Introduction
Colorectal cancer (CRC), which is also known as
bowel and colon cancer, originates from the colon or rectum
(1–3). According to the analysis of the
prevalence of malignant tumors in China in 2015, released by the
National Cancer Center in 2019, CRC ranks the third most common
cancer among the top 10 malignant tumors (4,5). The
number of patients with CRC and their mortality has reached a high
level worldwide in the past years (3). Although advances in surgery, radiation
therapy and chemotherapy have been recently made for the treatment
of CRC, due to the fast-growing nature and high rate of metastasis
of this cancer, the 5-year survival rate of CRC is still low, at
only ~14% (6–8). Recently, targeted therapy has been
revealed as a promising prospect for CRC treatment (9). Although several therapeutic targets of
CRC have been developed to combat this malignant tumor, more
effective therapeutic targets are urgently required (10).
Long non-coding RNAs (lncRNA) are defined as
transcripts that are not translated into proteins (11). LncRNAs have multiple cellular and
physiological functions, such as combating oxidative stress
(12,13). A large number of studies have
indicated that lncRNAs are associated with diverse human diseases,
such as myelodysplastic syndromes, Alzheimer's disease and fatty
liver disease (14–16). In addition, the lncRNA Neat1 may
contribute to the activation of inflammasomes in macrophages
(17).
Notably, the potential effects of lncRNAs on
multiple types of cancer have been revealed in recent decades
(18). Additionally, studies have
recently shown that lncRNAs may be abnormally expressed in tumor
tissues (19). Furthermore, lncRNAs
are involved in the progression of bladder cancer, breast cancer
and glioma (18,20). LncRNAs were also demonstrated to
affect the proliferation, apoptosis, migration and invasion of
tumor cells through different mechanisms (18). The lncRNA ABHD11-AS1 facilitates
thyroid carcinoma progression via the PI3K/AKT pathway (21). Additionally, the lncRNA GAS6-AS1
promotes gastric cancer progression by targeting GAS6 (22). However, novel lncRNAs that can
regulate the development of CRC still need to be identified.
Using a microarray analysis, the present study aimed
to identify upregulated genes in human CRC tissues, which may be
involved in the progression of CRC. Additionally, the present study
aimed to reveal the signaling pathways and the core lncRNAs with
potential to affect CRC progression. Through a series of in
vitro assays, a novel and potential oncogene involved in CRC
progression was identified.
Materials and methods
Samples
A total of 36 CRCs and the corresponding adjacent (2
cm away from the tumor border) surgical specimens in the present
study were collected between November 2018 and May 2019, with
complete clinicopathological data collected at The Second Hospital
of Shandong University (Jinan, China). In the present study,
patients had only received surgical treatment, and patients who had
received chemotherapy and/or targeted therapy were excluded. The
patients ranged in age between 35 and 65 years (mean age, 52
years), with 20 males and 16 females. All studies were approved by
the Ethics Committee of The Second Hospital of Shandong University,
and informed written consent was obtained from all patients.
Sequences of small interfering RNAs
(siRNAs) and quantitative PCR primers
The siRNA sequence targeting SCARNA9L was:
5′-CGGUCUACCUGAUGCAUGAUCUCUA-3′. The quantitative PCR primer
sequences were as follows: SCARNA9L forward,
5′-ATAAAGGTAGCAGTTGTAGGAATG-3′; SCARNA9L reverse,
5′-CTTCATAGTTACAAAGGTCAGTCG-3′; GAPDH forward,
5′-CGACCACTTTGTCAAGCTCA-3′; and GAPDH reverse,
5′-GGTTGAGCACAGGGTACTTTATT-3′.
Bioinformatics analysis
CRC gene expression microarrays were searched using
the search term ‘Colorectal cancer’ in the gene expression omnibus
(GEO) database (https://www.ncbi.nlm.nih.gov/geo/). GSE73360 and
GSE84984 datasets were used to screen the differentially expressed
lncRNAs in CRC (23–27). The process included: Comparison
analysis by limma R package (28)
(version:3.36.5; http://bioinf.wehi.edu.au/limma); Gene Ontology (GO;
http://geneontology.org/); analysis through
statistical method, by two side Fisher's exact test (29); pathway analysis through two side
Fisher's exact test (30)
(https://www.genome.jp/kegg/);
coexpression network analysis using R function (Cytoscape; version,
3.6.0; http://cytoscape.org/) (31); and global signal transduction network
(32) (version, 3.6.0; https://cytoscape.org/; http://www.genome.jp/kegg/).
Scatter diagrams were plotted. Additionally, the
limma package v3.36.5 (http://master.bioconductor.org/packages/release/bioc/html/limma.html)
was used to screen differentially expressed genes (DEGs), with
P<0.05 and log fold-change (FC) >2 set as the threshold. The
heatmap of differentially expressed lncRNAs was drawn using the
Heatmap Package v3 (https://CRAN.R-project.org/package=heatmap3). Pathway
enrichment analysis and coexpression analysis were performed,
according to the GSE73360 and GSE84984 datasets, with a false
discovery rate (FDR) <0.05 considered significant. Data on the
lncRNA expression were obtained from the Oncomine database
(www.oncomine.org). The expression data are
available from the GEO database (www.ncbi.nlm.nih.gov/geo) (23–27).
Cell culture and transfection
SW480 and SW620 human CRC CRC cells were obtained
from The Cell Bank of Type Culture Collection of the Chinese
Academy of Sciences. SW480 and SW620 cells were maintained in DMEM
culture medium (Gibco; Thermo Fisher Scientific, Inc.), and
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) in a 5% CO2 incubator at 37°C.
Both SW480 and SW620 CRC CRC cells were transfected
with control (scrambled siRNA) or the aforementioned siRNA
targeting SCARNA9L (50 nM; Guangzhou RiboBio Co., Ltd.) using
Lipofectamine™ 3000 (Thermo Fisher Scientific, Inc.), according to
the manufacturer's instructions. The in vitro assays were
performed 48 h after transfection.
Reverse transcription-quantitative
(RT-q)PCR assay
TRIzol reagent (cat. no. 9108; Takara Bio, Inc.) was
used to extract total RNA from both SW480 and SW620 cells. Total
RNA was subsequently reverse transcribed at 37°C for 1 h using
M-MLV reverse transcriptase (cat. no. M1701; Promega Corporation)
and using a cDNA synthesis system (cat. no. RR037A; Takara Bio,
Inc.), including oligo DT, Random 6-mers (100 µM), primers, 5X
PrimeScript buffer, PrimeScript RT Enzyme Mix I and DEPC water.
qPCR was performed using SYBR Premix Ex Taq™ (cat. no. RR820A;
Takara Bio, Inc.) with the following thermocycling conditions: 94°C
for 3 min, followed by 35 cycles of 94°C for 40 sec, 55°C for 1 min
and 72°C for 1 min, and finally 72°C for 10 min. The relative
expression level of the indicated RNAs was normalized to the
expression of GAPDH using the 2−ΔΔCq method (33).
Colony formation assays
Approximately 1,000 NC-transfected cells or human
CRC cells treated with the indicated siRNA treatment were seeded
into 6-well culture plates and incubated with 5% CO2 at
37°C for 7 days. The medium was replaced with fresh complete DMEM
(with 10% FBS) every two days. After 7 days, the cells were washed
with PBS twice, fixed with paraformaldehyde for 30 min at room
temperature, and stained with 1% crystal violet at room temperature
for 15 min, washed with PBS and observed using the Olympus CX31
light microscope (magnification, ×10). Subsequently, the number of
colonies was manually counted.
Cell cycle assays
The cell cycle distribution was detected by using a
cell cycle kit (Beijing 4A Biotech Co., Ltd.). In brief,
NC-transfected or indicated siRNA-treated CRC cells were fixed with
95% ethanol at −20°C for 24 h and incubated with 0.4 ml of
propidium iodide (200 µg/ml) for 30 min at room temperature prior
to analysis by flow cytometry.
Wound-closure assays
Both SW480 and SW620 CRC CRC cells were transfected
with siRNAs and grown to confluent monolayers. Subsequently, a
scratch was generated using a 10-µl pipette tip. Cell debris was
washed twice using PBS, and serum-free medium was added to induce
wound healing. Images of the wounds were captured at 0 and 24 h
using the Olympus CX31 light microscope (magnification, ×10), and
the percentage of wound closure was measured. The width of the
wound was indicated using lines at the edge of the wound, and the
closure percentage was measured as the healing width divided by the
original wound width.
Transwell assays
SW480 and SW620 cells were transfected with siRNAs
for 48 h and then trypsinized and resuspended in serum-free DMEM
(Gibco; Thermo Fisher Scientific, Inc.). A total of
1×105 cells in 100 µl DMEM without FBS were then added
to the upper chambers of the Transwell inserts (Millicell; Merck
KGaA) and allowed to migrate toward the bottom of the chambers,
which contained DMEM with 10% FBS. After 24 h, the remaining cells
in the upper chamber were removed, and cells on the underside were
fixed in 4% paraformaldehyde for 30 min at room temperature,
stained with 0.1% crystal violet for 30 min at room temperature and
captured using an Olympus type light microscope sz30.
Quantification of the migrated cells was performed by counting cell
numbers.
Statistical analysis
GraphPad Prism 6.0 software (GraphPad Software,
Inc.) was used for statistical analysis. All data in this study are
presented as the mean ± standard deviation (SD). Statistical
significance was calculated using a one-way analysis of variance
following by the Fisher's Least Significant Difference post hoc
test. Paired t-test was used to compare the expression of lncRNAs
in tumor tissues and normal tissues. Additionally, the expression
of SLMO2-ATP5E and LOC100132062, and the effects of siRNAs on CRC
cell proliferation and migration were analyzed using unpaired
t-tests. P<0.05 was used to indicate a statistically significant
difference.
Results
Identification of novel differentially
expressed lncRNAs in CRC via microarray analysis
Microarray analysis was used to screen
CRC-associated DEGs and predict the lncRNAs involved in CRC
progression. Analysis of the CRC gene expression datasets GSE73360
and GSE84984 was performed. The microarray data of all lncRNAs in
CRC tissues were extracted from the GEO dataset. R language was
used to screen for differentially expressed lncRNAs from the CRC
gene expression datasets, and the scatter diagrams of all lncRNAs
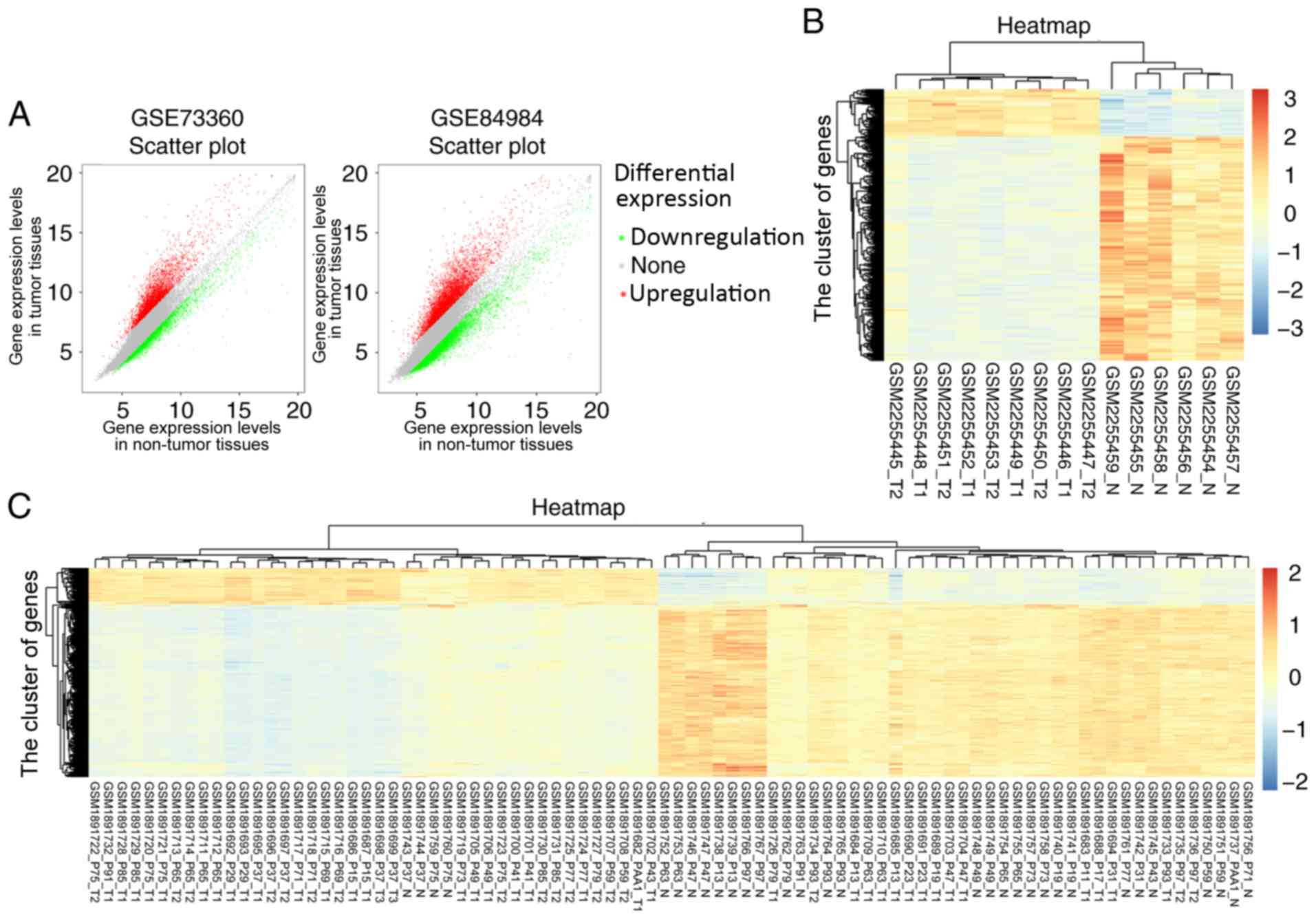
from the GSE73360 and GSE84984 datasets are shown in Fig. 1A. In total, 2233 differentially
expressed lncRNAs were screened from GSE73360, and 6749 DEGs were
screened from GSE84984, based on P<0.05 and FC >2.
Furthermore, the heatmaps of differentially
expressed lncRNAs that were screened from the GSE73360 and GSE84984
datasets are shown in Fig. 1B and C,
respectively. According to the results of the heatmaps, SCARNA9L,
SLMO2-ATP5E, and LOC100132062 were highly expressed in CRC, in both
the GSE73360 and GSE84984 datasets. To the best of our knowledge,
there are no studies on the effects of these lncRNAs on CRC;
therefore, the differential expression of SCARNA9L, SLMO2-ATP5E,
and LOC100132062 and their possible regulatory mechanisms in CRC
progression were investigated in the present study.
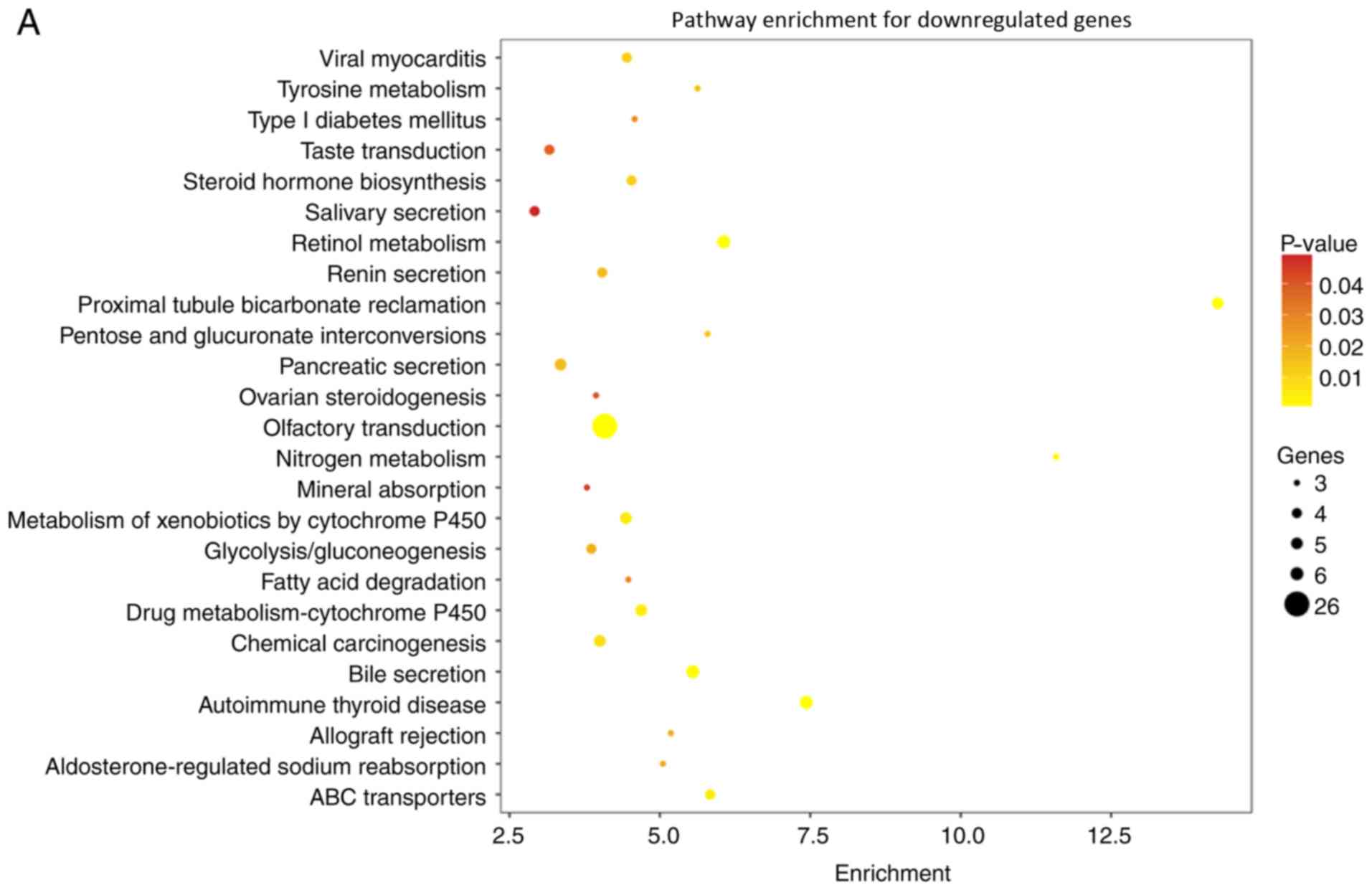
Interestingly, according to the KEGG pathway
analysis of GSE73360 and GSE84984, several pathways were notably
affected (including those upregulated and downregulated) in CRC
tissues (Fig. 2A and B). Several
genes in the indicated pathways, such as tyrosine metabolism and
fatty acid degradation pathways, were downregulated, suggesting
effects on cell metabolism and CRC progression (Fig. 2A). Additionally, in the analysis of
enrichment pathways for upregulated genes in the GSE73360 and
GSE84984 datasets, the cell cycle, adherent junctions, and mismatch
repair pathways, which have the potential to direct affect CRC
progression, were notably upregulated (Fig. 2B).
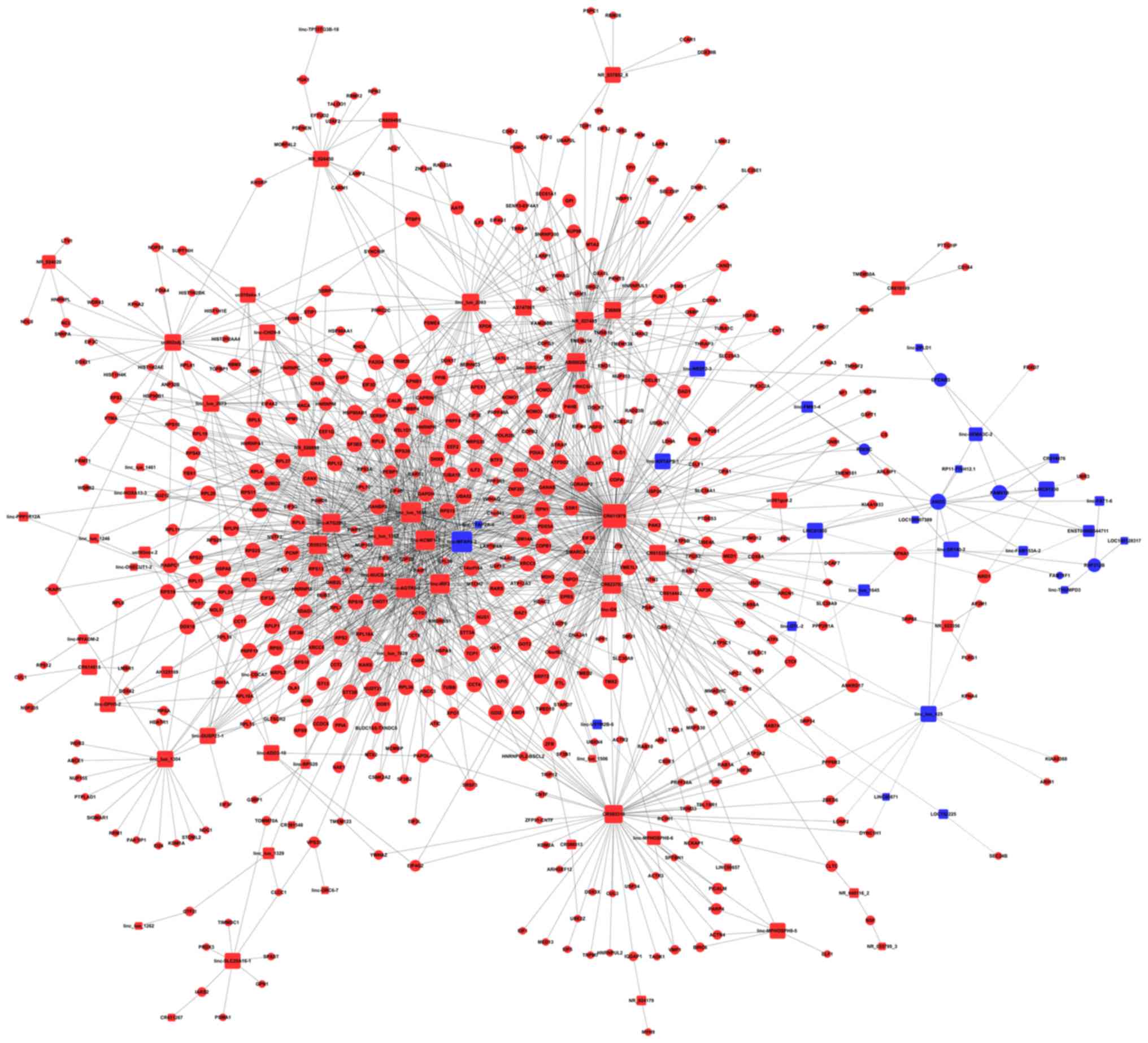
Subsequently, further coexpression network analysis
on GSE73360 and GSE84984 datasets were performed to identify the
key lncRNAs involved in the regulation of CRC progression (Fig. 3). Interestingly, the data revealed
that multiple core lncRNAs affected the expression of key proteins
involved in the regulation of cancer progression, such as
lnc-MFAP4-2, lnc-TFAP-2A-4, and lnc-KRTAP9-1 (Fig. 3), and the precise mechanisms need
further study. Furthermore, three obviously upregulated lncRNAs,
SCARNA9L, SLMO2-ATP5E, and LOC100132062, were identified from the
coexpression network analysis (Fig.
3).
Taken together, three lncRNAs, SCARNA9L, SLMO2-
ATP5E, and LOC100132062, were notably upregulated and have the
potential to be involved in CRC progression.
Knockdown of SCARNA9L inhibits the
proliferation of SW480 and SW620 cells
The potential involvement of the three identified
lncRNAs, SCARNA9L, SLMO2-ATP5E and LOC100132062, in the progression
of CRC was further explored.
siRNAs targeting the indicated lncRNAs were designed
and transfected into two human CRC cell lines, SW480 and SW620
cells, to inhibit its expression. Cell proliferation is critical in
cancer progression. Therefore, colony-formation assays were
performed to investigate whether these three lncRNAs affected the
proliferation of CRC cells. As was confirmed by RT-qPCR assays, the
expression levels of SLMO2-ATP5E and LOC100132062 in human CRC
tissues were notably higher compared with normal tissues (Fig. S1). The transfection of siRNAs
targeting SLMO2-ATP5E and LOC100132062 decreased the expression of
these lncRNAs, as confirmed by RT-qPCR assays (Fig. S2). However, the depletion of
SLMO2-ATP5E and LOC100132062 had no obvious effects on CRC cell
proliferation (Fig. S3), as shown
by the colony-forming assays.
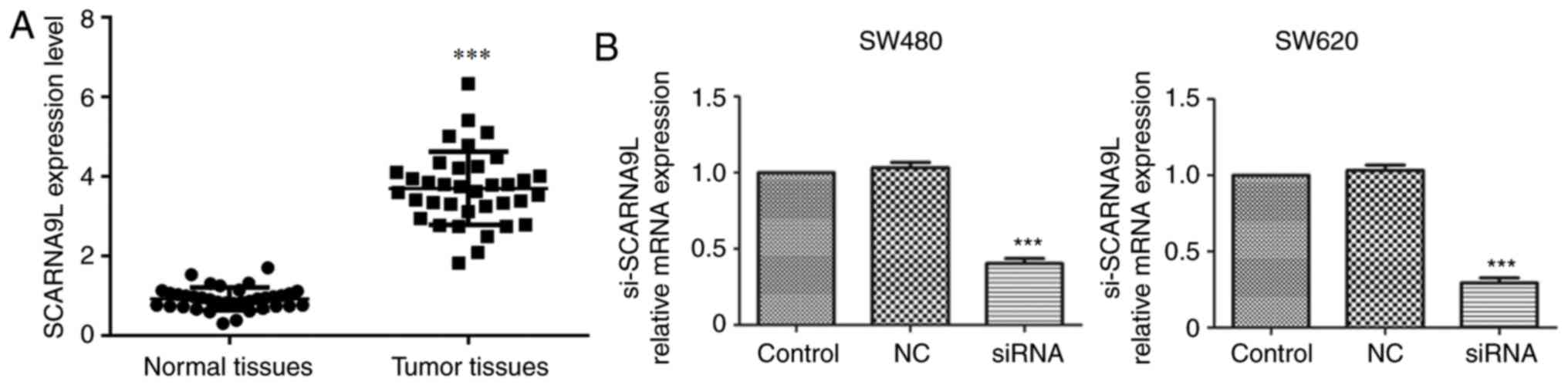
Subsequently, the effects of SCARNA9L on CRC
progression were studied. Firstly, high mRNA expression levels of
SCARNA9L were found in 36 human CRC tissues collected at The Second
Hospital of Shandong University (Fig.
4A). Also, through RT-qPCR assays, it was found that the
transfection of siRNAs targeting SCARNA9L effectively decreased the
expression levels of the indicated lncRNAs in both SW480 and SW620
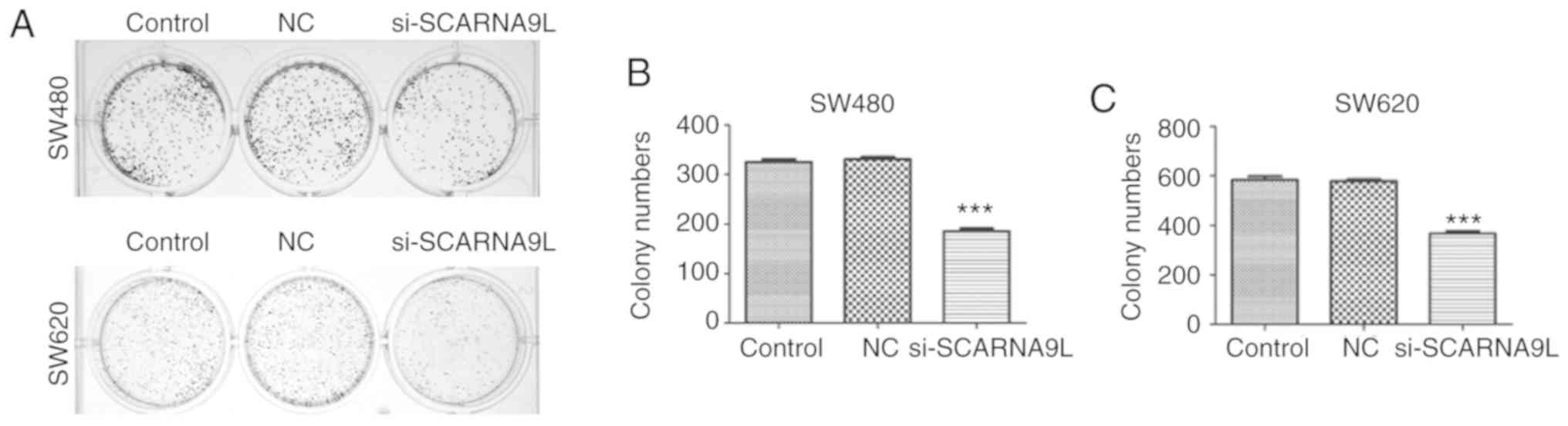
cells compared with the control group or the NC group (Fig. 4B). Interestingly, the depletion of
SCARNA9L dramatically suppressed the proliferation of SW480 and
SW620 cells, with a notable decrease in the number of colonies
(Fig. 5A and B). The aforementioned
results suggest that SCARNA9L could contribute to the proliferation
of CRC CRC cells.
Depletion of SCARNA9L induces cell
cycle arrest in CRC cells
The control of cell cycle is essential to maintain
the proliferation of cells, and disruption of the cell cycle will
result in abnormal proliferation and further promote tumorigenesis.
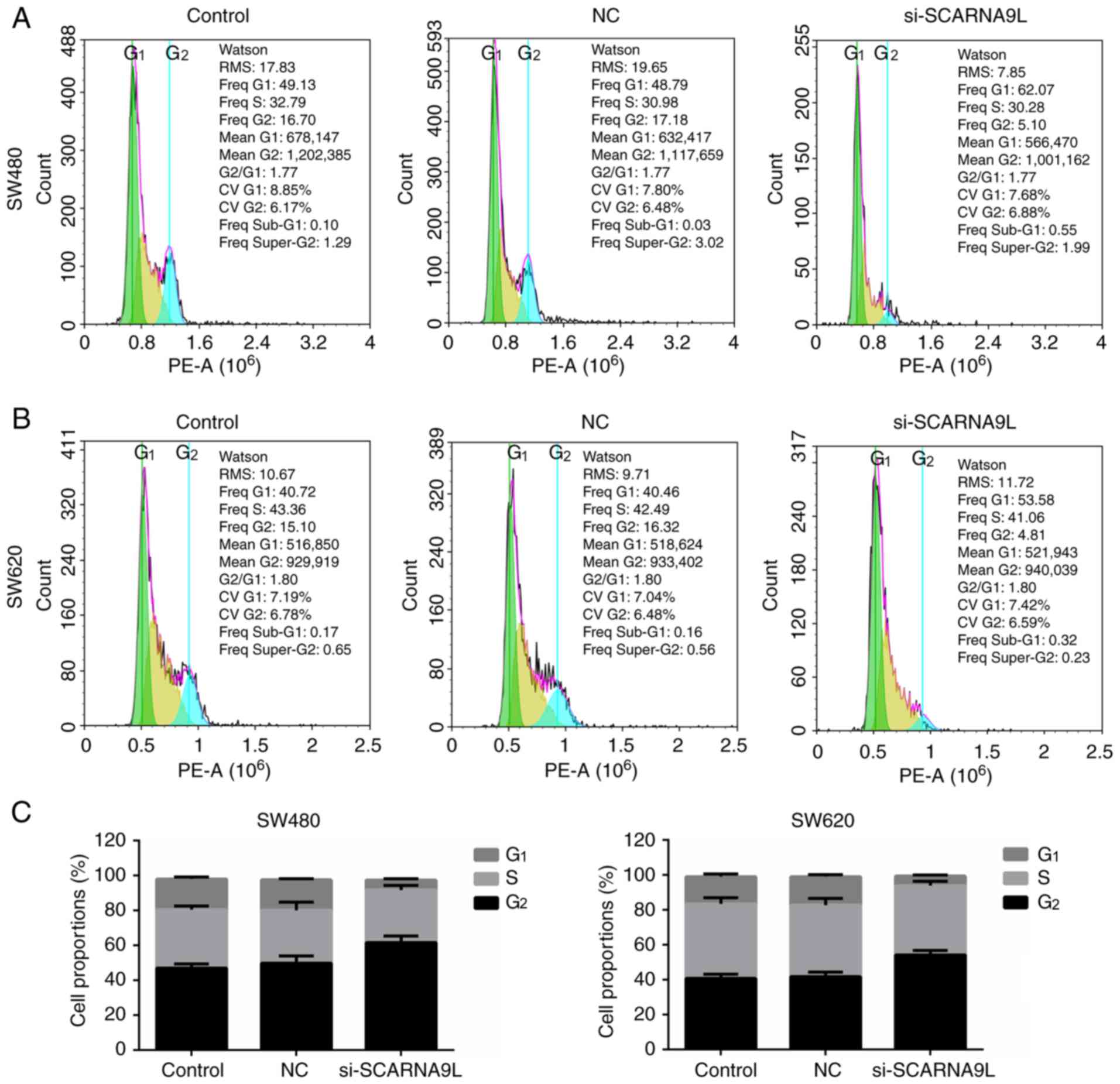
Therefore, differences in the cell cycle distribution between cells
with SCARNA9L knockdown and the control groups were detected.
Notably, the data indicated that the ablation of SCARNA9L
dramatically resulted in an increased percentage of cells at the
G1 phase and a decreased proportion of cells at the
G2/M phases (Fig. 6);
suggesting the arrest of cell cycle at G1 phase. Thus,
the ablation of SCARNA9L led to a significant cell cycle arrest and
may further block CRC CRC cell proliferation. However, the
depletion of SLMO2-ATP5E and LOC100132062 had no obvious effects on
cell cycle (data not shown).
SCARNA9L contributes to CRC cell
migration and invasion in vitro
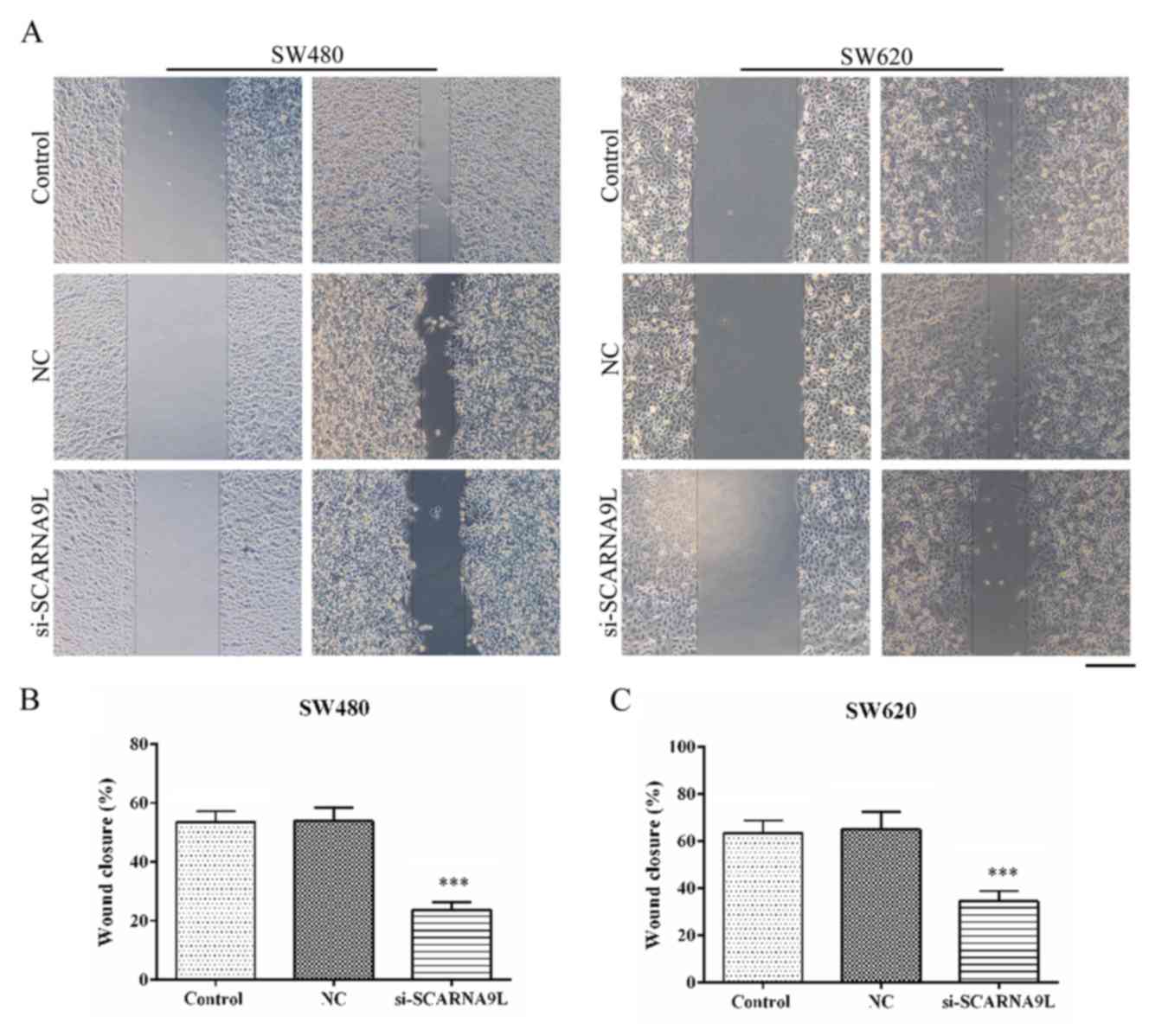
Wound-healing and transwell assays were performed to
evaluate the effects of SCARNA9L on the migration and invasion of
SW480 and SW620 cells. Interestingly, the depletion of SCARNA9L
significantly suppressed the extent of wound closure in both SW480
and SW620 cells (Fig. 7).
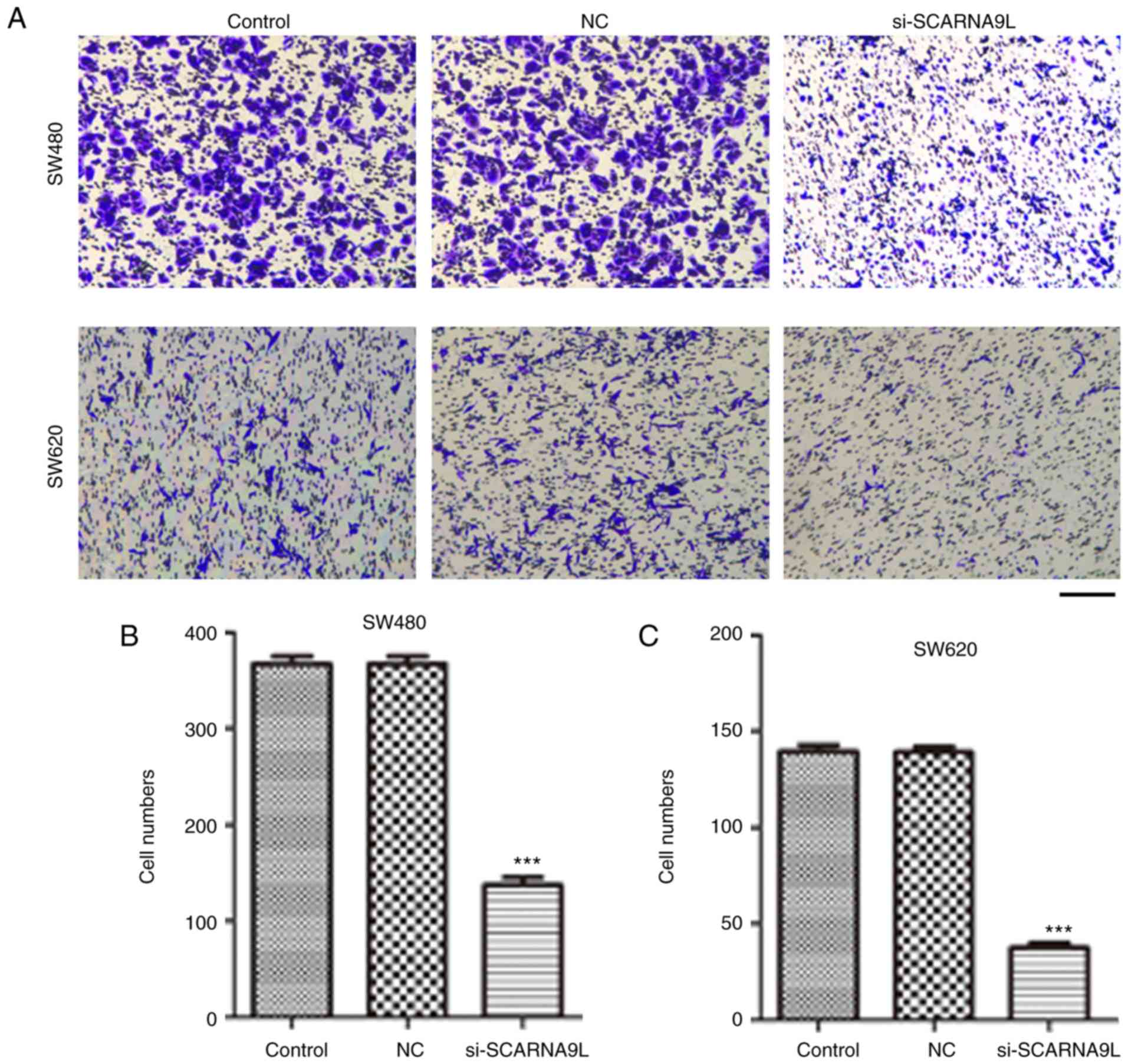
Additionally, according to the transwell assays, depletion of
SCARNA9L significantly inhibited the migration of SW480 and SW620
cells through membranes (Fig. 8). On
the other hand, SLMO2-ATP5E and LOC100132062 had no effects on CRC
cell migration (data not shown).
Overall, the lncRNA SCARNA9L contributes to cell
migration and invasion of CRC in vitro.
Discussion
CRC is a common malignancy of the digestive tract,
and its morbidity and mortality rates have notably increased
(34). The occurrence of CRC without
any obvious early symptoms also makes diagnosis difficult (35). Advanced CRC is highly metastatic and
grows rapidly, leading to a poor prognosis (7). Effective biomarkers and therapeutic
targets for CRC are urgently required (10). Notably, studies have indicated the
widespread involvement of lncRNAs in cancer progression (19). Several lncRNAs are abnormally
expressed in tumor tissues and correlate with the clinical features
of patients (36). Additionally,
these lncRNAs are usually involved in the regulation of cancer cell
proliferation, migration or apoptosis (19,37,38). The
present study identified three novel lncRNAs, SCARNA9L, SLMO2-ATP5E
and LOC100132062, which were highly expressed in CRC tissues
according to the microarray analysis. Furthermore, one of these
lncRNAs, SCARNA9L, was found to affect CRC cell proliferation,
migration and invasion. However, no significant effects were found
in SLMO2-ATP5E, and LOC100132062 groups on cell cycle and migration
(data not shown). These data indicate SCARNA9L as a novel and
potential biomarker for CRC.
In the present study, SCARNA9L was demonstrated to
be involved in CRC progression. Colony-formation assays showed
impaired proliferation capacity, following the depletion of
SCARNA9L. Flow cytometry assays confirmed that SCARNA9L affected
the cell cycle of CRC cells in vitro. Furthermore,
wound-closure and transwell assays found that SCARNA9L was also
involved in the regulation of CRC cell migration. These are the
first findings regarding the function of SCARNA9L in CRC
progression. Combined with the present study, abnormal expression
of SCARNA9L in CRC tissues was confirmed through microarray
analysis and in vitro cell experiments, and various
functions of CRC cells were affected. Thus, the present study
suggests that SCARNA9L may serve as a potential biomarker, although
further validation is required through mechanistic and animal
studies, and clinical trials.
In addition to SCARNA9L, various lncRNAs have been
identified to be abnormally expressed in CRC and affect its
occurrence and development (19).
The lncRNA LEF1-AS1 suppresses the progression of oral squamous
cell carcinoma through the Hippo signaling pathway (39). Another study indicated that the
lncRNA RP11 induces the dissemination of CRC cells via promoting
the expression of ZEB1 (23).
Similarly, the lncRNA MALAT1 also contributes to CRC progression
via sponging miR-363-3p to mediate the expression of EZH2 (40). These studies, together with the
findings of the present study, confirm the widespread involvement
of lncRNAs in CRC development. However, the present study only
found three lncRNAs that affected CRC proliferation, migration and
invasion; the targets of these lncRNAs were not identified. Given
that this study is the first to find that SCARNA9L can affect CRC
progression, and there is no other evidence that these lncRNAs play
key roles in the development of other tumors, the target genes of
SCARNA9L and the mechanisms that affect tumor cell proliferation,
migration and invasion should be further studied.
In the present study, SCARNA9L was identified to be
involved in CRC progression through microarray analysis.
Additionally, multiple signaling pathways and core lncRNAs that had
the potential to affect CRC progression, and were obviously
altered, were identified from the KEGG pathway analysis and
coexpression network analysis. The ablation of SCARNA9L
significantly restrained cell proliferation and induced cell cycle
arrest in SW480 and SW620 cells. However, no obvious effects of
SLMO2-ATP5E and LOC100132062 on CRC cell proliferation were
identified in the present study. Furthermore, the present study
identified SCARNA9L to significantly affect CRC cell migration.
Thus, SCARNA9L is suggested as a novel and potential therapeutic
target for the treatment of CRC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by The China Postdoctoral
Science Foundation (grant no. 2017M611177), The Shandong Province
Natural Science Foundation (grant nos. ZR2017MH129 and
ZR2013HL040), The China Natural Science Foundation (grant no.
81602578), and The Shandong Province Key Research and Development
Plan (grant no. GG201710070085).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article. The datasets generated and/or
analyzed during the current study are available in the Gene
Expression Omnibus (GEO) repository (https://www.ncbi.nlm.nih.gov/geo/).
Authors' contributions
Conception and design, JC and WM. Development of
methodology, JC, JZ, DH, WD, LH, LZ, BF, BL and WM. Acquisition of
data (provided animals, acquired and managed patients, provided
facilities), JC, JZ, DH, WD and BL. Analysis and interpretation of
data (statistical analysis, biostatistics, computational analysis),
HL and ZB. Writing, review, and/or revision of the manuscript, JC,
BL and WM. Administrative, technical, or material support
(reporting or organizing data, constructing databases), JC and JZ.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the ethics committee at
The Second Hospital of Shandong University (approval no.
201707003). Informed written consent was obtained from all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
lncRNA
|
long non-coding RNAs
|
|
GEO
|
Gene Expression Omnibus
|
|
DEGs
|
differentially expressed genes
|
|
NC
|
negative control
|
References
|
1
|
Stromberg U, Peterson S, Holmén A,
Holmberg E, Hultcrantz R, Martling A and Nilbert M: Rational
targeting of population groups and residential areas for colorectal
cancer screening. Cancer Epidemiol. 60:23–30. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jung C, Kim RS, Zhang H, Lee SJ, Sheng H,
Loehrer PJ, Gardner TA, Jeng MH and Kao C: HOXB13 is downregulated
in colorectal cancer to confer TCF4-mediated transactivation. Br J
Cancer. 92:2233–2239. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hosokawa A, Yamada Y, Shimada Y, Muro K,
Hamaguchi T, Morita H, Araake M, Orita H and Shirao K: Prognostic
significance of thymidylate synthase in patients with metastatic
colorectal cancer who receive protracted venous infusions of
5-fluorouracil. Int J ClinOncol. 9:388–392. 2004.
|
|
4
|
Tian Y, Xu B, Yu G, Li Y and Liu H:
Comorbidity and the risk of anastomotic leak in Chinese patients
with colorectal cancer undergoing colorectal surgery. Int J
Colorectal Dis. 32:947–953. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guo X, Zhang C, Ma W, Tian F, Xu G, Han X,
Sun P, Baklaushev VP, Bryukhovetskiy AS, Wang G, et al: Patterns of
bone metastases in newly diagnosed colorectal cancer: A real-world
analysis in the SEER database. Int J Colorectal Dis. 34:533–543.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ahmed S, Johnson K, Ahmed O and Iqbal N:
Advances in the management of colorectal cancer: From biology to
treatment. Int J Colorectal Dis. 29:1031–1042. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mele V, Sokol L, Kolzer VH, Pfaff D,
Muraro MG, Keller I, Stefan Z, Centeno I, Terracciano LM, Dawson H,
et al: The hyaluronan-mediated motility receptor RHAMM promotes
growth, invasiveness and dissemination of colorectal cancer.
Oncotarget. 8:70617–70629. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang KM, Park IJ, Lee JL, Kim CW, Yoon YS,
Lim SB, Yu CS and Kim JC: Benefits of repeated resections for liver
and lung metastases from colorectal cancer. Asian J Surg.
43:102–109. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Price TJ, Tang M, Gibbs P, Haller DG,
Peeters M, Arnold D, Segelov E, Roy A, Tebbutt N, Pavlakis N, et
al: Targeted therapy for metastatic colorectal cancer. Expert Rev
Anticancer Ther. 18:991–1006. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yamamoto H and Mori M: MicroRNAs as
therapeutic targets and colorectal cancer therapeutics. Adv Exp Med
Biol. 937:239–247. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun L, Zhang Y, Zhang Y, Gu Y, Xuan L, Liu
S, Zhao X, Wang N, Huang L, Huang Y, et al: Expression profile of
long non-coding RNAs in a mouse model of cardiac hypertrophy. Int J
Cardiol. 177:73–75. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hanly DJ, Esteller M and Berdasco M:
Interplay between long non-coding RNAs and epigenetic machinery:
Emerging targets in cancer? Philos Trans R Soc Lond B Biol Sci.
373:201700742018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dai L, Li J, Dong Z, Liu Y, Chen Y, Chen
N, Cheng L, Fang C, Wang H, Ji Y, et al: Temporal expression and
functional analysis of long non-coding RNAs in colorectal cancer
initiation. J Cell Mol Med. 23:4127–4138. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao X, Yin H, Li N, Zhu Y, Shen W, Qian
S, He G, Li J and Wang X: An integrated regulatory network based on
comprehensive analysis of mRNA expression, gene methylation and
expression of long non-coding RNAs (lncRNAs) in myelodysplastic
syndromes. Front Oncol. 9:2002019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Z: Long non-coding RNAs in
Alzheimer's disease. Curr Top Med Chem. 16:511–519. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hanson A, Wilhelmsen D and DiStefano JK:
The role of long non-coding RNAs (lncRNAs) in the development and
progression of fibrosis associated with nonalcoholic fatty liver
disease (NAFLD). Noncoding RNA. 4:E182018.PubMed/NCBI
|
|
17
|
Zhang P, Cao L, Zhou R, Yang X and Wu M:
The lncRNA Neat1 promotes activation of inflammasomes in
macrophages. Nat Commun. 10:14952019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chandra Gupta S and Nandan Tripathi Y:
Potential of long non-coding RNAs in cancer patients: From
biomarkers to therapeutic targets. Int J Cancer. 140:1955–1967.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xie X, Tang B, Xiao YF, Xie R, Li BS, Dong
H, Zhou JY and Yang SM: Long non-coding RNAs in colorectal cancer.
Oncotarget. 7:5226–5239. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou Y, Gong B, Jiang ZL, Zhong S, Liu XC,
Dong K, Wu HS, Yang HJ and Zhu SK: Microarray expression profile
analysis of long non-coding RNAs in pancreatic ductal
adenocarcinoma. Int J Oncol. 48:670–680. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wen J, Wang H, Dong T, Gan P, Fang H, Wu
S, Li J, Zhang Y, Du R and Zhu Q: STAT3-induced upregulation of
lncRNA ABHD11-AS1 promotes tumour progression in papillary thyroid
carcinoma by regulating miR-1301-3p/STAT3 axis and PI3K/AKT
signalling pathway. Cell Prolif. 52:e125692019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang P, Dong Q, Zhu H, Li S, Shi L and
Chen X: Long non-coding antisense RNA GAS6-AS1 supports gastric
cancer progression via increasing GAS6 expression. Gene. 696:1–9.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu Y, Yang X, Chen Z, Tian L, Jiang G,
Chen F, Li J, An P, Lu L, Luo N, et al: m6A-induced
lncRNA RP11 triggers the dissemination of colorectal cancer cells
via upregulation of Zeb1. Mol Cancer. 18:872019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Barresi V, Trovato-Salinaro A, Spampinato
G, Musso N, Castorina S, Rizzarelli E and Condorelli DF:
Transcriptome analysis of copper homeostasis genes reveals
coordinated upregulation of SLC31A1,SCO1, and COX11 in colorectal
cancer. FEBS Open Bio. 6:794–806. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Barresi V, Cinnirella G, Valenti G,
Spampinato G, Musso N, Castorina S and Condorelli DF: Gene
expression profiles in genome instability-based classes of
colorectal cancer. BMC Cancer. 18:12652018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Condorelli DF, Spampinato G, Valenti G,
Musso N, Castorina S and Barresi V: Positive caricature
transcriptomic effects associated with broad genomic aberrations in
colorectal cancer. Sci Rep. 8:148262018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang Y, Zhao Z, Xie CW and Zhao Y:
Dual-targeting liposome modified by glutamic hexapeptide and folic
acid for bone metastatic breast cancer. Chem Phys Lipids.
228:1048822020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Loennstedt I and Speed P: Replicated
microarray data. Statistica Sinica. 12:31–46. 2001.
|
|
29
|
Bauer S, Gagneur J and Robinson PN: GOing
Bayesian: Model-based gene set analysis of genome-scale data.
Nucleic Acids Res. 38:3523–3532. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Benjamini Y and Yekutieli D: The control
of the false discovery rate in multiple testing under dependency.
Annals of Statistics. 29:1165–1188. 2001.
|
|
31
|
Best D and Roberts E: Algorithm AS 89: The
upper tail probabilities of Spearman's rho. Applied Statistics.
24:377–379. 1975. View Article : Google Scholar
|
|
32
|
Kanehisa M, Araki M, Goto S, Hattori M,
Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T
and Yamanishi Y: KEGG for linking genomes to life and the
environment. Nucleic Acids Res. 36:D480–D484. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mateo-Lozano S, Bazzocco S, Rodrigues P,
Mazzolini R, Andretta E, Dopeso H, Fernández Y, Del Llano E, Bilic
J, Suárez-López L, et al: Loss of the EPH receptor B6 contributes
to colorectal cancer metastasis. Sci Rep. 7:437022017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cerdán-Santacruz C, Cano-Valderrama O,
Cárdenas-Crespo S, Torres-García AJ and Cerdán-Miguel J: Colorectal
cancer and its delayed diagnosis: Have we improved in the past 25
years? Rev Esp Enferm Dig. 103:458–463. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bai J, Yao B, Wang L, Sun L, Chen T, Liu
R, Yin G, Xu Q and Yang W: lncRNA A1BG-AS1 suppresses proliferation
and invasion of hepatocellular carcinoma cells by targeting
miR-216a-5p. J Cell Biochem. 120:10310–10322. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rossi MN and Antonangeli F: LncRNAs: New
players in apoptosis control. Int J Cell Biol. 2014:4738572014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gioia R, Drouin S, Ouimet M, Caron M,
St-Onge P, Richer C and Sinnett D: Lncrnasdownregulated in
childhood acute lymphoblastic leukemia modulate apoptosis, cell
migration, and DNA damage response. Oncotarget. 8:80645–80650.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jin Q, Dai Y, Wang Y, Zhang S and Liu G:
High kinesin family member 11 expression predicts poor prognosis in
patients with clear cell renal cell carcinoma. J ClinPathol.
72:354–362. 2019.
|
|
40
|
Xie JJ, Li WH, Li X, Ye W and Shao CF:
LncRNA MALAT1 promotes colorectal cancer development by sponging
miR-363-3p to regulate EZH2 expression. J Biol Regul Homeost
Agents. 33:331–343. 2019.PubMed/NCBI
|