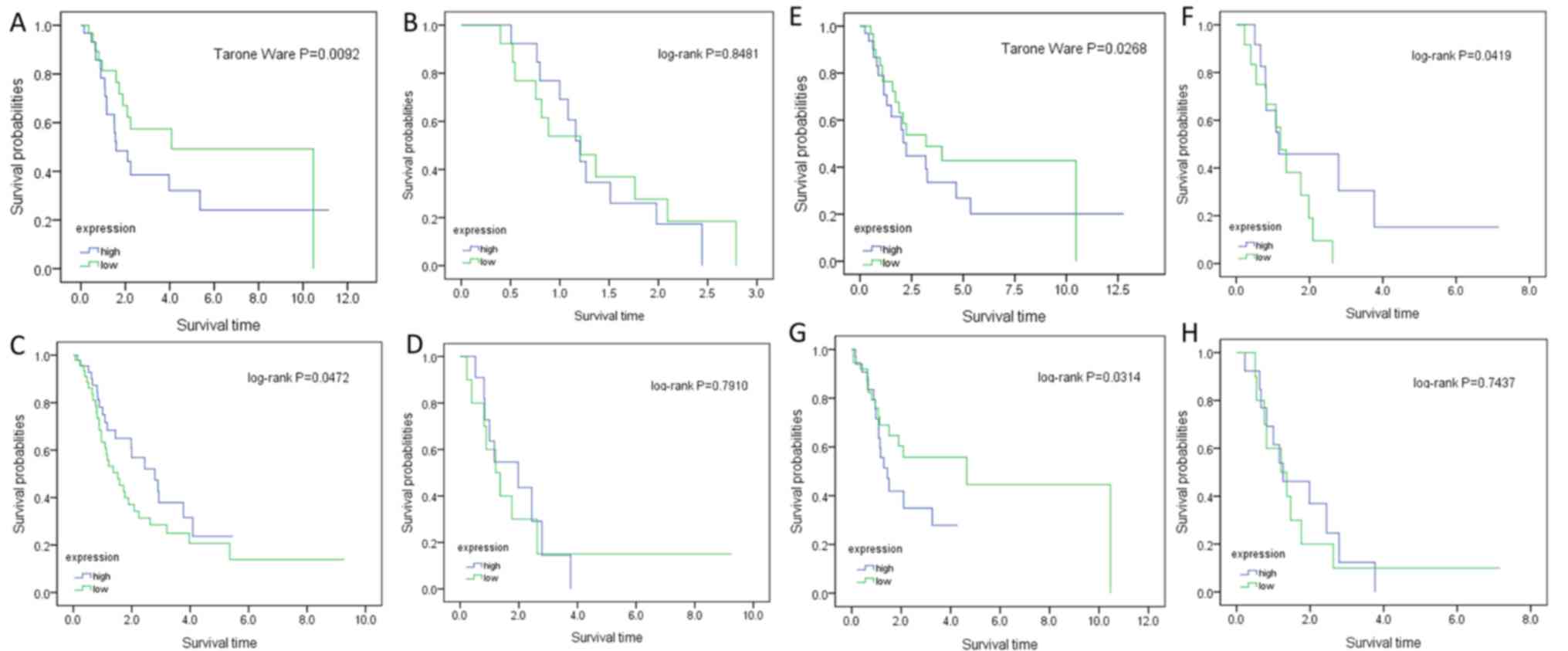
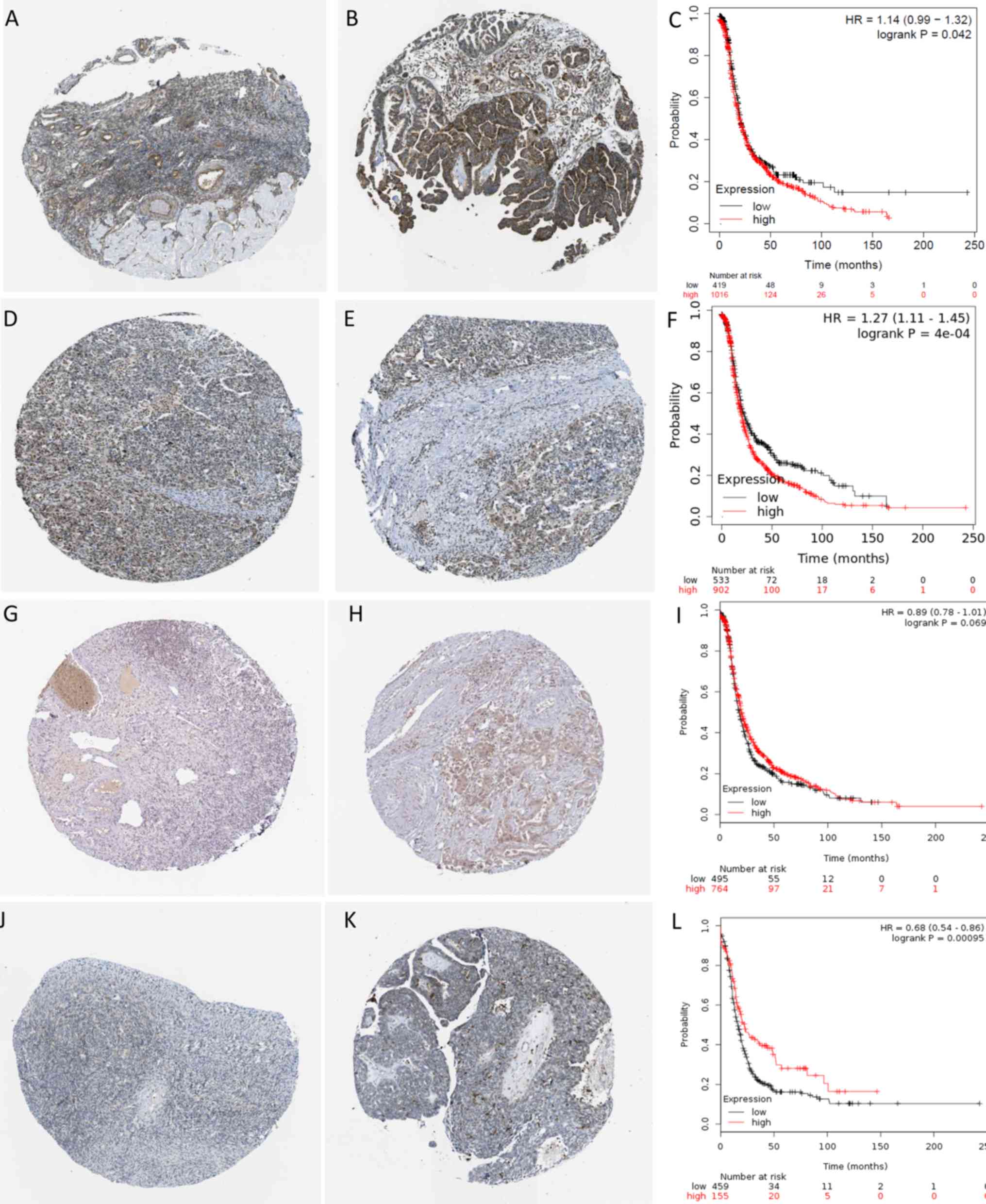
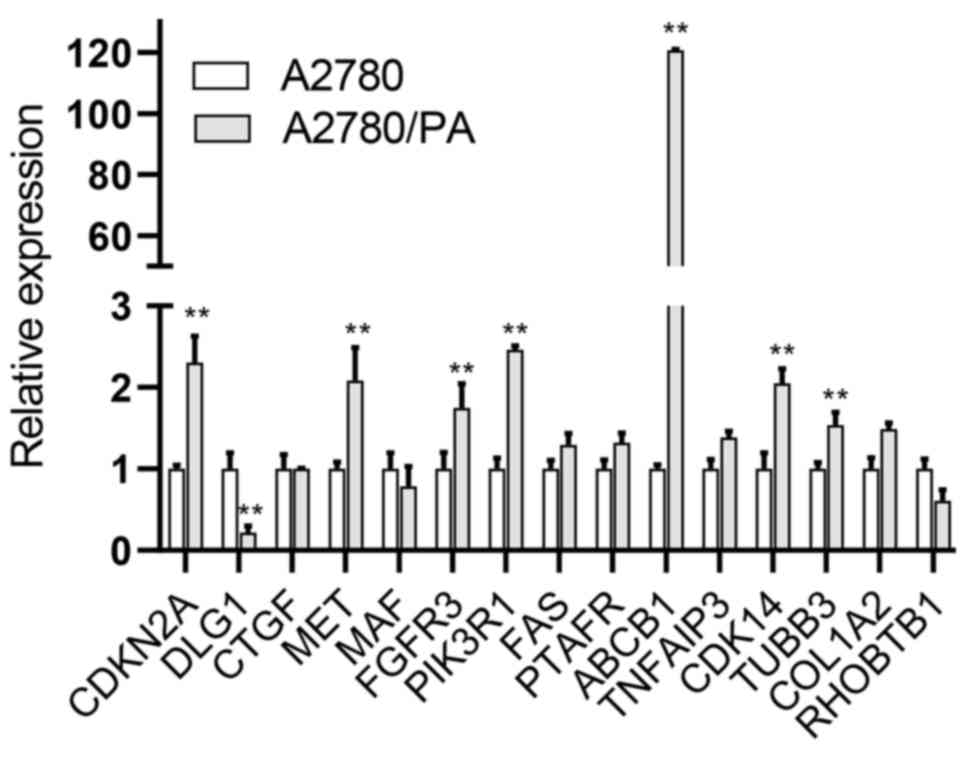
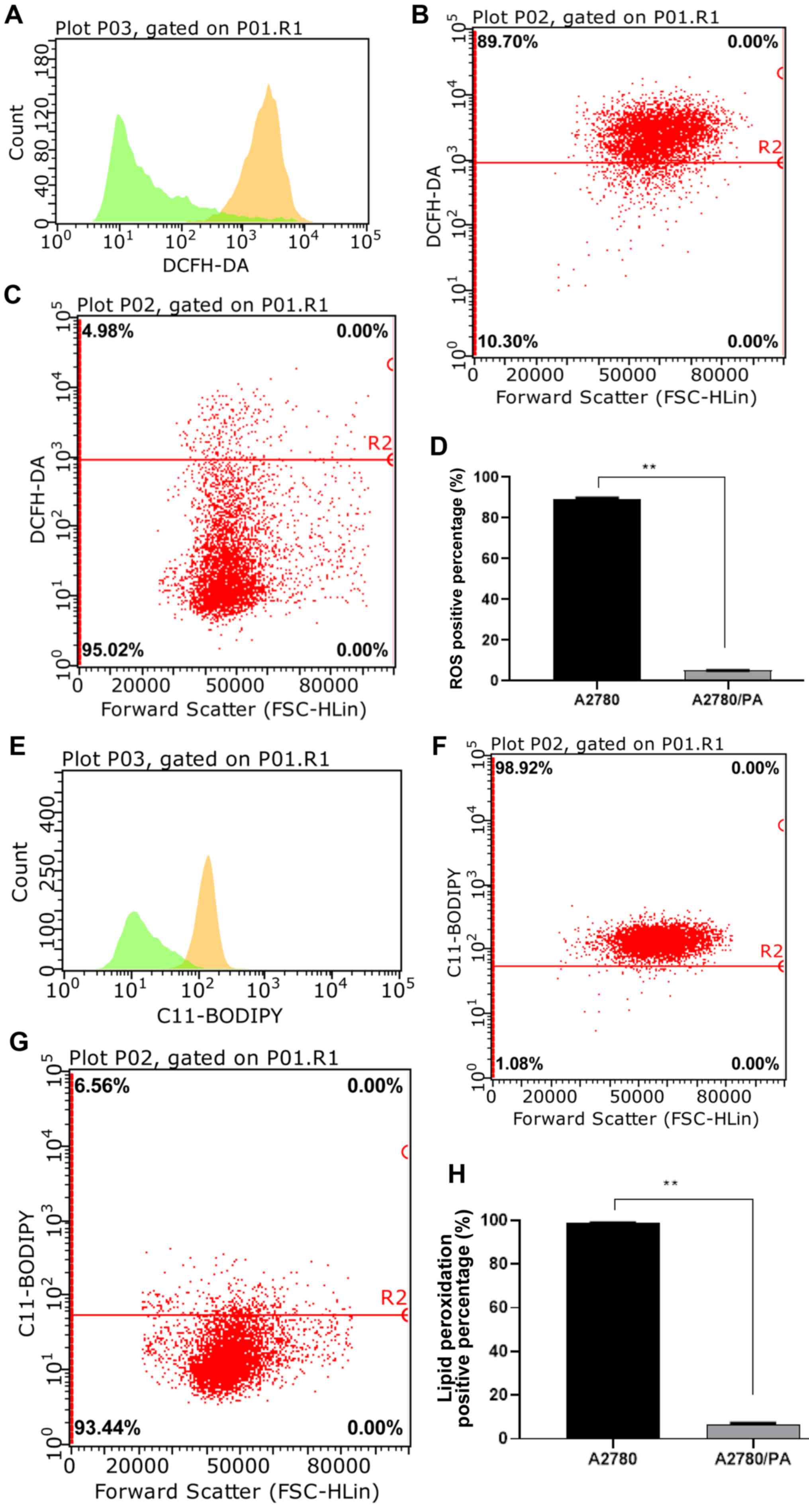
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Terraneo N, Jacob F, Dubrovska A and
Grünberg J: Novel therapeutic strategies for ovarian cancer stem
cells. Front Oncol. 10:3192020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marth C, Reimer D and Zeimet AG:
Front-line therapy of advanced epithelial ovarian cancer: Standard
treatment. Ann Oncol. 28 (Suppl_8):viii36–viii39. 2017. View Article : Google Scholar
|
|
4
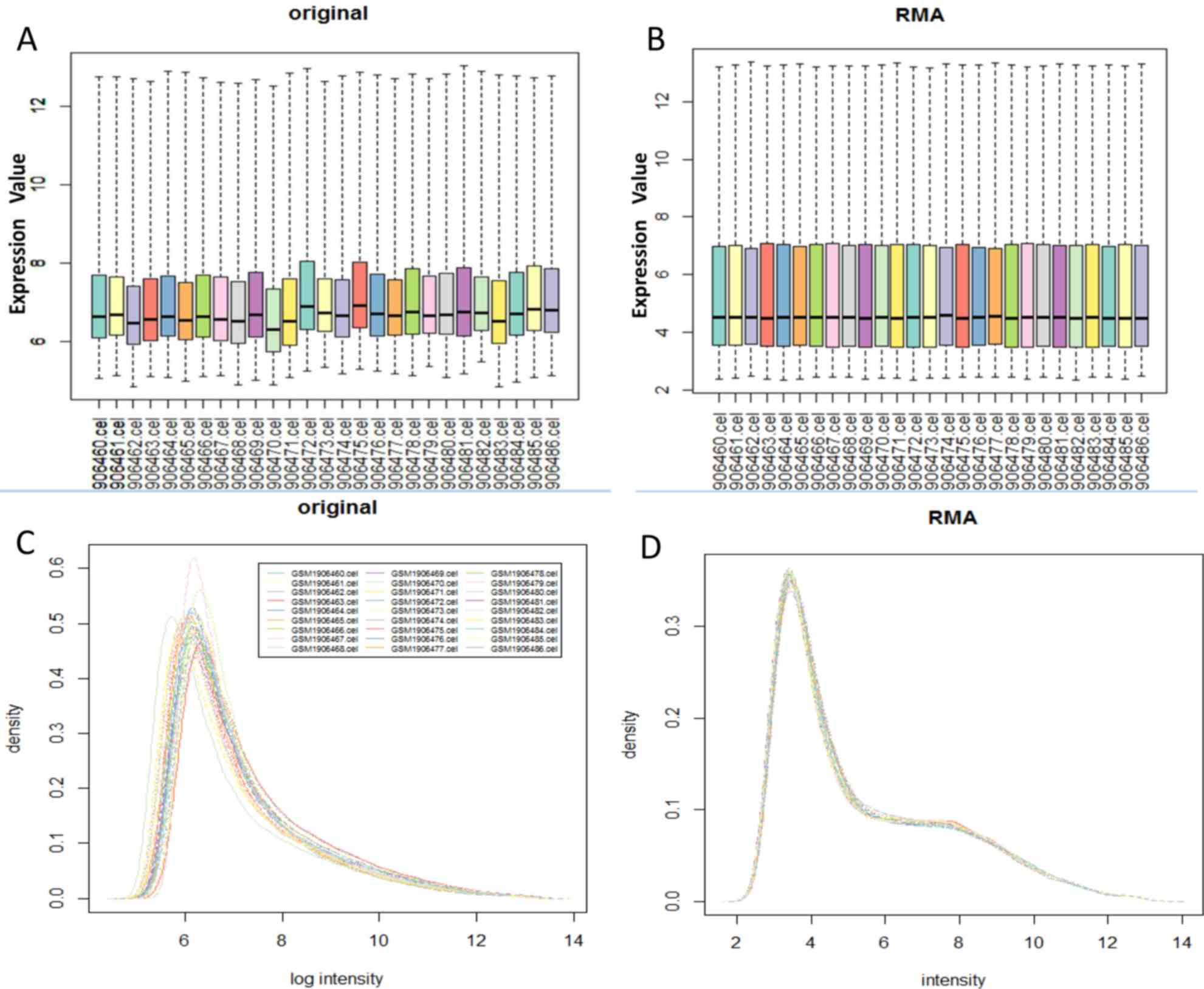
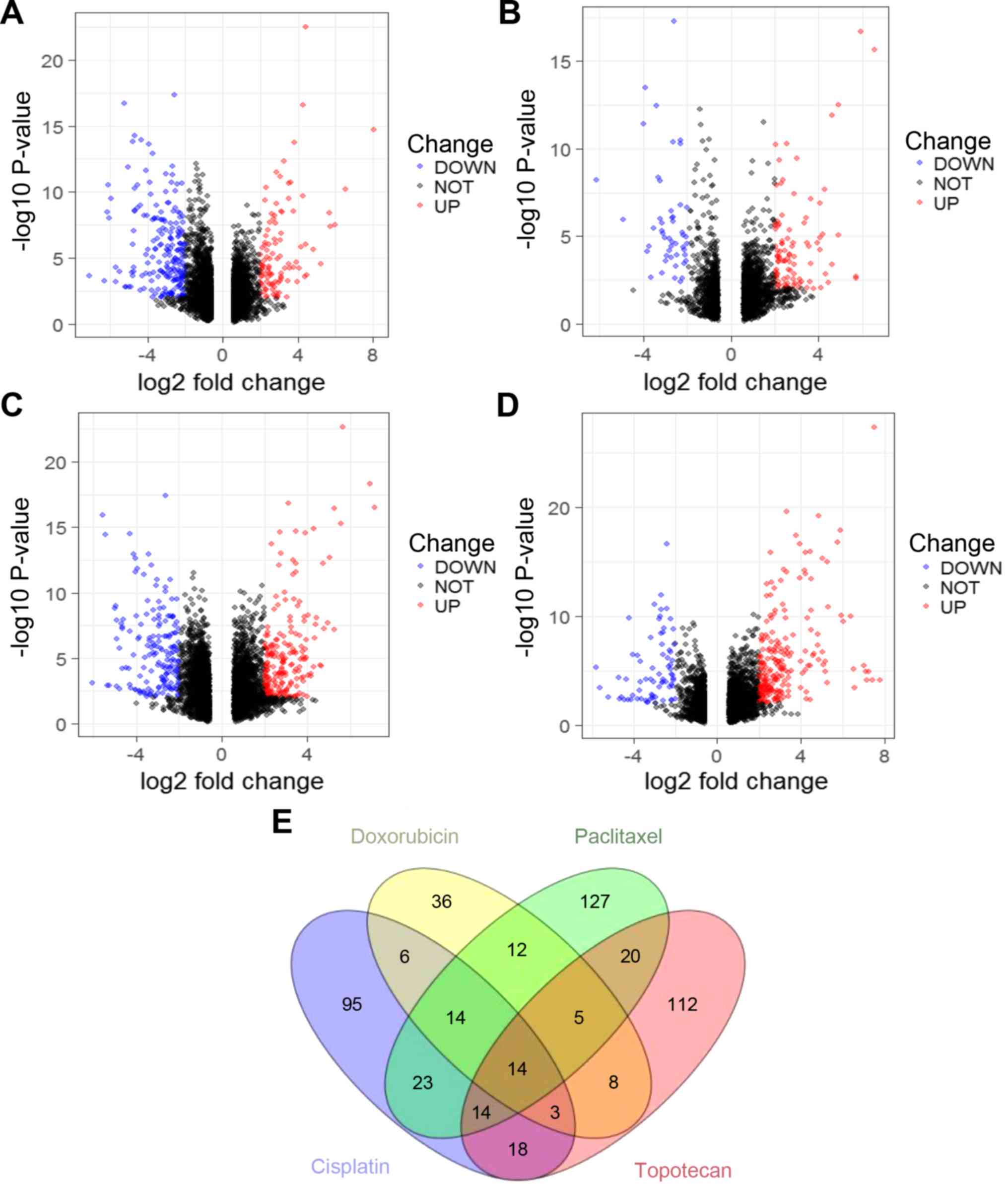
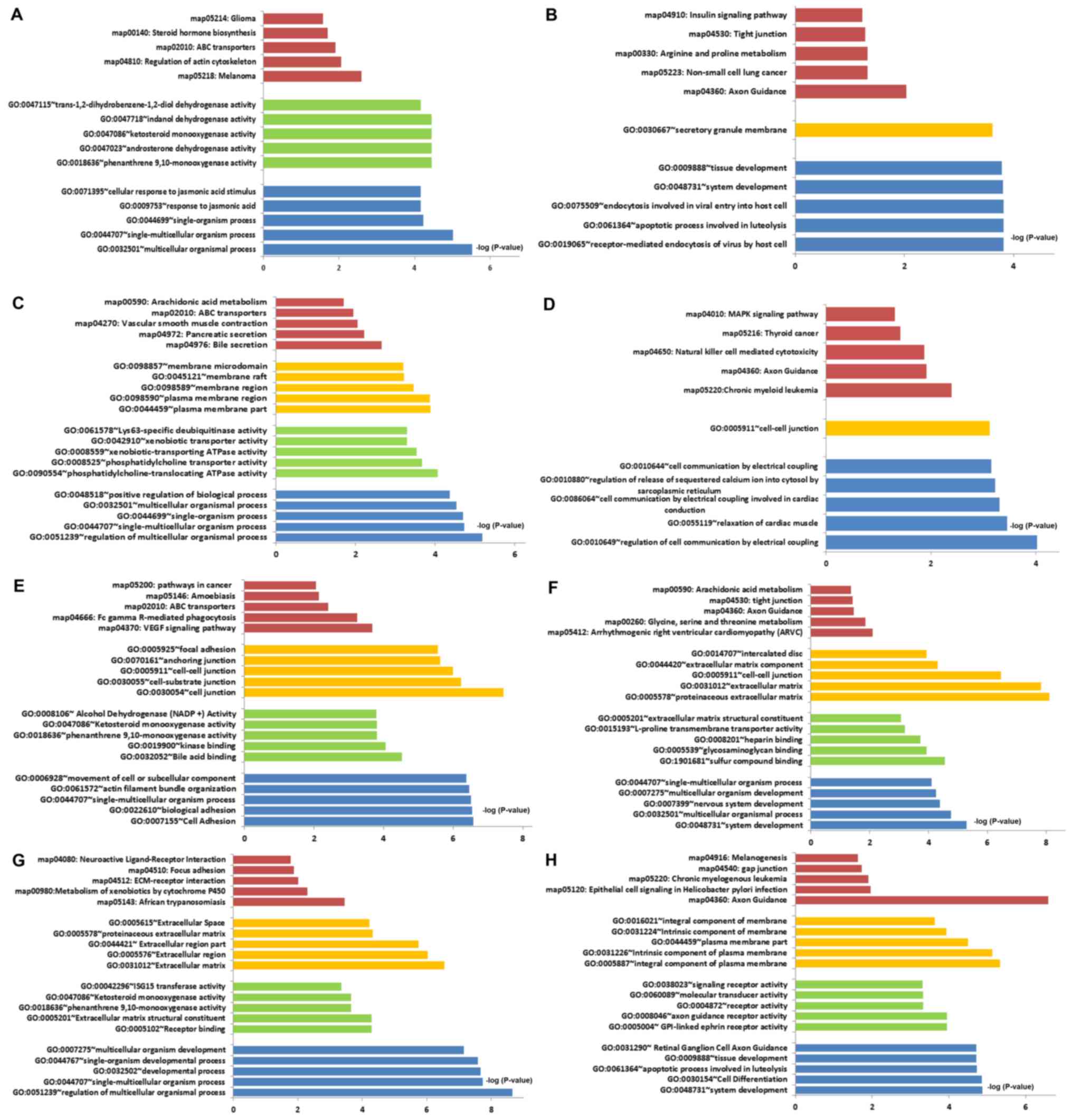
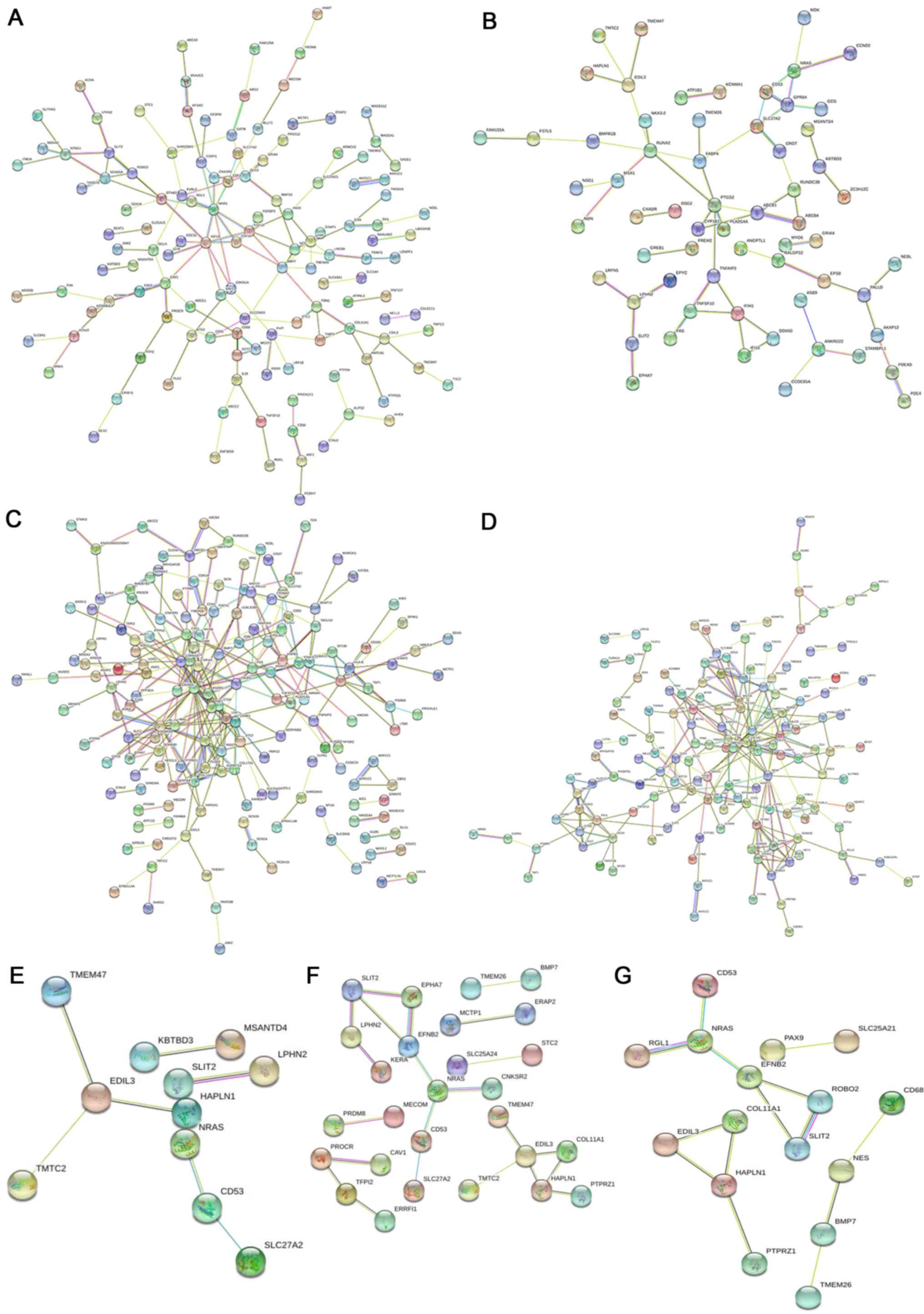
|
Pignata S, Lauraine EP, du Bois A and
Pisano C: Pegylated liposomal doxorubicin combined with
carboplatin: A rational treatment choice for advanced ovarian
cancer. Crit Rev Oncol Hematol. 73:23–30. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Morgan RJ Jr, Armstrong DK, Alvarez RD,
Bakkum-Gamez JN, Behbakht K, Chen LM, Copeland L, Crispens MA,
DeRosa M, Dorigo O, et al: Ovarian cancer, version 1.2016, NCCN
clinical practice guidelines in oncology. J Natl Compr Canc Netw.
14:1134–1163. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rojas V, Hirshfield KM, Ganesan S and
Rodriguez-Rodriguez L: Molecular characterization of epithelial
ovarian Cancer: Implications for diagnosis and treatment. Int J Mol
Sci. 17(pii): E21132016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thomas A and Pommier Y: Targeting
Topoisomerase I in the Era of Precision Medicine. Clin Cancer Res.
25:6581–6589. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dasari S and Tchounwou PB: Cisplatin in
cancer therapy: Molecular mechanisms of action. Eur J Pharmacol.
740:364–378. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wenningmann N, Knapp M, Ande A, Vaidya TR
and Ait-Oudhia S: Insights into doxorubicin-induced cardiotoxicity:
Molecular mechanisms, preventive strategies, and early monitoring.
Mol Pharmacol. 96:219–232. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhu L and Chen L: Progress in research on
paclitaxel and tumor immunotherapy. Cell Mol Biol Lett. 24:402019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hoskins P, Eisenhauer E, Vergote I,
Dubuc-Lissoir J, Fisher B, Grimshaw R, Oza A, Plante M, Stuart G
and Vermorken J: Phase II feasibility study of sequential couplets
of Cisplatin/Topotecan followed by paclitaxel/cisplatin as primary
treatment for advanced epithelial ovarian cancer: A National Cancer
Institute of Canada Clinical Trials Group Study. J Clin Oncol.
18:4038–4044. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brotto L, Brundage M, Hoskins P, Vergote
I, Cervantes A, Casado HA, Poveda A, Eisenhauer E and Tu D;
Gynecologic Cancer Intergroup Study of NCIC Clinical Trials Group
(NCIC CTG); European Organization for Research and Treatment of
Cancer, : Randomized study of sequential
cisplatin-topotecan/carboplatin-paclitaxel versus
carboplatin-paclitaxel: Effects on quality of life. Support Care
Cancer. 24:1241–1249. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hoskins P, Vergote I, Cervantes A, Tu D,
Stuart G, Zola P, Poveda A, Provencher D, Katsaros D, Ojeda B, et
al: Advanced ovarian cancer: Phase III randomized study of
sequential cisplatin-topotecan and carboplatin-paclitaxel vs
carboplatin-paclitaxel. J Natl Cancer Inst. 102:1547–1556. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Antman E, Weiss S and Loscalzo J: Systems
pharmacology, pharmacogenetics, and clinical trial design in
network medicine. Wiley Interdiscip Rev Syst Biol Med. 4:367–383.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Garralda E, Dienstmann R, Piris-Giménez A,
Braña I, Rodon J and Tabernero J: New clinical trial designs in the
era of precision medicine. Mol Oncol. 13:549–557. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pignata S, Scambia G, Ferrandina G,
Savarese A, Sorio R, Breda E, Gebbia V, Musso P, Frigerio L, Del
Medico P, et al: Carboplatin plus paclitaxel versus carboplatin
plus pegylated liposomal doxorubicin as first-line treatment for
patients with ovarian cancer: the MITO-2 randomized phase III
trial. J Clin Oncol. 29:3628–3635. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kurtz JE, Kaminsky MC, Floquet A, Veillard
AS, Kimmig R, Dorum A, Elit L, Buck M, Petru E, Reed N, et al:
Ovarian cancer in elderly patients: Carboplatin and pegylated
liposomal doxorubicin versus carboplatin and paclitaxel in late
relapse: A Gynecologic Cancer Intergroup (GCIG) CALYPSO sub-study.
Ann Oncol. 22:2417–2423. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sehouli J, Chekerov R, Reinthaller A,
Richter R, Gonzalez-Martin A, Harter P, Woopen H, Petru E, Hanker
LC, Keil E, et al: Topotecan plus carboplatin versus standard
therapy with paclitaxel plus carboplatin (PC) or gemcitabine plus
carboplatin (GC) or pegylated liposomal doxorubicin plus
carboplatin (PLDC): A randomized phase III trial of the
NOGGO-AGO-Study Group-AGO Austria and GEICO-ENGOT-GCIG intergroup
study (HECTOR). Ann Oncol. 27:2236–2241. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bolis G, Scarfone G, Raspagliesi F,
Mangili G, Danese S, Scollo P, Lo Russo D, Villa A, Aimone PD and
Scambia G: Paclitaxel/carboplatin versus
topotecan/paclitaxel/carboplatin in patients with FIGO suboptimally
resected stage III–IV epithelial ovarian cancer a multicenter,
randomized study. Eur J Cancer. 46:2905–2912. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Galluzzi L, Vitale I, Michels J, Brenner
C, Szabadkai G, Harel-Bellan A, Castedo M and Kroemer G: Systems
biology of cisplatin resistance: Past, present and future. Cell
Death Dis. 5:e12572014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Armstrong DK, Alvarez RD, Bakkum-Gamez JN,
Barroilhet L, Behbakht K, Berchuck A, Berek JS, Chen LM, Cristea M,
DeRosa M, et al: NCCN Guidelines Insights: Ovarian Cancer, Version
1.2019. J Natl Compr Canc Netw. 17:896–909. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng X and Li H: TKTL1 modulates the
response of paclitaxel-resistant human ovarian cancer cells to
paclitaxel. Biochem Biophys Res Commun. 503:572–579. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Okon IS and Zou MH: Mitochondrial ROS and
cancer drug resistance: Implications for therapy. Pharmacol Res.
100:170–174. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen G, Wang F, Trachootham D and Huang P:
Preferential killing of cancer cells with mitochondrial dysfunction
by natural compounds. Mitochondrion. 10:614–625. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Okon IS, Coughlan KA, Zhang M, Wang Q and
Zou MH: Gefitinib-mediated reactive oxygen specie (ROS) instigates
mitochondrial dysfunction and drug resistance in lung cancer cells.
J Biol Chem. 290:9101–9110. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Środa-Pomianek K, Michalak K, Świątek P,
Poła A, Palko-Łabuz A and Wesołowska O: Increased lipid
peroxidation, apoptosis and selective cytotoxicity in colon cancer
cell line LoVo and its doxorubicin-resistant subline LoVo/Dx in the
presence of newly synthesized phenothiazine derivatives. Biomed
Pharmacother. 106:624–636. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Harter P, Hilpert F, Mahner S, Heitz F,
Pfisterer J and du Bois A: Systemic therapy in recurrent ovarian
cancer: Current treatment options and new drugs. Expert Rev
Anticancer Ther. 10:81–88. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tempfer CB, Hartmann F, Hilal Z and
Rezniczek GA: Intraperitoneal cisplatin and doxorubicin as
maintenance chemotherapy for unresectable ovarian cancer: A case
report. BMC Cancer. 17:262017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tempfer CB, Giger-Pabst U, Seebacher V,
Petersen M, Dogan A and Rezniczek GA: A phase I, single-arm,
open-label, dose escalation study of intraperitoneal cisplatin and
doxorubicin in patients with recurrent ovarian cancer and
peritoneal carcinomatosis. Gynecol Oncol. 150:23–30. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Feng MX, Ma MZ, Fu Y, Li J, Wang T, Xue F,
Zhang JJ, Qin WX, Gu JR, Zhang ZG and Xia Q: Elevated autocrine
EDIL3 protects hepatocellular carcinoma from anoikis through
RGD-mediated integrin activation. Mol Cancer. 13:2262014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiang SH, Wang Y, Yang JY, Li J, Feng MX,
Wang YH, Yang XM, He P, Tian GA, Zhang XX, et al: Overexpressed
EDIL3 predicts poor prognosis and promotes anchorage-independent
tumor growth in human pancreatic cancer. Oncotarget. 7:4226–4240.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Etemadmoghadam D, Azar WJ, Lei Y, Moujaber
T, Garsed DW, Kennedy CJ, Fereday S, Mitchell C, Chiew YE, Hendley
J, et al: EIF1AX and NRAS mutations co-occur and
cooperate in low-grade serous ovarian carcinomas. Cancer Res.
77:4268–4278. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Althawadi H, Alfarsi H, Besbes S, Mirshahi
S, Ducros E, Rafii A, Pocard M, Therwath A, Soria J and Mirshahi M:
Activated protein C upregulates ovarian cancer cell migration and
promotes unclottability of the cancer cell microenvironment. Oncol
Rep. 34:603–609. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Polyak K and Metzger Filho O: SnapShot:
Breast cancer. Cancer Cell. 22:562–562.e1. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang D, Liu C, Wang J, Jia Y, Hu X, Jiang
H, Shao ZM and Zeng YA: Protein C receptor stimulates multiple
signaling pathways in breast cancer cells. J Biol Chem.
293:1413–1424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Huynh M, Pak C, Markovina S, Callander NS,
Chng KS, Wuerzberger-Davis SM, Bakshi DD, Kink JA, Hematti P, Hope
C, et al: Hyaluronan and proteoglycan link protein 1 (HAPLN1)
activates bortezomib-resistant NF-κB activity and increases drug
resistance in multiple myeloma. J Biol Chem. 293:2452–2465. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ecker BL, Kaur A, Douglass SM, Webster MR,
Almeida FV, Marino GE, Sinnamon AJ, Neuwirth MG, Alicea GM, Ndoye
A, et al: Age-related Changes in HAPLN1 increase lymphatic
permeability and affect routes of melanoma metastasis. Cancer
Discov. 9:82–95. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yau C, Esserman L, Moore DH, Waldman F,
Sninsky J and Benz CC: A multigene predictor of metastatic outcome
in early stage hormone receptor-negative and triple-negative breast
cancer. Breast Cancer Res. 12:R852010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Eckstein N, Servan K, Hildebrandt B,
Pölitz A, von Jonquières G, Wolf-Kümmeth S, Napierski I, Hamacher
A, Kassack MU, Budczies J, et al: Hyperactivation of the
insulin-like growth factor receptor I signaling pathway is an
essential event for cisplatin resistance of ovarian cancer cells.
Cancer Res. 69:2996–3003. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Du J, Shi HR, Ren F, Wang JL, Wu QH, Li X
and Zhang RT: Inhibition of the IGF signaling pathway reverses
cisplatin resistance in ovarian cancer cells. BMC Cancer.
17:8512017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang Y, Huang S, Guo Y and Li L: MiR-1294
confers cisplatin resistance in ovarian Cancer cells by targeting
IGF1R. Biomed Pharmacother. 106:1357–1363. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang L, Zhan X, Shen X, Li M, Yang J, Yu
W, Chen H, Jin B and Mao Z: P16 promotes the growth and mobility
potential of breast cancer both in vitro and in vivo: the key role
of the activation of IL-6/JAK2/STAT3 signaling. Mol Cell Biochem.
446:137–148. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zuidscherwoude M, Dunlock VE, van den
Bogaart G, van Deventer SJ, van der Schaaf A, van Oostrum J,
Goedhart J, In't Hout J, Hämmerling GJ, Tanaka S, et al:
Tetraspanin microdomains control localized protein kinase C
signaling in B cells. Sci Signal. 10(pii): eaag27552017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Schaper F and van Spriel AB: Antitumor
immunity is controlled by tetraspanin proteins. Front Immunol.
9:11852018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jiang Z, Liang G, Xiao Y, Qin T, Chen X,
Wu E, Ma Q and Wang Z: Targeting the SLIT/ROBO pathway in tumor
progression: molecular mechanisms and therapeutic perspectives.
Ther Adv Med Oncol. 11:17588359198552382019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Dickinson RE and Duncan WC: The SLIT-ROBO
pathway: A regulator of cell function with implications for the
reproductive system. Reproduction. 139:697–704. 2010. View Article : Google Scholar : PubMed/NCBI
|