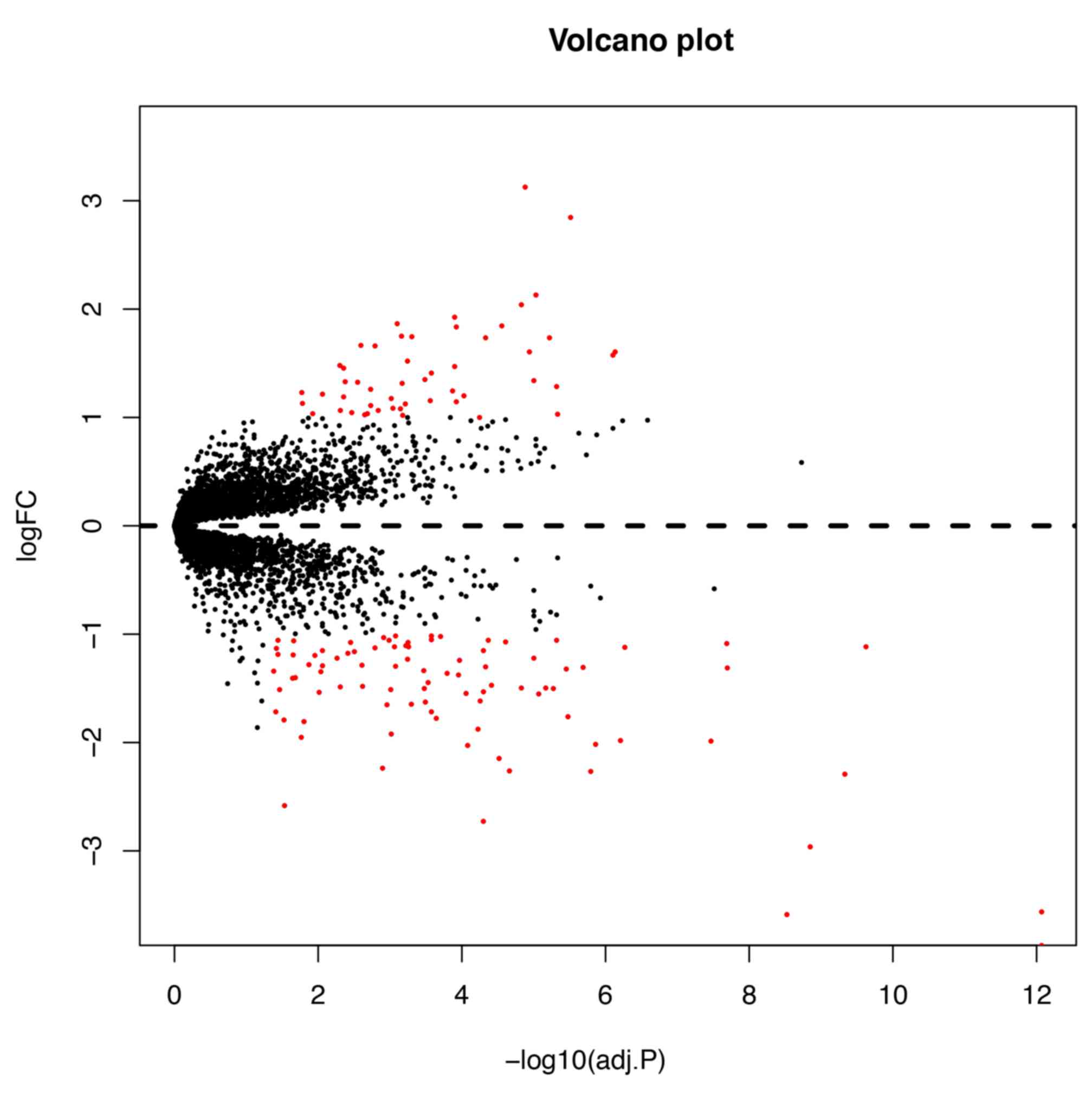
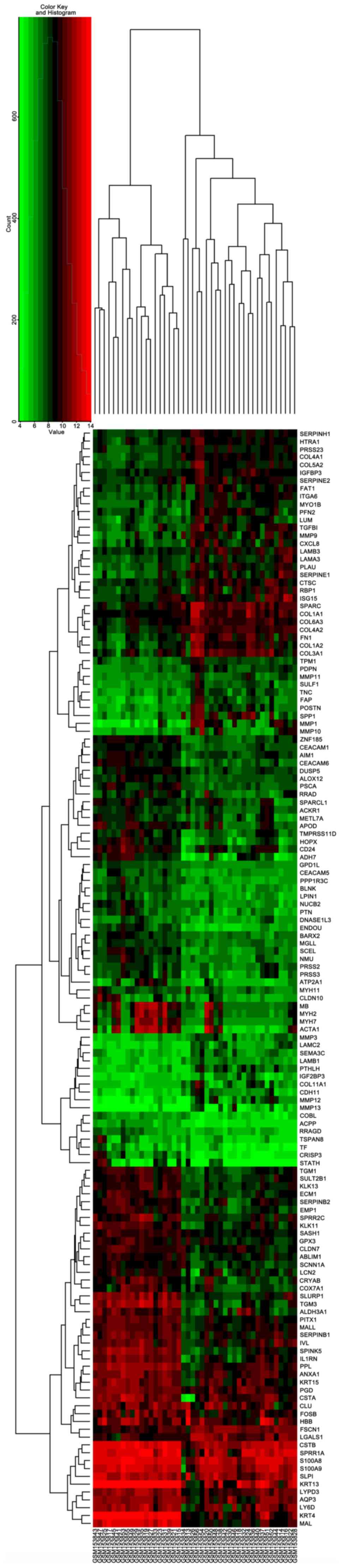
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yamazaki H, Suzuki G, Nakamura S, Hirano
S, Yoshida K, Konish K, Teshima T and Ogawa K: Radiotherapy for
locally advanced resectable T3-T4 laryngeal cancer-does laryngeal
preservation strategy compromise survival? J Radiat Res. 59:77–90.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Marioni G, Marchese-Ragona R, Cartei G,
Marchese F and Staffieri A: Current opinion in diagnosis and
treatment of laryngeal carcinoma. Cancer Treat Rev. 32:504–515.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yilmaz T, Suslu N, Atay G, Gunaydin RO,
Bajin MD and Ozer S: The effect of midline crossing of lateral
supraglottic cancer on contralateral cervical lymph node
metastasis. Acta Otolaryngol. 135:484–488. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Elegbede AI, Rybicki LA, Adelstein DJ,
Kaltenbach JA, Lorenz RR, Scharpf J and Burkey BB: Oncologic and
functional outcomes of surgical and nonsurgical treatment of
advanced squamous cell carcinoma of the supraglottic larynx. JAMA
Otolaryngol Head Neck Surg. 141:1111–1117. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Koirala K: Epidemiological study of
laryngeal carcinoma in Western Nepal. Asian Pac J Cancer Prev.
16:6541–6544. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fan CY: Genetic alterations in head and
neck cancer: Interactions among environmental carcinogens, cell
cycle control, and host DNA repair. Curr Oncol Rep. 3:66–71. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Conrad DF, Pinto D, Redon R, Feuk L,
Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, et
al: Origins and functional impact of copy number variation in the
human genome. Nature. 464:704–712. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu X, Moon JW, Li S, Xu W, Wang X, Liu Y
and Lee JY: Amplification and overexpression of CTTN and CCND1 at
chromosome 11q13 in Esophagus squamous cell carcinoma (ESCC) of
North Eastern Chinese population. Int J Med Sci. 13:868–874. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Beroukhim R, Mermel CH, Porter D, Wei G,
Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J,
Urashima M, et al: The landscape of somatic copy-number alteration
across human cancers. Nature. 463:899–905. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Edge SB and Compton CC: The American joint
committee on cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kuriakose MA, Chen WT, He ZM, Sikora AG,
Zhang P, Zhang ZY, Qiu WL, Hsu DF, McMunn-Coffran C, Brown SM, et
al: Selection and validation of differentially expressed genes in
head and neck cancer. Cell Mol Life Sci. 61:1372–1383. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wickham H: ggplot2: Elegant graphics for
data analysis. Springer-Verlag; New York: ISBN 978-3-319-24277-4.
2016
|
|
15
|
Karatzanis AD, Psychogios G, Waldfahrer F,
Kapsreiter M, Zenk J, Velegrakis GA and Iro H: Management of
locally advanced laryngeal cancer. J Otolaryngol Head Neck Surg.
43:42014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Myllykangas S, Tikka J, Bohling T,
Knuutila S and Hollmen J: Classification of human cancers based on
DNA copy number amplification modeling. BMC Med Genomics. 1:152008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vissers LE, de Vries BB, Osoegawa K,
Janssen IM, Feuth T, Choy CO, Straatman H, van der Vliet W, Huys
EH, van Rijk A, et al: Array-based comparative genomic
hybridization for the genomewide detection of submicroscopic
chromosomal abnormalities. Am J Hum Genet. 73:1261–1270. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shaw-Smith C, Redon R, Rickman L, Rio M,
Willatt L, Fiegler H, Firth H, Sanlaville D, Winter R, Colleaux L,
et al: Microarray based comparative genomic hybridisation
(array-CGH) detects submicroscopic chromosomal deletions and
duplications in patients with learning disability/mental
retardation and dysmorphic features. J Med Genet. 41:241–248. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Riazimand SH, Welkoborsky HJ, Bernauer HS,
Jacob R and Mann WJ: Investigations for fine mapping of
amplifications in chromosome 3q26.3–28 frequently occurring in
squamous cell carcinomas of the head and neck. Oncology.
63:385–392. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Singh B, Stoffel A, Gogineni S, Poluri A,
Pfister DG, Shaha AR, Pathak A, Bosl G, Cordon-Cardo C, Shah JP and
Rao PH: Amplification of the 3q26.3 locus is associated with
progression to invasive cancer and is a negative prognostic factor
in head and neck squamous cell carcinomas. Am J Pathol.
161:365–371. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ashman JN, Patmore HS, Condon LT, Cawkwell
L, Stafford ND and Greenman J: Prognostic value of genomic
alterations in head and neck squamous cell carcinoma detected by
comparative genomic hybridisation. Br J Cancer. 89:864–869. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang Q, Yu GP, McCormick SA, Mo J, Datta
B, Mahimkar M, Lazarus P, Schaffer AA, Desper R and Schantz SP:
Genetic differences detected by comparative genomic hybridization
in head and neck squamous cell carcinomas from different tumor
sites: Construction of oncogenetic trees for tumor progression.
Genes Chromosomes Cancer. 34:224–233. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jarmuz-Szymczak M, Pelinska K,
Kostrzewska-Poczekaj M, Bembnista E, Giefing M, Brauze D,
Szaumkessel M, Marszalek A, Janiszewska J, Kiwerska K, et al:
Heterogeneity of 11q13 region rearrangements in laryngeal squamous
cell carcinoma analyzed by microarray platforms and fluorescence in
situ hybridization. Mol Biol Rep. 40:4161–4171. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Patmore HS, Ashman JN, Stafford ND,
Berrieman HK, MacDonald A, Greenman J and Cawkwell L: Genetic
analysis of head and neck squamous cell carcinoma using comparative
genomic hybridisation identifies specific aberrations associated
with laryngeal origin. Cancer Lett. 258:55–62. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fields AP, Justilien V and Murray NR: The
chromosome 3q26 OncCassette: A multigenic driver of human cancer.
Adv Biol Regul. 60:47–63. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ambrosio EP, Silveira CG, Drigo SA,
Sacomano Vde S, Molck MC, Rocha RM, Domingues MA, Soares FA,
Kowalski LP and Rogatto SR: Chromosomal imbalances exclusively
detected in invasive front area are associated with poor outcome in
laryngeal carcinomas from different anatomical sites. Tumour Biol.
34:3015–3026. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jarvinen AK, Autio R, Haapa-Paananen S,
Wolf M, Saarela M, Grenman R, Leivo I, Kallioniemi O, Makitie AA
and Monni O: Identification of target genes in laryngeal squamous
cell carcinoma by high-resolution copy number and gene expression
microarray analyses. Oncogene. 25:6997–7008. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Singh B, Gogineni SK, Sacks PG, Shaha AR,
Shah JP, Stoffel A and Rao PH: Molecular cytogenetic
characterization of head and neck squamous cell carcinoma and
refinement of 3q amplification. Cancer Res. 61:4506–4513.
2001.PubMed/NCBI
|
|
29
|
Gen Y, Yasui K, Zen Y, Zen K, Dohi O, Endo
M, Tsuji K, Wakabayashi N, Itoh Y, Naito Y, et al: SOX2 identified
as a target gene for the amplification at 3q26 that is frequently
detected in esophageal squamous cell carcinoma. Cancer Genet
Cytogenet. 202:82–93. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hussenet T, Dali S, Exinger J, Monga B,
Jost B, Dembele D, Martinet N, Thibault C, Huelsken J, Brambilla E
and du Manoir S: SOX2 is an oncogene activated by recurrent 3q26.3
amplifications in human lung squamous cell carcinomas. PLoS One.
5:e89602010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Weina K and Utikal J: SOX2 and cancer:
Current research and its implications in the clinic. Clin Transl
Med. 3:192014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Badura M, Braunstein S, Zavadil J and
Schneider RJ: DNA damage and eIF4G1 in breast cancer cells
reprogram translation for survival and DNA repair mRNAs. Proc Natl
Acad Sci USA. 109:18767–18772. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bauer C, Brass N, Diesinger I, Kayser K,
Grasser FA and Meese E: Overexpression of the eukaryotic
translation initiation factor 4G (eIF4G-1) in squamous cell lung
carcinoma. Int J Cancer. 98:181–185. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jin X, Zhai B, Fang T, Guo X and Xu L:
FXR1 is elevated in colorectal cancer and acts as an oncogene.
Tumour Biol. 37:2683–2690. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen XQ, Jiang J, Wang XT, Zhang CL, Ji AY
and Chen XJ: Role and mechanism of Dvl3 in the esophageal squamous
cell carcinoma. Eur Rev Med Pharmacol Sci. 22:7716–7725.
2018.PubMed/NCBI
|
|
36
|
Shuang Y, Li C, Zhou X, Huang Y and Zhang
L: MicroRNA-195 inhibits growth and invasion of laryngeal carcinoma
cells by directly targeting DCUN1D1. Oncol Rep. 38:2155–2165. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ye S, Song W, Xu X, Zhao X and Yang L:
IGF2BP2 promotes colorectal cancer cell proliferation and survival
through interfering with RAF-1 degradation by miR-195. FEBS Lett.
590:1641–1650. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu W, Li Z, Xu W, Wang Q and Yang S:
Humoral autoimmune response to IGF2 mRNA-binding protein (IMP2/p62)
and its tissue-specific expression in colon cancer. Scand J
Immunol. 77:255–260. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Barghash A, Golob-Schwarzl N, Helms V,
Haybaeck J and Kessler SM: Elevated expression of the IGF2 mRNA
binding protein 2 (IGF2BP2/IMP2) is linked to short survival and
metastasis in esophageal adenocarcinoma. Oncotarget. 7:49743–49750.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu W, Li Y, Wang B, Dai L, Qian W and
Zhang JY: Autoimmune response to IGF2 mRNA-Binding Protein 2
(IMP2/p62) in breast cancer. Scand J Immunol. 81:502–507. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Manor E, Tetro S and Bodner L:
Translocation (12;14) and other chromosome abnormalities in
squamous cell carcinoma of the tongue. Eur Arch Otorhinolaryngol.
267:1273–1276. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tai AL, Mak W, Ng PK, Chua DT, Ng MY, Fu
L, Chu KK, Fang Y, Qiang Song Y, Chen M, et al: High-throughput
loss-of-heterozygosity study of chromosome 3p in lung cancer using
single-nucleotide polymorphism markers. Cancer Res. 66:4133–4138.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jiang D, Li X, Wang H, Shi Y, Xu C, Lu S,
Huang J, Xu Y, Zeng H, Su J, et al: The prognostic value of EGFR
overexpression and amplification in Esophageal squamous cell
carcinoma. BMC Cancer. 15:3772015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hu N, Wang C, Su H, Li WJ, Emmert-Buck MR,
Li G, Roth MJ, Tang ZZ, Lu N, Giffen C, et al: High frequency of
CDKN2A alterations in esophageal squamous cell carcinoma from a
high-risk Chinese population. Genes Chromosomes Cancer. 39:205–216.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Willem P, Brown J and Schouten J: A novel
approach to simultaneously scan genes at fragile sites. BMC Cancer.
6:2052006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Escudero-Esparza A, Bartoschek M, Gialeli
C, Okroj M, Owen S, Jirstrom K, Orimo A, Jiang WG, Pietras K and
Blom AM: Complement inhibitor CSMD1 acts as tumor suppressor in
human breast cancer. Oncotarget. 7:76920–76933. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jung AR, Eun YG, Lee YC, Noh JK and Kwon
KH: Clinical significance of CUB and Sushi multiple domains 1
inactivation in head and neck squamous cell carcinoma. Int J Mol
Sci. 19(pii): E39962018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang R and Song C: Loss of CSMD1 or 2 may
contribute to the poor prognosis of colorectal cancer patients.
Tumour Biol. 35:4419–4423. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lv J, Zhu P, Yang Z, Li M, Zhang X, Cheng
J, Chen X and Lu F: PCDH20 functions as a tumour-suppressor gene
through antagonizing the Wnt/β-catenin signalling pathway in
hepatocellular carcinoma. J Viral Hepat. 22:201–211. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Imoto I, Izumi H, Yokoi S, Hosoda H,
Shibata T, Hosoda F, Ohki M, Hirohashi S and Inazaw J: Frequent
silencing of the candidate tumor suppressor PCDH20 by epigenetic
mechanism in non-small-cell lung cancers. Cancer Res. 66:4617–4626.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang Q, Lv Q, Bian H, Yang L, Guo KL, Ye
SS, Dong XF and Tao LL: A novel tumor suppressor SPINK5 targets
Wnt/β-catenin signaling pathway in esophageal cancer. Cancer Med.
8:2360–2371. 2019. View Article : Google Scholar : PubMed/NCBI
|