Province,
China (grant no. BK20140288), the Science Technology Project of
Suzhou Xiangcheng District (grant nos. XJ201456 and XJ201532), the
Livelihood Science and Technology of Soochow (grant nos. SYS201752
and SS2018062), the Industry-University-Research Cooperation,
Prospective Joint Research Project of Jiangsu Province (grant no.
BY2015039-01), the Science Foundation of The First People's
Hospital of Wujiang District, Suzhou (grant nos. 201310 and 201806)
and the Project in Science and Education of Wujiang District (grant
no. wwk201808).
Availability of data and materials
The datasets generated and/or analyzed during the
present study are available from the corresponding author upon
reasonable request. The datasets generated and/or analyzed during
the current study are also available in the specified repository
and the web links are listed in the Materials and methods.
Authors' contributions
CC and XY designed the study and wrote the
manuscript. FZ collected specimens and patient information, and
revised the initial manuscript. ZL and ZQN performed the
experiments. THC and HL conducted the statistical analysis. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The First People's Hospital of Wujiang District.
Written informed consent was obtained from all the patients
recruited.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
lncRNA
|
long non-coding RNA
|
|
SCLC
|
small-cell lung cancer
|
|
RFPL3S
|
RFPL3 antisense
|
|
lncRNA
|
long non-coding RNA
|
|
OS
|
overall survival
|
|
PPS
|
post-progression survival
|
|
FP
|
first progression
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Salem A, Asselin MC, Reymen B, Jackson A,
Lambin P, West CML, O'Connor JPB and Faivre-Finn C: Targeting
hypoxia to improve non-small cell lung cancer outcome. J Natl
Cancer Inst. 110:2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lin DD, Shen Y, Qiao S, Liu WW, Zheng L,
Wang YN, Cui N, Wang YF, Zhao S and Shi JH: Upregulation of OTUD7B
(Cezanne) promotes tumor progression via AKT/VEGF pathway in lung
squamous carcinoma and adenocarcinoma. Front Oncol. 9:8622019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang J, Lu Q, Cai J, Wang Y, Lai X, Qiu Y,
Huang Y, Ke Q, Zhang Y, Guan Y, et al: Nestin regulates cellular
redox homeostasis in lung cancer through the Keap1-Nrf2 feedback
loop. Nat Commun. 10:50432019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang N, Guo W, Ren K, Li W, Jiang Y, Sun
J, Dai W and Zhao W: LncRNA AFAP1-AS1 supresses miR-139-5p and
promotes cell proliferation and chemotherapy resistance of
non-small cell lung cancer by competitively upregulating RRM2.
Front Oncol. 9:11032019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pfannschmidt J, Muley T, Bulzebruck H,
Hoffmann H and Dienemann H: Prognostic assessment after surgical
resection for non-small cell lung cancer: Experiences in 2083
patients. Lung Cancer. 55:371–377. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Reungwetwattana T, Weroha SJ and Molina
JR: Oncogenic pathways, molecularly targeted therapies, and
highlighted clinical trials in non-small-cell lung cancer (NSCLC).
Clin Lung Cancer. 13:252–266. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Karuppasamy R, Veerappapillai S, Maiti S,
Shin WH and Kihara D: Current progress and future perspectives of
polypharmacology: From the view of non-small cell lung cancer.
Semin Cancer Biol. Nov 4–2019.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hirashima T, Satouchi M, Hida T, Nishio M,
Kato T, Sakai H, Imamura F, Kiura K, Okamoto I, Kasahara K, et al:
Osimertinib for Japanese patients with T790M-positive advanced
non-small-cell lung cancer: A pooled subgroup analysis. Cancer Sci.
110:2884–2893. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Balekian AA, Wisnivesky JP and Gould MK:
Surgical disparities among patients with stage i lung cancer in the
national lung screening trial. Chest. 155:44–52. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Davis JN, Medbery C, Sharma S, Pablo J,
Kimsey F, Perry D, Muacevic A and Mahadevan A: Stereotactic body
radiotherapy for centrally located early-stage non-small cell lung
cancer or lung metastases from the RSSearch(®) patient
registry. Radiat Oncol. 10:1132015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
ENCODE Project Consortium: An integrated
encyclopedia of DNA elements in the human genome. Nature.
489:57–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shaughnessy AF: High false-positive rate
with lung cancer screening. Am Fam Physician. 96:128–129.
2017.PubMed/NCBI
|
|
15
|
Belanger AR and Akulian JA: An update on
the role of advanced diagnostic bronchoscopy in the evaluation and
staging of lung cancer. Ther Adv Respir Dis. 11:211–221. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jiang M, Huang O, Xie Z, Wu S, Zhang X,
Shen A, Liu H, Chen X, Wu J, Lou Y, et al: A novel long non-coding
RNA-ARA: Adriamycin resistance-associated. Biochem Pharmacol.
87:254–283. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yan B and Wang Z: Long noncoding RNA: Its
physiological and pathological roles. DNA Cell Biol. 31 (Suppl
1):S34–S41. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fang XY, Pan HF, Leng RX and Ye DQ: Long
noncoding RNAs: Novel insights into gastric cancer. Cancer Lett.
356:357–366. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kaikkonen MU, Lam MT and Glass CK:
Non-coding RNAs as regulators of gene expression and epigenetics.
Cardiovasc Res. 90:430–440. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li X, Wu Z, Fu X and Han W: Long noncoding
RNAs: Insights from biological features and functions to diseases.
Med Res Rev. 33:517–553. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guttman M and Rinn JL: Modular regulatory
principles of large non-coding RNAs. Nature. 482:339–346. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Martianov I, Ramadass A, Serra Barros A,
Chow N and Akoulitchev A: Repression of the human dihydrofolate
reductase gene by a non-coding interfering transcript. Nature.
445:666–670. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gong Z, Zhang S, Zhang W, Huang H, Li Q,
Deng H, Ma J, Zhou M, Xiang J, Wu M, et al: Long non-coding RNAs in
cancer. Sci China Life Sci. 55:1120–1124. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu W, Gius D, Onyango P, Muldoon-Jacobs K,
Karp J, Feinberg AP and Cui H: Epigenetic silencing of tumour
suppressor gene p15 by its antisense RNA. Nature. 451:202–206.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hung T and Chang HY: Long noncoding RNA in
genome regulation: Prospects and mechanisms. RNA Biol. 7:582–585.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Satish S, Sourav G, Vaibhav J, Vinod S and
Shantanu S: Genome-wide analysis reveals distinct patterns of
epigenetic features in long non-coding RNA loci. Nucleic Acids Res.
40:10018–10031. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Volders PJ, Helsens K, Wang X, Menten B,
Martens L, Gevaert K, Vandesompele J and Mestdagh P: LNCipedia: A
database for annotated human lncRNA transcript sequences and
structures. Nucleic Acids Res. 41((Database Issue)): D246–D251.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Niazi F and Valadkhan S: Computational
analysis of functional long noncoding RNAs reveals lack of
peptide-coding capacity and parallels with 3′ UTRs. RNA.
18:825–843. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Moran VA, Perera RJ and Khalil AM:
Emerging functional and mechanistic paradigms of mammalian long
non-coding RNAs. Nucleic Acids Res. 40:6391–6400. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Brunner AL, Beck AH, Edris B, Sweeney RT,
Zhu SX, Li R, Montgomery K, Varma S, Gilks T, Guo X, et al:
Transcriptional profiling of long non-coding RNAs and novel
transcribed regions across a diverse panel of archived human
cancers. Genome Biol. 13:R752012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Seroussi E, Kedra D, Pan HQ, Peyrard M,
Schwartz C, Scambler P, Donnai D, Roe BA and Dumanski JP:
Duplications on human chromosome 22 reveal a novel Ret Finger
Protein-like gene family with sense and endogenous antisense
transcripts. Genome Res. 9:803–814. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Travis WD, Brambilla E, Nicholson AG,
Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E,
Flieder DB, et al: The 2015 world health organization
classification of lung tumors: Impact of Genetic, Clinical and
radiologic advances since the 2004 classification. J Thorac Oncol.
10:1243–1260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Anna W and Jacek J: The new TNM
classification in lung cancer. Pneumonol Alergol Pol. 78:407–417.
2010.(In Polish). PubMed/NCBI
|
|
36
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lin HY, Zeng, Liang YK, Wei XL and Chen
CF: GATA3 and TRPS1 are distinct biomarkers and prognostic factors
in breast cancer: Database mining for GATA family members in
malignancies. Oncotarget. 8:34750–34761. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pao W, Miller VA, Politi KA, Riely GJ,
Somwar R, Zakowski MF, Kris MG and Varmus H: Acquired resistance of
lung adenocarcinomas to gefitinib or erlotinib is associated with a
second mutation in the EGFR kinase domain. PLoS Med. 2:e732005.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Engelman JA, Zejnullahu K, Mitsudomi T,
Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen
J, et al: MET amplification leads to gefitinib resistance in lung
cancer by activating ERBB3 signaling. Science. 316:1039–1043. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Choi YL, Soda M, Yamashita Y, Ueno T,
Takashima J, Nakajima T, Yatabe Y, Takeuchi K, Hamada T, Haruta H,
et al: EML4-ALK mutations in lung cancer that confer resistance to
ALK inhibitors. N Engl J Med. 363:1734–1739. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mittal V: Epithelial mesenchymal
transition in aggressive lung cancers. Adv Exp Med Biol. 890:37–56.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J
and Fang G: Up-regulated long non-coding RNA H19 contributes to
proliferation of gastric cancer cells. FEBS J. 279:3159–3165. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Qi D, Li J, Que B, Su J, Li M, Zhang C,
Yang M, Zhou G and Ji W: Long non-coding RNA DBCCR1-003 regulate
the expression of DBCCR1 via DNMT1 in bladder cancer. Cancer Cell
Int. 16:812016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Qiu MT, Hu JW, Yin R and Xu L: Long
noncoding RNA: An emerging paradigm of cancer research. Tumour
Biol. 34:613–620. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sun J, Song Y, Chen X, Zhao J, Gao P,
Huang X, Xu H and Wang Z: Novel long non-coding RNA RP11-119F7.4 as
a potential biomarker for the development and progression of
gastric cancer. Oncol Lett. 10:115–120. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tian Y, Zheng Y and Dong X: AGAP2-AS1
serves as an oncogenic lncRNA and prognostic biomarker in
glioblastoma multiforme. J Cell Biochem. 120:9056–9062. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kunej T, Obsteter J, Pogacar Z, Horvat S
and Calin GA: The decalog of long non-coding RNA involvement in
cancer diagnosis and monitoring. Crit Rev Clin Lab Sci. 51:344–357.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang S, Ma H, Zhang D, Xie S, Wang W, Li
Q, Lin Z and Wang Y: LncRNA KCNQ1OT1 regulates proliferation and
cisplatin resistance in tongue cancer via miR-211-5p mediated
Ezrin/Fak/Src signaling. Cell Death Dis. 9:7422018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lu Z, Xiao Z, Liu F, Cui M, Li W, Yang Z,
Li J, Ye L and Zhang X: Long non-coding RNA HULC promotes tumor
angiogenesis in liver cancer by up-regulating sphingosine kinase 1
(SPHK1). Oncotarget. 7:241–254. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liang H, Tong Y, Yue H, Jiang H, Wang C,
You T, Zhao X, Shan H, Yang R, Yang L, et al: LncRNA PTAR promotes
EMT and invasion-metastasis in serous ovarian cancer by
competitively binding miR-101-3p to regulate ZEB1 expression. Mol
Cancer. 17:1192018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Su M, Xiao Y, Ma J, Cao D, Zhou Y, Wang H,
Liao Q and Wang W: Long non-coding RNAs in esophageal cancer:
Molecular mechanisms, functions, and potential applications. J
Hematol Oncol. 11:1182018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Barletta JA, Yeap BY and Chirieac LR:
Prognostic significance of grading in lung adenocarcinoma. Cancer.
116:659–669. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tabata K, Tanaka T, Hayashi T, Hori T,
Nunomura S, Yonezawa S and Fukuoka J: Ki-67 is a strong prognostic
marker of non-small cell lung cancer when tissue heterogeneity is
considered. BMC Clin Pathol. 14:232014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Woodard GA, Jones KD and Jablons DM: Lung
cancer staging and prognosis. Cancer Treat Res. 170:47–75. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang H and Cai B: The impact of tobacco
on lung health in China. Respirology. 8:17–21. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Levine AJ, Momand J and Finlay CA: The p53
tumour suppressor gene. Nature. 351:453–456. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Fujino M, Dosaka-Akita H, Harada M,
Hiroumi H, Kinoshita I, Akie K and Kawakami Y: Prognostic
significance of p53 and ras p21 expression in nonsmall cell lung
cancer. Cancer. 76:2457–2463. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Pfeifer GP, Denissenko MF, Olivier M,
Tretyakova N, Hecht SS and Hainaut P: Tobacco smoke carcinogens,
DNA damage and p53 mutations in smoking-associated cancers.
Oncogene. 21:7435–7451. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
The Medical Research Council: Tobacco
smoking and lung cancer. Br Med J. 1:1523–1524. 1957. View Article : Google Scholar
|
|
62
|
Chen W, Lu J, Qin Y, Wang J, Tian Y, Shi
D, Wang S, Xiao Y, Dai M, Liu L, et al: Ret finger protein-like 3
promotes tumor cell growth by activating telomerase reverse
transcriptase expression in human lung cancer cells. Oncotarget.
5:11909–11923. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lin C, Qin Y, Zhang H, Gao MY and Wang YF:
EGF upregulates RFPL3 and hTERT via the MEK signaling pathway in
nonsmall cell lung cancer cells. Oncol Rep. 40:29–38.
2018.PubMed/NCBI
|
|
64
|
Yu Q, Wangbing C, Yao X, Yu W, Cai X, Dai
M, Xu T, Huang W, Guo W, Deng W and Wu T: RFPL3 and CBP
synergistically upregulate hTERT activity and promote lung cancer
growth. Oncotarget. 6:27130–27145. 2015. View Article : Google Scholar : PubMed/NCBI
|