Introduction
Colorectal cancer (CRC) is a common malignancy
associated with mutations in multiple genes, such as KRAS
(1) and SDC2 (2). Progression from a benign tumor to CRC
takes up to 10 years in 80% of affected individuals (3). Therefore, CRC screening is critical for
early detection and treatment of CRC. The fecal occult blood test
(FOBT) and colonoscopy are the mainstay of CRC screening. However,
the FOBT has a low diagnostic performance, particularly for
colorectal adenoma (3–5). Colonoscopy is the gold standard for
diagnosing CRC with good sensitivity and specificity; however, it
is associated with a high risk of complications and low compliance
(5). Cancer cells from early-stage
CRC are continuously shed into the colonic lumen and mixed into
stool (6). Tests for genetic and
epigenetic alterations in fecal DNA have been considered as a
possible method for the early detection of CRC (7).
KRAS is a common oncogene in malignant
tumors. KRAS mutations are detected in 30–40% of CRCs
(1). There are seven mutation
hotspots that account for >90% of the KRAS mutations,
including Gly12Asp, Gly12Val, Gly12Ser, Gly12Cys, Gly12Ala,
Gly12Arg and Gly13Asp (8). It has
been demonstrated that mutations in codons 12 and 13 of exon 2 of
KRAS are closely associated with the development of CRC, and
mutations in codon 12 are associated with a less favorable
prognosis compared with mutations in codon 13 (9).
Hypermethylation of CpG islands in gene promoter
regions suppresses specific gene expression and promotes
tumorigenesis in various types of cancer (1). The early occurrence of CRC is closely
associated with the methylation of CRC-related gene promoter
regions (10–12). The N-myc downstream-regulated gene
(NDRG) 4 is a member of the NDRG family of tumor suppressor genes
(13). It has been demonstrated that
the 5′ regulatory region of NDRG4 contains CpG islands,
which are often methylated during the development of CRC (13). Methylation of NDRG4 is
considered to be an important biological characteristic of CRC
(14). Therefore, NDRG4 may
be a potential diagnostic biomarker for CRC screening.
Syndecan-2 (SDC2), also known as fibroglycan,
encodes a transmembrane (type I) heparan sulfate proteoglycan and
regulates adhesion and proliferation of colon carcinoma cells
(15). Hypermethylation of
SDC2 has been detected at high frequency in the blood of
patients with CRC (2). As a
molecular marker of potential CRC, SDC2 methylation
demonstrates a high degree of specificity for the diagnosis of
early-stage tumors (16).
The tissue factor pathway inhibitor (TFPI) 2
belongs to a previously described group in embryonic cells of
polycomb group-marked genes that may be predisposed to aberrant DNA
methylation in the early stages of CRC carcinogenesis (17). It has been demonstrated that TFPI2
levels determined by fecal DNA testing are associated with CRC
recurrence and early-stage CRC (17–23).
In the present study, a multitarget stool DNA
(mt-sDNA) test was designed, including quantitative molecular
assays for KRAS mutations and aberrant NDRG4, SDC2
and TFPI2 methylation for the diagnosis of CRC. The
diagnostic performance of the mt-sDNA test was compared with a
commercially available FOBT in the detection of carcinoma and large
adenoma (≥1 cm in diameter).
Materials and methods
Participants and stool collection
The present study included 151 participants who
underwent colonoscopy at Shanghai Jiao Tong University, School of
Medicine, Ruijin Hospital North (Shanghai, China) between January
2016 and January 2017. The inclusion criteria were as follows: i)
Colorectal adenoma (≥1 cm in diameter; smaller or diminutive polyps
excluded); or ii) colorectal carcinoma; and iii) age >18 years.
Patients with the following conditions were excluded: i)
Contraindications to colonoscopy; ii) severe gastrointestinal
bleeding; and iii) hemorrhoids. A total of 50 participants who were
free of colorectal polyps or tumors were selected from individuals
who were receiving routine medical examinations.
Stool was collected from all participants prior to
bowel purgation and colonoscopy, or otherwise 1 week after the
colonoscopy but prior to neoplasm resection. The FOBT was performed
before addition of the preservative buffer to the stool. The
homogenized stools were stored at −20°C for the subsequent mt-sDNA
test. Colorectal adenoma or carcinoma tissues and adjacent normal
tissues within 1 cm of the tumor were biopsied and stored in −196°C
liquid nitrogen prior to DNA extraction.
FOBT
The FOBT was performed using a guaiac-based
immunochemical kit according to the manufacturer's protocol
(Hemosure T1-CK50, W.H.P.M. Bioresearch & Technology) with a
minimum detection limit of 0.2 µg/ml. Depending on whether a large
adenoma or tumor was identified during colonoscopy, the test
results were classified as follows: A positive result was a true
positive if a neoplasm was detected, or a false positive if no
neoplasm was detected; a negative result was a false negative if a
neoplasm was detected, or a true negative if no neoplasm was
detected. Sensitivity and specificity were expressed as
percentages.
DNA extraction
Frozen or fresh colorectal tissues were collected
from 50 patients with CRC and 51 patients with colorectal adenoma,
and were used to determine whether mutated KRAS and
hypermethylated NDRG4, SDC2 and TFPI2 could be used
to detect CRC and large adenoma. DNA was extracted using a TIANamp
DNA kit (Tiangen Biotech Co., Ltd.) according to the manufacturer's
protocol. The methylation levels of NDRG4, SDC2 and
TFPI2 were detected using quantitative (q)PCR.
Stool samples were thawed at room temperature and
homogenized. The aliquots were transferred to tubes and centrifuged
at 14,000 × g for 5 min at room temperature. The supernatant was
used as the source of mt-sDNA. The mt-sDNA markers were enriched
using sequence-specific DNA captures and magnetic beads-based
oligonucleotides, and purified using magnetic separation.
Bisulfite treatment
The tissue DNA and stool DNA were treated with
bisulfite using an EZ DNA methylation kit (Zymo Research Corp.)
according to the manufacturer's protocol. For tissue DNA, 600 ng
genomic DNA was added into the bisulfite reaction and eluted in 30
µl Tris/EDTA (TE) buffer. For stool DNA, 30 µl captured DNA was
added into the reaction and eluted in 20 µl TE buffer.
Methylation-specific qPCR
The test panel included three methylated (m) genes
(mNDRG4, mSDC2 and mTFPI2). The mutant forms of
KRAS and β-actin were used as references. The mt-sDNA for
the methylation assay was treated with bisulfite. The genomic DNA
was used for the KRAS mutation assay. Multiplex qPCR was
used to detect the mutation and methylation profile of candidate
genes (ABI 7500 Real-Time PCR system; Applied Biosystems; Thermo
Fisher Scientific, Inc.). Each run consisted of sDNA samples,
positive controls (mNDRG4, mSDC2, mTFPI2, mutant KRAS
and internal control), and negative controls (water blanks).
Briefly, each multiplex PCR assay was performed with a final
reaction mixture volume of 20 µl containing 0.5 U Phusion
polymerase (Thermo Fisher Scientific, Inc.) in high-fidelity
Phusion buffer with a final concentration of 200 µM deoxynucleotide
triphosphates and 3 mM MgCl2. The primers were used at a
final concentration of 0.2–0.4 µM. Primer sequences are listed in
Table I. SYBR-Green I (Cambrex Bio
Science Rockland, Inc.) was diluted as recommended by the
manufacturer. The hot-start technique was used to prevent
non-specific amplification (24).
The amplification cycles consisted of incubation at 98°C for 30
sec, 65°C for 30 sec, 72°C for 30 sec and 72°C for 10 sec. After 30
cycles, a melting curve was determined using SYBR-Green
fluorescence with a ramp speed of 0.2°C/sec between 72 and 98°C,
with a reading every 0.2°C. The cycle threshold (Ct) value of each
gene was used to evaluate the result of each sample using the
2−ΔΔCq method (25). All
assays were performed in a blinded manner.
 | Table I.Primer sequence for the DNA
methylation and KRAS mutation assays. |
Table I.
Primer sequence for the DNA
methylation and KRAS mutation assays.
| Name | Primer
sequences |
|---|
| NDRG4 | MS:
5′-TTTAGGTTCGGTATCGTTTCGC-3′ |
|
|
3′-CGAACTAAAAACGATACGCCG-5′ |
|
| US:
5′-GATTAGTTTTAGGTTTGGTATTGTTTTGT-3′ |
|
|
3′-AAAACCAAACTAAAAACAATACACCA-5′ |
| TFPI2 | MS:
ATTTTTTAGGTTTCGTTTCGGC |
|
|
5′-GCCTAACGAAAAAAAATACGCG-3′ |
|
| US:
TTAGTTATTTTTTAGGTTTTGTTTTGGT |
|
|
3′-AAAAACACCTAACAAAAAAAAATACACA-5′ |
| SDC2 | MS:
5′-AAAGATTCGGCGACCACCGAACGACTCAAACTCGAAAACTCG-3′ |
|
|
3′-GACTCAAACTCGAAAACTCGAA-5′ |
|
| US:
5′-TTCGGGGCGTAGTTGCGGGCGG-3′ |
|
|
3′-TTCGGGGCGTAGTTGCGGGCGG-5′ |
| KRAS |
5′-CTGGTGCAGTATTTGATAGTGTA-3′ |
|
|
3′-TGAAAATGGTCAGAGAAACCTTTA-5′ |
| β-actin |
5′-GCTAAGTGTGCTGGGGTCTTGGGAT-3′ |
|
|
3′-GCTCTTTTTCTGGTGTTTGTCTCTC-5′ |
Statistical analysis
The strand number of each marker output from the ABI
7500 was quantified according to the Ct value. If there was no
amplification, the maximum amplification cycle number (Ct=45) was
considered. A logistic regression with specific boundary conditions
was developed to evaluate the performance of each biomarker. The
single marker cut-off was identified by a logistic regression
algorithm that produced dichotomous (positive/negative) results for
each sample. A threshold was defined for each marker in the mt-sDNA
panel that optimally separated the tumor samples from the control
samples. The logistic regression assigned a weight to each
component assay result and subsequently aggregated these individual
marker results to obtain a logistic score. Boundary conditions for
each of the methylation and mutation markers were defined on the
basis of a single value for each marker above which a positive
result could be inferred. A positive result for the logistic score
or a value exceeding any of the boundary conditions resulted in a
positive result for the mt-sDNA test. Colonoscopy-based findings
were compared with the mt-sDNA test results. Sensitivity and
specificity were calculated as percentages for comparison with
FOBT. Receiver operating characteristic (ROC) curves were
constructed to analyze the diagnostic performances of the
biomarkers.
Continuous data were presented as the mean ± SD and
compared using the one-way ANOVA test followed by Tukey's post hoc
test. Categorical data were presented as percentages and compared
using the χ2 test. All statistical analyses were
performed using SPSS v21.0 (IBM Corp.). P<0.05 was considered to
indicate a statistically significant difference.
Results
Patient characteristics
To determine the performance of biomarkers
(NDRG4, SDC2, TFPI2 and KRAS) in detecting CRC and
adenoma, an independent tissue study was performed. A total of 211
frozen or fresh colorectal tissues, including 64 pairs of CRC and
adjacent normal tissues (median age, 63 years; range, 43–79 years;
48.4% women) and 83 colorectal adenomas (≥1 cm in diameter; median
age, 57 years; range, 39–72; 41% women) were included in the
present study. Age and sex distributions were similar between
patients with CRC, patients with colorectal adenomas and normal
controls (Table II). In the
carcinoma and adenoma tissues, 38.0% (19/50) and 43.1% (22/51) of
the neoplasms were located in the colon, respectively. The
demographic and clinical characteristics of the subjects are shown
in Table II.
 | Table II.Clinical characteristics of the
participants. |
Table II.
Clinical characteristics of the
participants.
| Variable | CRC (n=50) | Adenoma (n=51) | Control (n=50) | P-value |
|---|
| Age, years (mean ±
SD) | 60.2±13.6 | 58.5±10.7 | 64.9±11.0 | >0.05 |
| Female | 52.0% (26/50) | 60.8% (31/51) | 46.0% (23/50) | >0.05 |
| Colon
neoplasms | 38.0% (19/50) | 43.1% (22/51) | – | >0.05 |
| Rectum
neoplasms | 62.0% (31/50) | 56.9% (29/51) | – | >0.05 |
Detection of the DNA markers
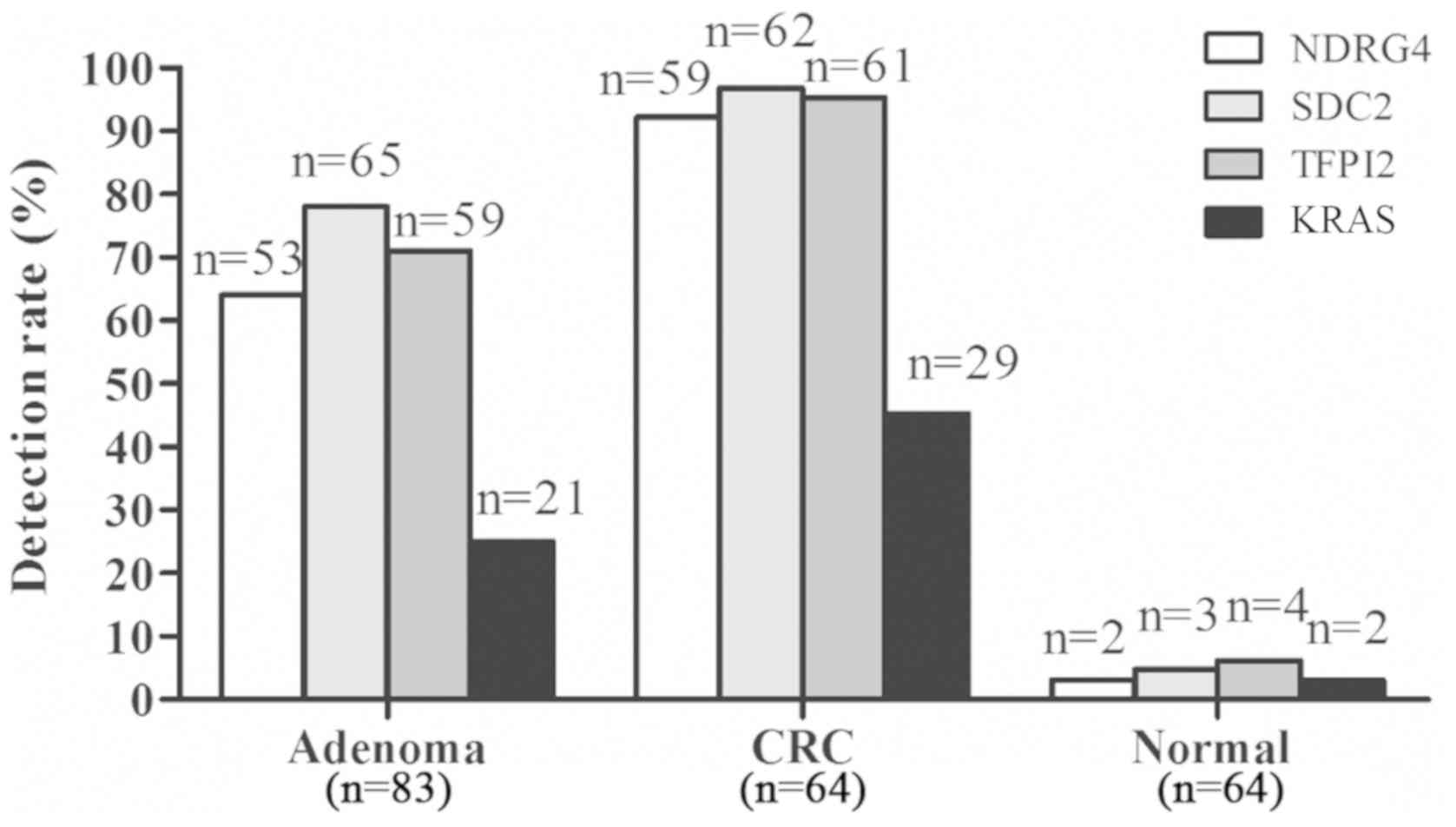
mNDRG4 was detected in 92.2% (59/64) of the
carcinoma tissues, 63.9% (53/83) of the adenoma tissues and 3.1%
(2/64) of the adjacent normal tissues (Fig. 1). mSDC2 was detected in 96.9%
(62/64) of the carcinoma tissues and 78.3% (65/83) of the adenoma
samples, with a specificity of 95.3% (61/64). mTFPI2 was
detected in 95.3% (61/64) of the carcinoma tissues and 71.1%
(59/83) of the adenoma samples, with a specificity of 93.8%
(60/64). KRAS mutations were detected in 45.3% (29/64) of
the carcinoma samples, 25.3% (21/83) of the adenoma samples and
3.1% (2/64) of the adjacent normal tissues (Fig. 1).
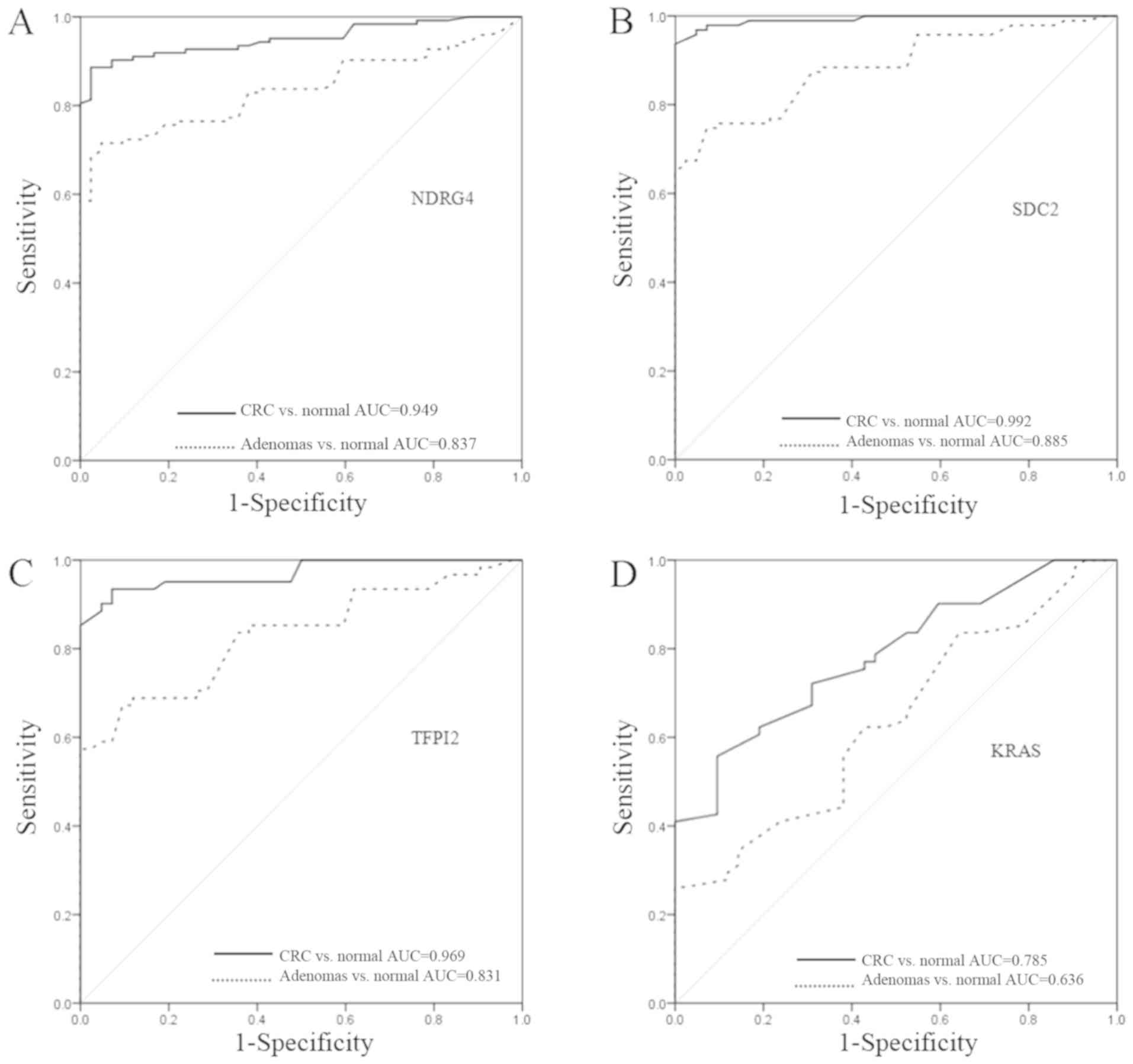
ROC curves were constructed for each of the four
genes (Fig. 2). When comparing the
cancer tissues with the adjacent normal tissues, the area under the
curve (AUC) values were 0.949, 0.992, 0.969 and 0.785 for NDRG4,
SDC2, TFPI2 and KRAS, respectively. When comparing
adenoma tissues with adjacent normal tissues, the AUC values were
0.837, 0.885, 0.831 and 0.636 for NDRG4, SDC2, TFPI2 and
KRAS, respectively.
Comparison of the mt-sDNA test with
FOBT in CRC detection
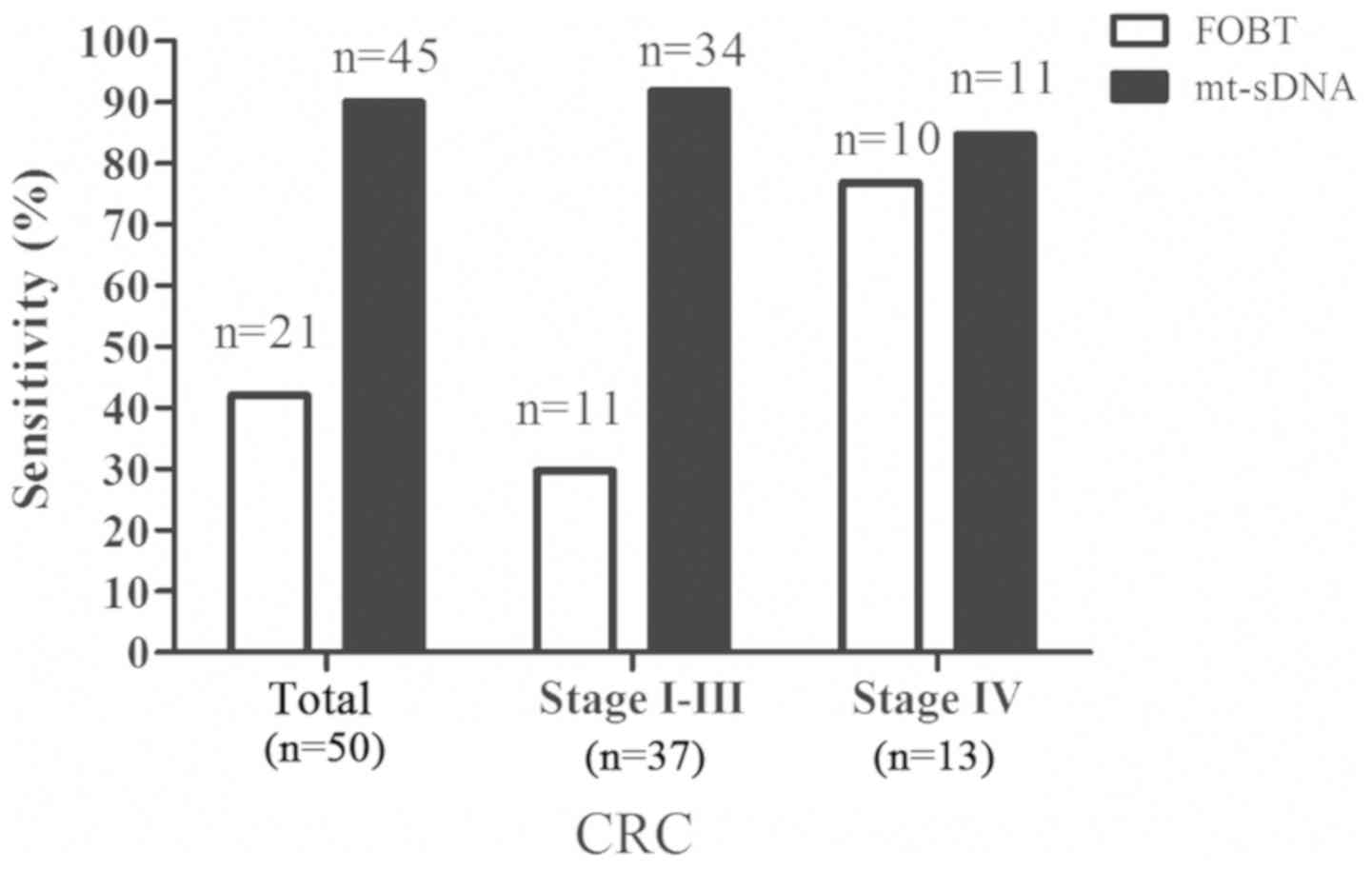
A total of 50 patients with CRC underwent the
mt-sDNA test. The sensitivity of the mt-sDNA test for CRC was 90.0%
(45/50), with a 91.9% (34/37) sensitivity for stage I–III CRC and
an 84.6% (11/13) sensitivity for stage IV CRC (Fig. 3). The FOBT had a sensitivity of 42.0%
(21/50) for CRC in the same samples, with a 29.7% (11/37)
sensitivity for stage I–III CRC and a 76.9% (10/13) sensitivity for
stage IV CRC (Fig. 3). These results
demonstrated that the mt-sDNA test outperformed the FOBT in
detecting CRC. In addition, the specificity of the mt-sDNA test
(94.0%; 47/50) was higher than that of the FOBT (90.0%; 45/50)
(data not shown).
Comparison of the mt-sDNA test with
FOBT for the detection of large adenoma
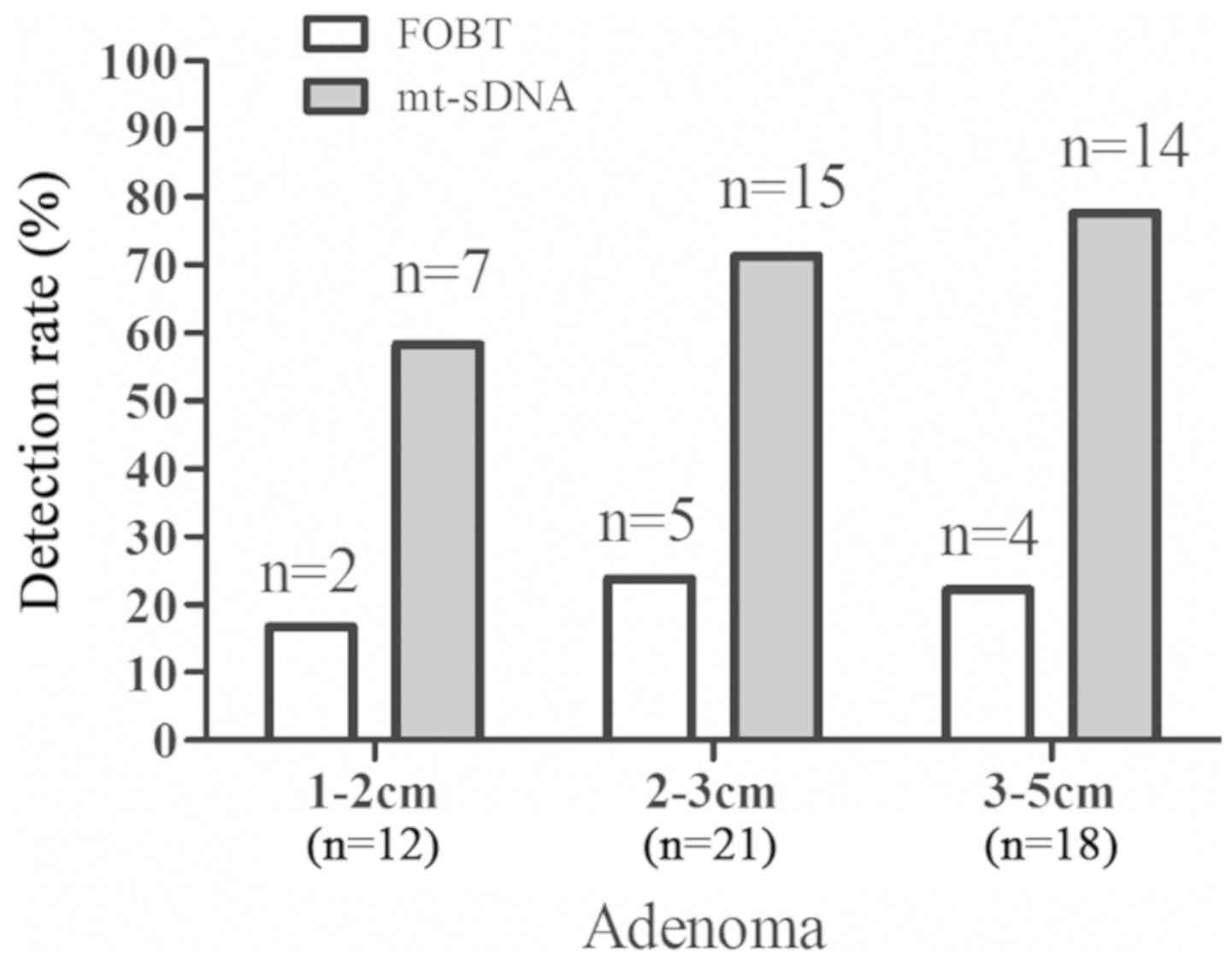
A total of 51 individuals were diagnosed with
advanced adenoma by colonoscopy. The size of the adenoma was 1–2 cm
in 12 samples, 2–3 cm in 21 samples and 3–5 cm in 18 samples. The
mt-sDNA test detected 7 adenomas of 1–2 cm, 15 adenomas of 2–3 cm
and 14 adenomas of 3–5 cm, whereas FOBT detected 2 adenomas of 1–2
cm, 5 adenoma of 2–3 cm and 4 adenomas of 3–5 cm (Fig. 4). The mt-sDNA test outperformed the
FOBT in detecting advanced adenomas with a sensitivity of 70.6%
(36/51) vs. 19.6% (10/51) (data not shown).
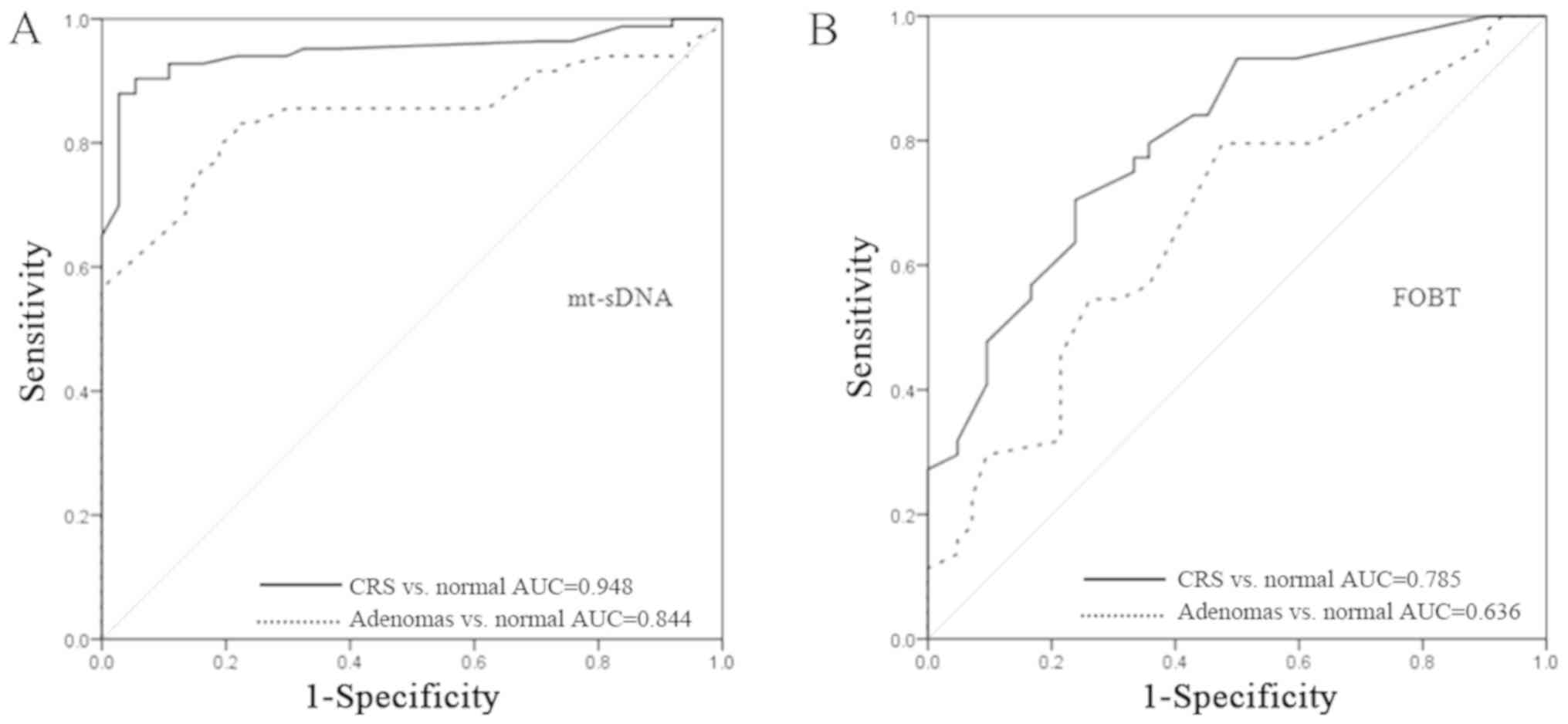
For the mt-sDNA test, the area under the ROC curve
was 0.948 (95% CI, 0.98–1) for detecting CRC and 0.844 (95% CI,
0.83–0.93) for detecting adenomas (Fig.
5A). For FOBT, the area under the ROC curve was 0.785 (95% CI,
0.69–0.87) for detecting CRC and 0.636 (95% CI, 0.53–0.74) for
detecting adenoma (Fig. 5B).
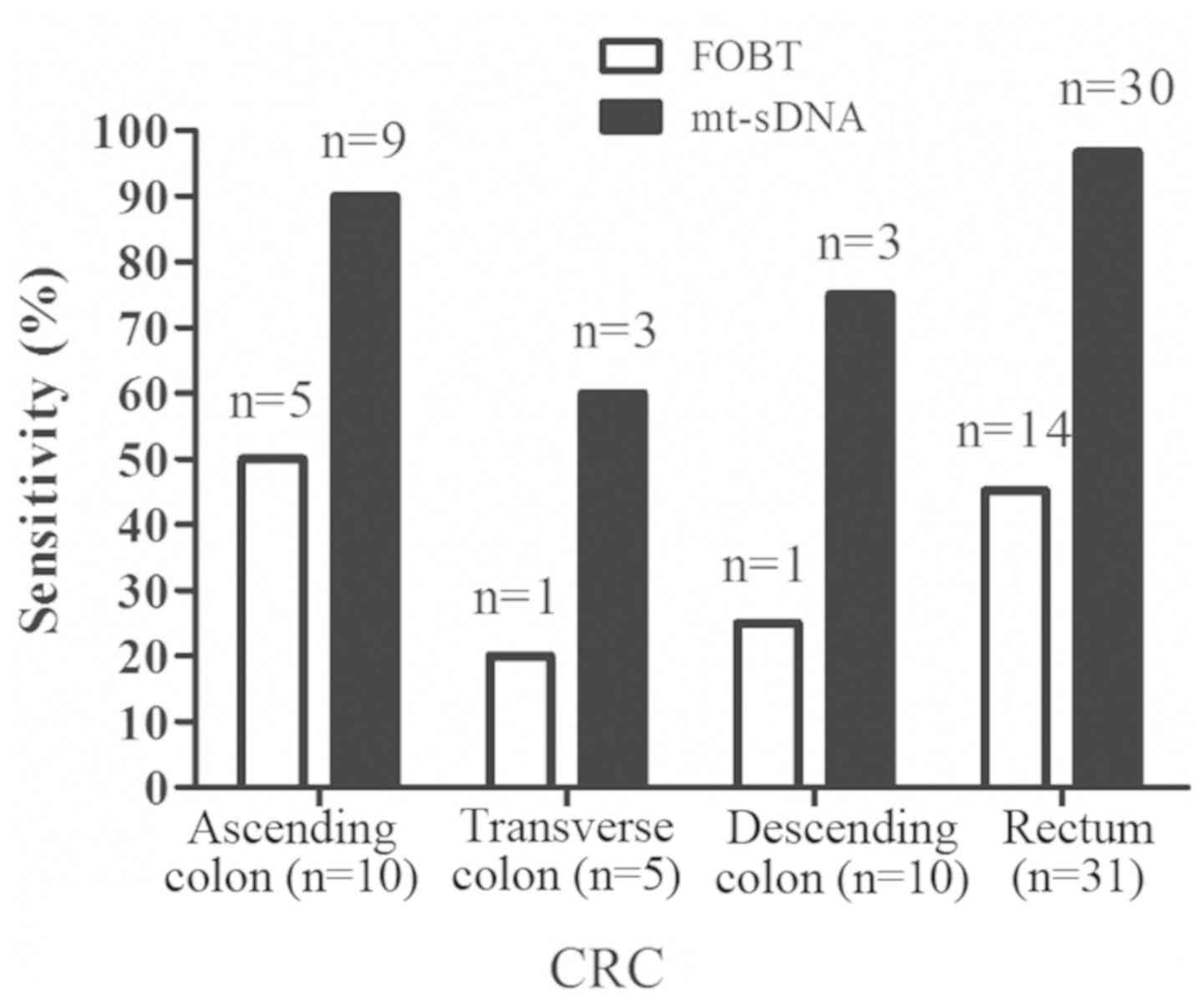
Detecting CRC at different sites
In terms of tumor location, the 50 CRC samples
included 10 samples in the ascending colon, 5 samples in the
transverse colon, 4 samples in the descending colon and 31 samples
in the rectum. The sensitivity of the mt-sDNA test for detecting
CRC was 90.0% (9/10) for the ascending colon, 60.0% (3/5) for the
transverse colon, 75.0% (3/4) for the descending colon and 96.8%
(30/31) for the rectum, whereas the sensitivity of FOBT for
detecting CRC was 50.0% (5/10) for the ascending colon, 20.0% (1/5)
for the transverse colon, 25.0% (1/4) for the descending colon and
45.2% (14/31) for the rectum (Fig.
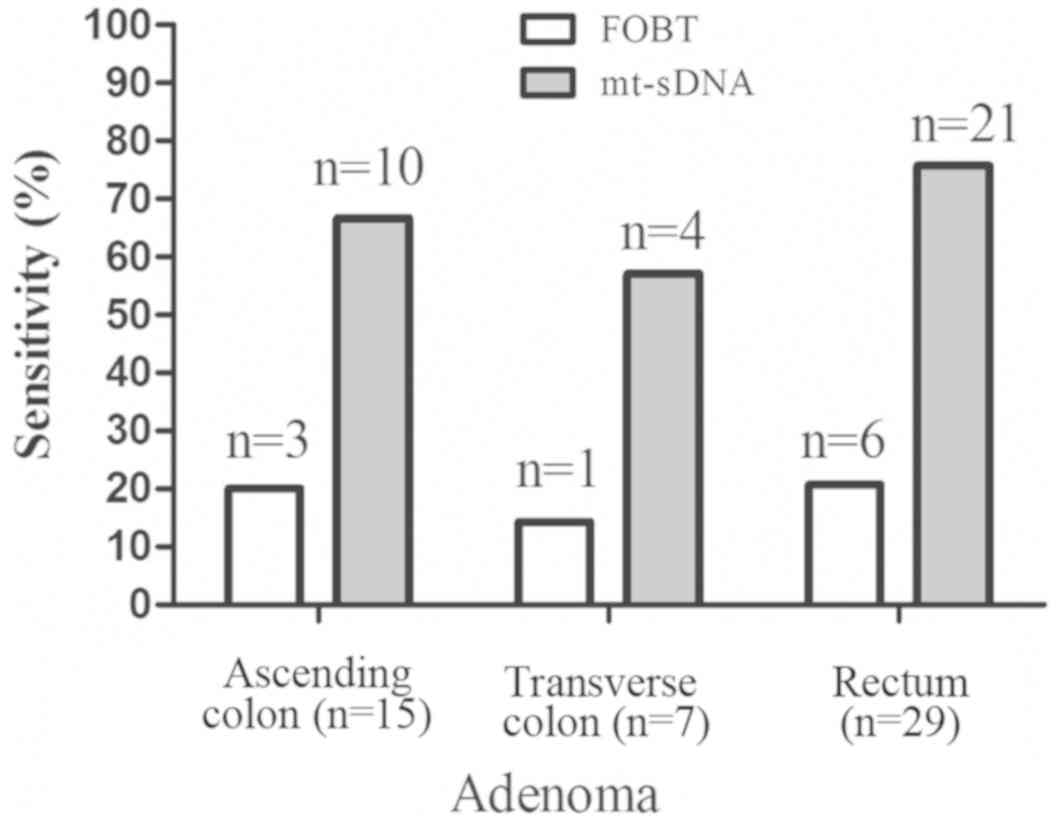
6). The 51 adenoma samples comprised 15 in the ascending colon,
7 in the transverse colon and 29 in the rectum. The sensitivity of
the FOBT for detecting adenoma was 20.0% (3/15) for the ascending
colon, 14.3% (1/7) for the transverse colon and 20.7% (6/29) for
the rectum, whereas the sensitivity of the mt-sDNA test for
detecting adenoma was 66.7% (10/15) for the ascending colon, 57.1%
(4/7) for the transverse colon and 72.4% (21/29) for the rectum
(Fig. 7).
Discussion
The present study demonstrated that the mt-sDNA test
was superior to FOBT in detecting both CRC and large adenoma with a
specificity of 93.0% vs. 91.0%. The current findings suggested that
the mt-sDNA test may be a feasible and promising approach for early
detection of CRC. FOBT is a traditional screening tool for CRC.
However, it is not widely used for CRC screening in China,
partially due to its inherent low sensitivity for detecting
colorectal neoplasms, particularly advanced adenomas in
asymptomatic patients (26,27). The present study demonstrated that
FOBT had a sensitivity of 19.6% for advanced adenoma and 29.7% for
stage I–III CRC. The mt-sDNA test had a 50% higher sensitivity for
adenomas and a 60% higher sensitivity for stage I–III CRC. In
addition, the sensitivity of the mt-sDNA test for detecting CRC was
90.0% (9/10) in the ascending colon, 60.0% (3/5) in the transverse
colon, 75.0% (3/4) in the descending colon and 96.8% (30/31) in the
rectum. However, due to the small sample size, the current results
did not support any conclusion concerning the performance of
mt-sDNA for diagnosing CRC at any specific stage or location.
Colonoscopy is considered the gold standard for CRC
diagnosis, but its application in CRC screening has been hindered
by a number of factors, such as the requirement of a visible
lesion, the risk of complications and the invasiveness of the
procedure, resulting in low patient compliance (28). The novel multitarget panel presented
in the current study had an improved performance compared with
previous findings (29). A dozen of
exfoliated markers, including mutated KRAS and
hypermethylated NDRG4, SDC2 and TFPI2, were analyzed
in tissue and stool assays in our study. NDRG4, SDC2 and
TFPI2 were highly methylated in CRC tissues, distinguishing
them from normal colon mucosal tissues, and KRAS mutated
tumors were more likely to develop on the right side of the colon,
in accordance with a previous study (30). Stool observations were consistent
with the tissue assays, and the analyzed biomarkers exhibited high
sensitivity and discrimination between CRC lesions and normal
tissues. In the present study, the mt-sDNA panel exhibited no
differences among the diverse tumor sites with 90% or higher
sensitivity.
As of 2018, CRC is the third most prevalent cancer
worldwide, and its morbidity and mortality in China has gradually
increased (31). In the United
States, the incidence and mortality of CRC have gradually
decreased, mainly due to large-scale population screening,
interventions for precancerous lesions for primary prevention and
early detection of CRC (31). In
China, screening rates for CRC remain low and there is a shortage
of medical resources. The present study offered a non-invasive
approach for CRC diagnosis and screening with high sensitivity and
specificity. In a previous study, a number of average-risk
participants were recruited to investigate their compliance with
fecal DNA testing via questionnaires, with >90% of these
individuals being prone to the mt-sDNA test, indicating that the
mt-sDNA test is patient-friendly to the average-risk population
(32). In addition, the mt-sDNA test
may have the potential to detect CRC at an earlier stage of tumor
development compared with FOBT. However, the relatively high cost
of the mt-sDNA test may limit its popularity.
In conclusion, the mt-sDNA test had a higher
sensitivity and specificity in diagnosing both CRC and advanced
adenoma compared with FOBT. Considering its molecular diagnostic
capability and its broad accessibility at clinical laboratories,
the mt-sDNA test may be a valuable addition to current CRC
screening options.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
Health and Family Planning Commission of Jiading District (grant
no. 2018-KY-01) and the Shanghai Jiao Tong University School of
Medicine, Ruijin Hospital North (grant no. 2018ZY16).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CY analyzed the samples and drafted the manuscript.
WW, JS, XYa, KL, HY, YY, SJ, XYu, YS, YZ, SZ, YX, YD, LX, BC and XX
collected the samples. PC, WZ and RZ analyzed the data and designed
the study. YW conceived the study and critically revised the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Shanghai Jiao Tong University School of Medicine,
Ruijin Hospital North. All participants provided written informed
consent.
Patient consent for publication
All participants gave permission for
publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Imamura Y, Morikawa T, Liao X, Lochhead P,
Kuchiba A, Yamauchi M, Qian ZR, Nishihara R, Meyerhardt JA, Haigis
KM, et al: Specific mutations in KRAS codons 12 and 13, and patient
prognosis in 1075 BRAF wild-type colorectal cancers. Clin Cancer
Res. 18:4753–4763. 2012.PubMed/NCBI
|
|
2
|
Xiao W, Zhao H, Dong W, Li Q, Zhu J, Li G,
Zhang S and Ye M: Quantitative detection of methylated NDRG4 gene
as a candidate biomarker for diagnosis of colorectal cancer. Oncol
Lett. 9:1383–1387. 2015.PubMed/NCBI
|
|
3
|
Luo SL, Hu RY, Gong WW, Wang H, Pan J, Fei
FR and Yu M: Survival rate of colorectal cancer patients during
2005–2010 in Zhejiang province, China. Zhonghua Liu Xing Bing Xue
Za Zhi. 34:1194–1197. 2013.(In Chinese). PubMed/NCBI
|
|
4
|
Song LL and Li YM: Current noninvasive
tests for colorectal cancer screening: An overview of colorectal
cancer screening tests. World J Gastrointest Oncol. 8:793–800.
2016.PubMed/NCBI
|
|
5
|
Iannone A, Losurdo G, Pricci M, Girardi B,
Massaro A, Principi M, Barone M, Ierardi E and Di Leo A: Stool
investigations for colorectal cancer screening: From occult blood
test to DNA analysis. J Gastrointest Cancer. 47:143–151.
2016.PubMed/NCBI
|
|
6
|
Ahlquist DA, Harrington JJ, Burgart LJ and
Roche PC: Morphometric analysis of the ‘mucocellular layer’
overlying colorectal cancer and normal mucosa: Relevance to
exfoliation and stool screening. Hum Pathol. 31:51–57.
2000.PubMed/NCBI
|
|
7
|
Brenner H, Hoffmeister M, Arndt V,
Stegmaier C, Altenhofen L and Haug U: Protection from right- and
left-sided colorectal neoplasms after colonoscopy: Population-based
study. J Natl Cancer Inst. 102:89–95. 2010.PubMed/NCBI
|
|
8
|
Lu H, Huang S, Zhang X, Wang D, Zhang X,
Yuan X, Zhang Q and Huang Z: DNA methylation analysis of SFRP2,
GATA4/5, NDRG4 and VIM for the detection of colorectal cancer in
fecal DNA. Oncol Lett. 8:1751–1756. 2014.PubMed/NCBI
|
|
9
|
Renaud S, Guerrera F, Seitlinger J,
Costardi L, Schaeffer M, Romain B, Mossetti C, Claire-Voegeli A,
Filosso PL, Legrain M, et al: KRAS exon 2 codon 13 mutation is
associated with a better prognosis than codon 12 mutation following
lung metastasectomy in colorectal cancer. Oncotarget. 8:2514–2524.
2017.PubMed/NCBI
|
|
10
|
Ross JP, Rand KN and Molloy PL:
Hypomethylation of repeated DNA sequences in cancer. Epigenomics.
2:245–269. 2010.PubMed/NCBI
|
|
11
|
Rasmussen SL, Krarup HB, Sunesen KG,
Pedersen IS, Madsen PH and Thorlacius-Ussing O: Hypermethylated DNA
as a biomarker for colorectal cancer: A systematic review.
Colorectal Dis. 18:549–561. 2016.PubMed/NCBI
|
|
12
|
Zou H, Harrington JJ, Shire AM, Rego RL,
Wang L, Campbell ME, Oberg AL and Ahlquist DA: Highly methylated
genes in colorectal neoplasia: Implications for screening. Cancer
Epidemiol Biomarkers Prev. 16:2686–2696. 2007.PubMed/NCBI
|
|
13
|
Hinoue T, Weisenberger DJ, Lange CP, Shen
H, Byun HM, Van Den Berg D, Malik S, Pan F, Noushmehr H, van Dijk
CM, et al: Genome-scale analysis of aberrant DNA methylation in
colorectal cancer. Genome Res. 22:271–282. 2012.PubMed/NCBI
|
|
14
|
Mitchell SM, Ross JP, Drew HR, Ho T, Brown
GS, Saunders NF, Duesing KR, Buckley MJ, Dunne R, Beetson I, et al:
A panel of genes methylated with high frequency in colorectal
cancer. BMC Cancer. 14:542014.PubMed/NCBI
|
|
15
|
Melotte V, Lentjes MH, van den Bosch SM,
Hellebrekers DM, de Hoon JP, Wouters KA, Daenen KL,
Partouns-Hendriks IE, Stessels F, Louwagie J, et al: N-Myc
downstream-regulated gene 4 (NDRG4): A candidate tumor suppressor
gene and potential biomarker for colorectal cancer. J Natl Cancer
Inst. 101:916–927. 2009.PubMed/NCBI
|
|
16
|
Park H, Kim Y, Lim Y, Han I and Oh ES:
Syndecan-2 mediates adhesion and proliferation of colon carcinoma
cells. J Biol Chem. 277:29730–29736. 2002.PubMed/NCBI
|
|
17
|
Glöckner SC, Dhir M, Yi JM, McGarvey KE,
Van Neste L, Louwagie J, Chan TA, Kleeberger W, de Bruïne AP, Smits
KM, et al: Methylation of TFPI2 in stool DNA: A potential novel
biomarker for the detection of colorectal cancer. Cancer Res.
69:4691–4699. 2009.PubMed/NCBI
|
|
18
|
Oh TJ, Oh HI, Seo YY, Jeong D, Kim C, Kang
HW, Han YD, Chung HC, Kim NK and An S: Feasibility of quantifying
SDC2 methylation in stool DNA for early detection of colorectal
cancer. Clin Epigenetics. 9:1262017.PubMed/NCBI
|
|
19
|
Lao VV and Grady WM: Epigenetics and
colorectal cancer. Nat Rev Gastroenterol Hepatol. 8:686–700.
2011.PubMed/NCBI
|
|
20
|
Gerecke C, Scholtka B, Löwenstein Y, Fait
I, Gottschalk U, Rogoll D, Melcher R, Kleuser B, et al:
Hypermethylation of ITGA4, TFPI2 and VIMENTIN promoters is
increased in inflamed colon tissue: Putative risk markers for
colitis-associated cancer. J Cancer Res Clin Oncol. 141:2097–2107.
2015.PubMed/NCBI
|
|
21
|
Kadiyska T and Nossikoff A: Stool DNA
methylation assays in colorectal cancer screening. World J
Gastroenterol. 21:10057–10061. 2015.PubMed/NCBI
|
|
22
|
Zou H, Taylor WR, Harrington JJ, Hussain
FT, Cao X, Loprinzi CL, Levine TR, Rex DK, Ahnen D, Knigge KL, et
al: High detection rates of colorectal neoplasia by stool DNA
testing with a novel digital melt curve assay. Gastroenterology.
136:459–470. 2009.PubMed/NCBI
|
|
23
|
Ahlquist DA, Zou H, Domanico M, Mahoney
DW, Yab TC, Taylor WR, Butz ML, Thibodeau SN, Rabeneck L, Paszat
LF, et al: Next-generation stool DNA test accurately detects
colorectal cancer and large adenomas. Gastroenterology.
142:248–256. 2012.PubMed/NCBI
|
|
24
|
Green MR and Sambrook J: Hot start
polymerase chain reaction (PCR). Cold Spring Harb Protoc.
2018:2018.
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI
|
|
26
|
Jiang X, Harrington JJ, Mahoney DW, Oberg
AL, Devens ME, Simonson J, Ahlquist DA and Zou H: T1102 detection
of colorectal neoplasia by stool DNA testing: High discrimination
with multi-marker quantitation. Gastroenterology. 134 (Suppl
1):A–484. 2008.
|
|
27
|
Huang Z, Li L and Wang J: Hypermethylation
of SFRP2 as a potential marker for stool-based detection of
colorectal cancer and precancerous lesions. Dig Dis Sci.
52:2287–2291. 2007.PubMed/NCBI
|
|
28
|
Baek YH, Chang E, Kim YJ, Kim BK, Sohn JH
and Park DI: Stool methylation-specific polymerase chain reaction
assay for the detection of colorectal neoplasia in Korean patients.
Dis Colon Rectum. 52:1452–1459. 2009.PubMed/NCBI
|
|
29
|
Bleeker WA, Hayes VM, Karrenbeld A,
Hofstra RM, Hermans J, Buys CC and Plukker JT: Impact of KRAS and
TP53 mutations on survival in patients with left- and right-sided
Dukes' C colon cancer. Am J Gastroenterol. 95:2953–2957.
2000.PubMed/NCBI
|
|
30
|
Zauber AG: The impact of screening on
colorectal cancer mortality and incidence: Has it really made a
difference? Dig Dis Sci. 60:681–691. 2015.PubMed/NCBI
|
|
31
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI
|
|
32
|
Yang D, Hillman SL, Harris AM, Sinicrope
PS, Devens ME and Ahlquist DA: Patient perceptions of stool DNA
testing for pan-digestive cancer screening: A survey questionnaire.
World J Gastroenterol. 20:4972–4979. 2014.PubMed/NCBI
|