Introduction
Colorectal cancer (CRC) is one of the most common
malignancies and the second leading cause of cancer-associated
mortality worldwide accounting for 9.2% of total cases in 2018
(1). The prognosis of patients with
CRC varies according to tumor stage at the time of diagnosis,
whereby ~90% of mortalities are preventable if patients are
diagnosed at an early stage (2).
However, the currently available fecal occult blood test and serum
tumor biomarkers, such as carcinoembryonic antigen (CEA), are
neither highly sensitive nor specific for early diagnosis of CRC
(3). Colonoscopy and tissue biopsy
remain the gold standard for detecting and diagnosing CRC; however,
the invasiveness of colonoscopy limits its use in scanning patients
with CRC (4). Thus, novel promising
diagnostic biomarkers for CRC are required.
microRNAs (miRNAs/miR) are short single-stranded
non-coding RNAs that degrade target mRNA or inhibit its translation
by directly binding to the 3ʹ-untranslated region of targets
(5). Dysregulated miRNAs have been
implicated in several types of cancer, including CRC, and are
associated with tumor development and progression (6–9).
Increasing evidence indicates that cancer cells secrete
intracellular miRNAs into the peripheral blood of patients and the
circulating miRNAs may persist in serum when protected by
particles, such as exosomes (10,11),
which makes circulating miRNAs novel promising diagnostic molecules
of different types of cancer (12–14). For
example, Abu-Duhier et al (15) reported that plasma miR-21 expression
is notably upregulated in patients with lung cancer compared with
healthy controls, thus confirming circulating miR-21 as an
efficient non-invasive biomarker for the screening of patients with
lung cancer. Furthermore, Imaoka et al (16) demonstrated that elevated circulating
miR-1290 may be developed as a novel diagnostic and prognostic
biomarker in human CRC, suggesting that tumor-derived miRNAs used
for diagnosis may improve the specificity of biomarkers.
Overall, the present study aimed to identify novel
circulating miRNAs that differentiate between patients with CRC and
advanced colorectal adenomas (AAs) from healthy individuals, with
notable diagnostic precision.
Materials and methods
Study design
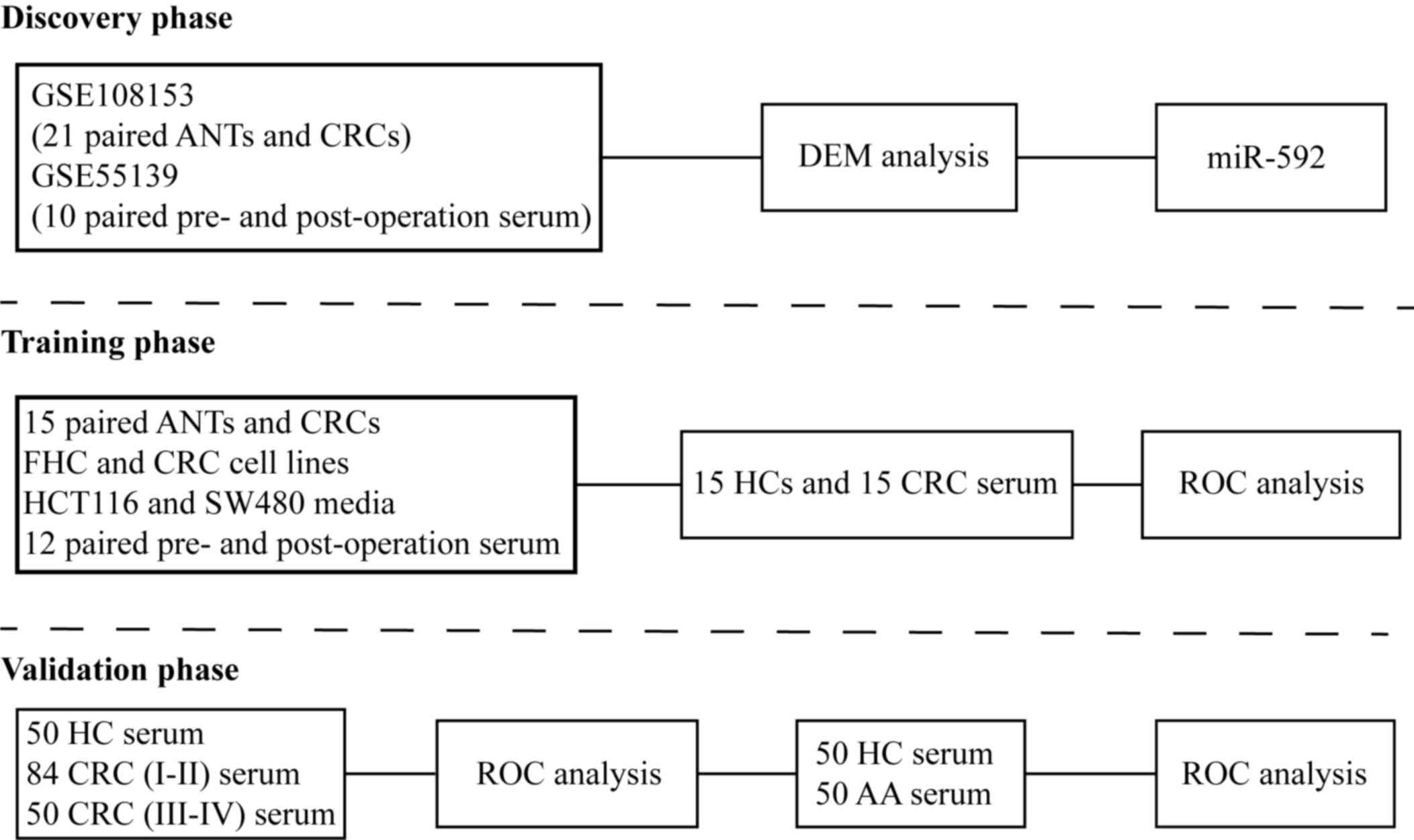
The present study consisted of three phases
(Fig. 1). The discovery phase used
the GSE108153 (17) and GSE55139
(18) datasets, downloaded from the
Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov), in order to identify
CRC tissues and pre-operation serum samples with upregulated
miR-592 expression, which decreased following surgical excision of
the tumor. The GSE108153 dataset consisted of 21-paired CRC tissues
and adjacent normal tissues (ANTs), while the GSE55139 dataset
included 10 paired pre- and post-operative serum samples.
Dysregulated miRNAs were analyzed by using online tool GEO2R
(https://www.ncbi.nlm.nih.gov/geo/geo2r/) in GEO
database according to the instructions. In the training phase,
miR-592 expression was identified in 15 paired CRC tissues and
ANTs, as well as CRC cell lines and fetal human colon (FHC) cells.
Furthermore, 12 paired pre- and post-operative serum samples of
patients with CRC (7 males and 5 females) whose mean age was 63
(range from 45 to 79) were implemented to validate the source of
serum miR-592. Subsequently, 30 serum samples collected from 15
healthy individuals (8 males and 7 females) and 15 patients with
CRC (9 males and 6 females) were used to measure serum miR-592
expression and its diagnostic value. The mean age of healthy
individuals and CRC patients were 59 (range from 43 to 78) and 61
(range from 44 to 80), respectively. In the validation phase,
another independent cohort with a larger number of serum samples
collected from; 50 healthy controls (HCs) (34 males and 26
females), 84 patients with stages I–II CRC (51 males and 33
females), 50 patients with stages III–IV CRC (37 males and 13
females) and 50 patients with advanced colorectal adenomas (32
males and 18 females) was implemented to confirm the diagnostic
value of serum miR-592. The mean age of HCs, patients with CRC and
patients with advanced colorectal adenomas were 59 (range from 41
to 79), 62 (range from 43 to 81) and 60 (range from 41 to 78),
respectively.
Study population
The present study was approved by the Research and
Ethical Committee at the Second Affiliated Hospital of Nanjing
Medical University (Nanjing, China) and written informed consent
was provided by all patients prior to the study start. Diagnosis of
CRC and AAs was histologically confirmed by two independent
pathologists from Department of Pathology of the Second Affiliated
Hospital of Nanjing Medical University via analysis of resected
tumors following surgery and colonoscopy examination, and tumor
stage was determined according to the tumor-node-metastasis (TNM)
system (19). Patients with any
anti-tumor treatment before specimen collection, such as
chemoradiotherapy, were excluded. The detailed characteristics of
patients with CRC were downloaded from the medical record system of
the Second Affiliated Hospital of Nanjing Medical University and
are presented in Table I. The
post-operative serum samples of patients with CRC were collected
one week after surgical excision of the tumors. HCs were collected
from age-matched volunteers who participated in the routine
physical examination. There was no difference in age and sex among
patients with CRC, AAs and the HCs.
 | Table I.Association between serum miR-592
expression and clinicopathological characteristics in patients with
colorectal cancer (n=134). |
Table I.
Association between serum miR-592
expression and clinicopathological characteristics in patients with
colorectal cancer (n=134).
|
|
| miR-592
expression |
|
|---|
|
|
|
|
|
|---|
| Characteristic | Patient, n | High | Low | P-value |
|---|
| Age, years |
|
|
|
|
|
<65 | 53 | 24 | 29 |
|
| ≥65 | 81 | 43 | 38 | 0.480 |
| Sex |
|
|
|
|
| Male | 88 | 46 | 42 |
|
|
Female | 46 | 21 | 25 | 0.590 |
| Location |
|
|
|
|
|
Colon | 98 | 51 | 47 |
|
|
Rectum | 36 | 16 | 20 | 0.560 |
| Tumor size, cm |
|
|
|
|
|
<5 | 60 | 23 | 37 |
|
| ≥5 | 74 | 44 | 30 | 0.024 |
|
Differentiation |
|
|
|
|
|
High/middle | 65 | 27 | 38 |
|
|
Low | 69 | 40 | 29 | 0.110 |
| TNM stage |
|
|
|
|
|
I–II | 84 | 35 | 49 |
|
| III–IV | 50 | 32 | 18 | 0.020 |
| Lymphatic
metastasis |
|
|
|
|
| No | 65 | 29 | 36 |
|
|
Yes | 69 | 38 | 31 | 0.300 |
| Distant
metastasis |
|
|
|
|
| No | 63 | 25 | 38 |
|
|
Yes | 71 | 42 | 29 | 0.027 |
Sample processing
The CRC cell lines (HCT8, HT-29, HCT116, SW480 and
SW620) and normal FHC cells were purchased from Shanghai Institute
of Biological Sciences. Cells were cultured in DMEM supplemented
with 10% FBS (Thermo Fisher Scientific, Inc.) at 37°C in 5%
CO2. Following histological confirmation, tissue samples
were immediately stored in liquid nitrogen (−196°C), while serum
samples collected from venous blood were centrifuged at 3,500 × g
at 4°C for 10 min and stored at −80°C for subsequent
experimentation. All cell lines were authenticated via the Short
Tandem Repeat profiling method.
Cell transfection
miR-592 inhibitor (5′-ACATCATCGCATATTGACACAA-3′) and
corresponding non-targeting sequence (5′-TTCTCCGAACGTGTCACGTTTC-3′)
were obtained from Shanghai GenePharma Co., Ltd. HCT116 and SW480
cells at 5×105 density were transfected with miR-592
inhibitor or corresponding non-targeting sequence at a final
concentration of 100 µM by using Lipofectamine™ 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instruction. CRC cells that were transfected with
miR-592 inhibitor and corresponding non-targeting sequence were
classified as the inhibitor group and negative control (NC) group,
respectively. After transfection, the expression of miR-592 in the
media of CRC cells were assessed by RT-qPCR every 12 h. The
differential expression of miR-592 between these two groups was
analyzed 48 h after transfection.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from CRC tissues, sera and
media using TRIzol® LS reagent (Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol.
Caenorhabditis elegans miR-39 (Shanghai GenePharma Co.,
Ltd.) was added to each serum sample at a final concentration of
1×10−4 pmol/µl, which served as the external reference.
RT-qPCR was performed using the Hairpin-it™ miRNA RT-PCR
Quantitation kit (Shanghai GenePharma Co., Ltd.), according to the
manufacturer's protocol. The primer sequences were as follows:
miR-592, forward: 5′-ACGTTGTGTCAATATGCGATGA-3′ and reverse:
5′-GTGCAGGGTCCGAGGT-3′; miR-39, forward:
5′-ATATCATCTCACCGGGTGTAATC-3′, and reverse:
5′-TATGGTTTTGACGACTGTGTGAT-3′. The following thermocycling
conditions used for qPCR were as follows: Initial denaturation at
95°C for 3 min, 40 cycles of denaturation at 95°C for 15 sec,
annealing and elongation at 62°C for 34 sec. Relative miR-592
expression was measured using the 2−ΔΔCq method
(20) and normalized to the internal
reference gene, U6 small nuclear RNA. The primer sequences of U6
were as follows: Forward: 5′-CTCGCTTCGGCAGCACA-3′ and reverse:
5′-AACGCTTCACGAATTTGCGT-3′.
Statistical analysis
Statistical analysis was performed using SPSS
(version 22.0; SPSS, Inc.) and GraphPad Prism (version 8; GraphPad
Software, Inc.) software. The association between miR-592
expression and clinicopathological characteristics was assessed
using the χ2 test. Differential expression of miR-592
was determined using Student's paired or unpaired t-test. The
comparisons among multiple groups were analyzed using Tukey's post
hoc test. The receiver operating characteristic (ROC) curve and
area under the curve (AUC) were established to determine the
diagnostic value of serum miR-592. Cut-off values of serum miR-592
were determined using Youden's index. P<0.05 was considered to
indicate a statistically significant difference.
Results
Dysregulated serum miR-592 expression
may be a tumor-derived miRNA in patients with CRC
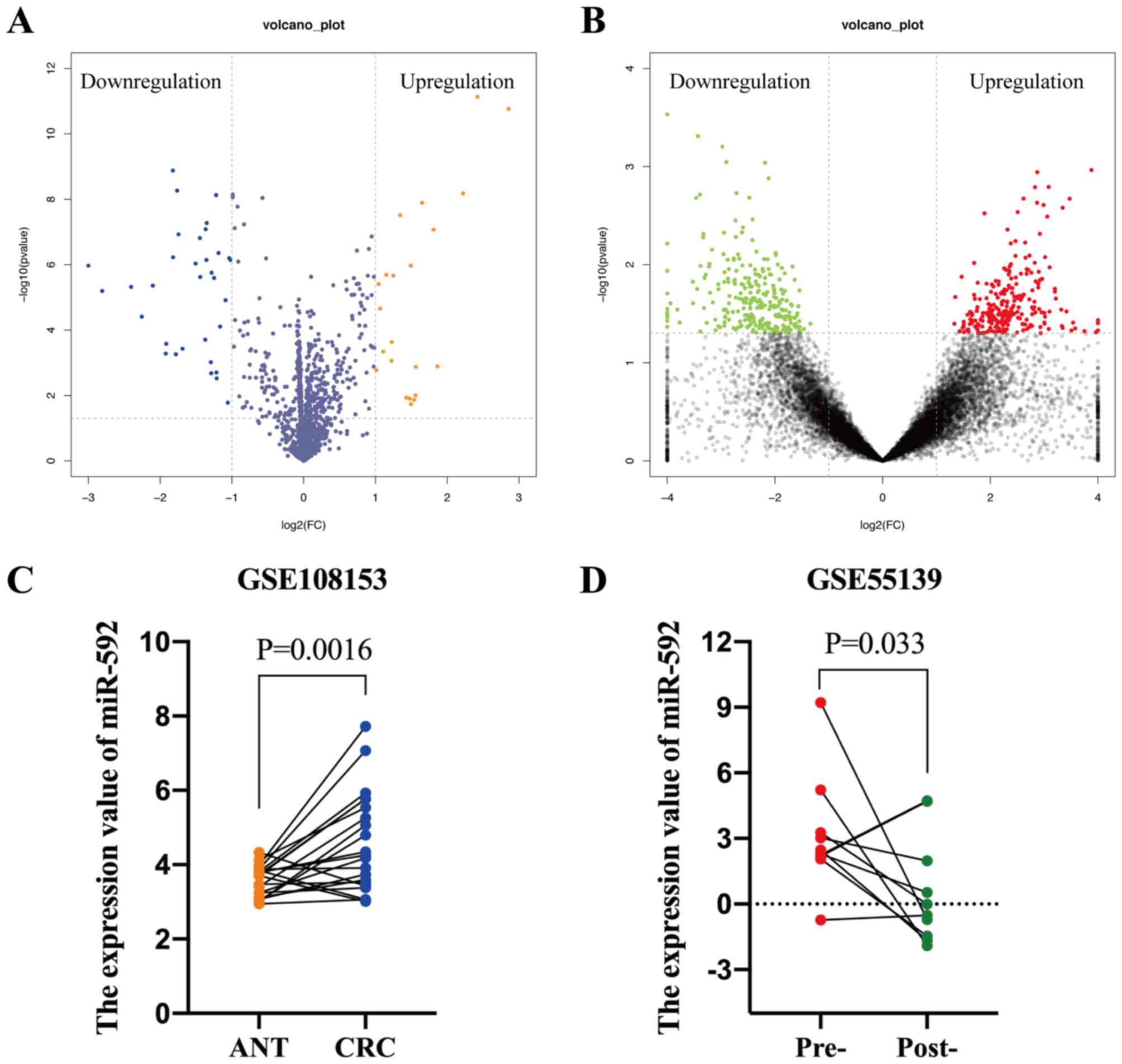
The GEO database was searched using keywords, such
as ‘miRNA’ and ‘colorectal cancer’, in order to identify notably
differentially expressed miRNAs (DEMs) in CRC, of which two
datasets were acquired. The GSE108153 dataset [Agilent-046064
Unrestricted_Human_miRNA_V19.0_Microarray (GPL19730) platform]
(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE108153)
contained 21-paired CRC tissues and ANTs. The GEO2R online tool was
used to analyze the DEMs, which identified 22 upregulated and 33
downregulated miRNAs in CRC tissues compared with ANTs,
respectively (Fig. 2A). The GSE55139
dataset [Agilent-021827 Human miRNA Microarray G4470C (GPL14767)
platform] (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi), which
consisted of 10-paired pre- and post-operative serum samples of
patients with CRC, was used to determine whether the elevated
miRNAs were secreted into the peripheral blood by tumor cells.
Notably, 248 miRNAs were downregulated following surgical resection
of the tumor tissues (Fig. 2B).
Furthermore, miR-592 expression levels were significantly
upregulated in CRC tissues (P=0.0016) and pre-operative serum
samples (P=0.033), which decreased following surgical excision
(Fig. 2C and D). Taken together,
these results suggest that dysregulated serum miR-592 may be a
tumor-derived miRNA in patients with CRC.
Tumor-derived miR-592 is notably
elevated in the serum of patients with CRC
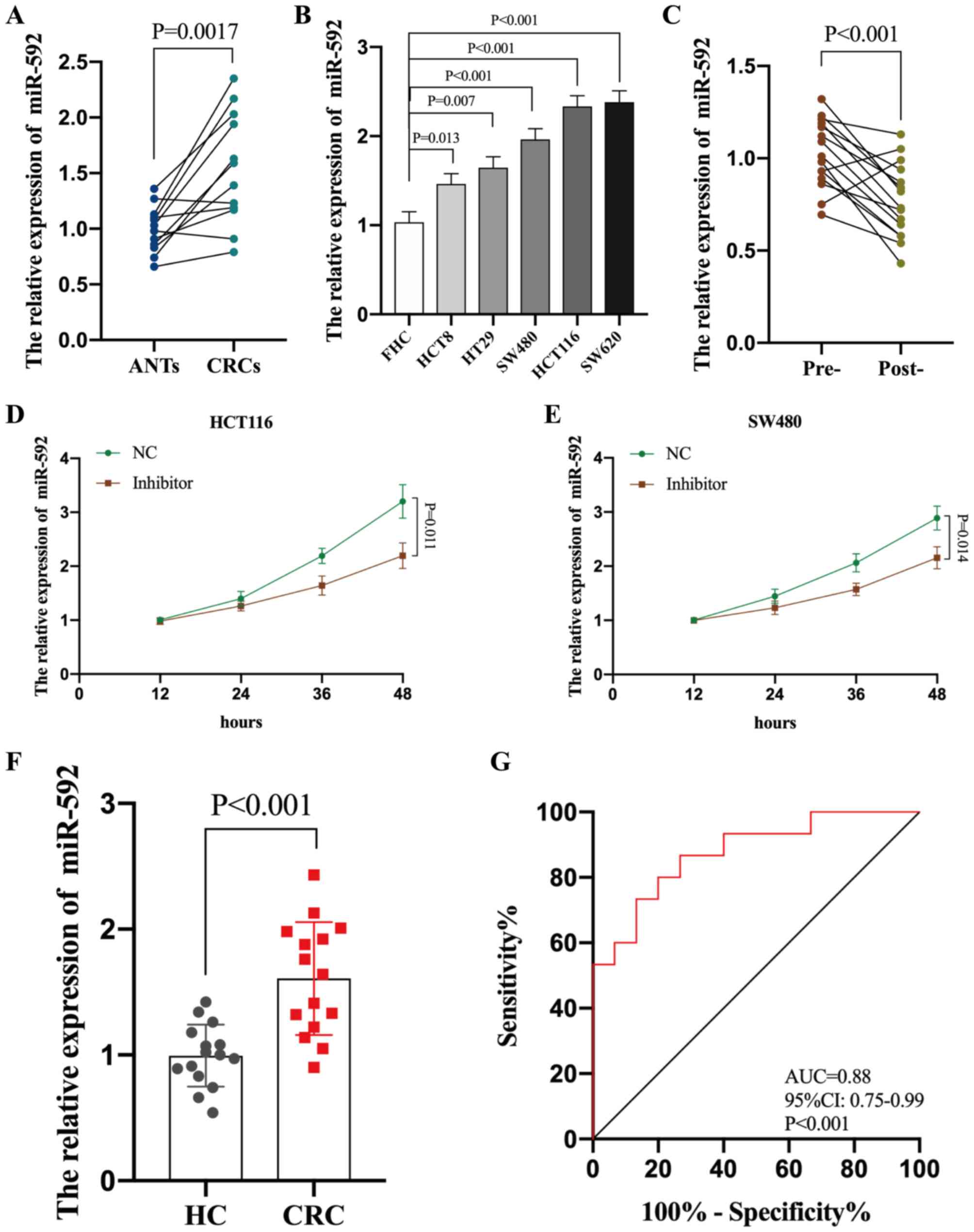
miR-592 expression was determined across all CRC
tissues and cell lines. RT-qPCR analysis demonstrated that miR-592
expression was significantly upregulated in both CRC tissues and
cell lines compared with ANTs and FHC cells, respectively (all
P<0.05) (Fig. 3A and B).
Subsequently, miR-592 expression was assessed in the 10 paired pre-
and post-operative serum samples of patients with CRC (P<0.001),
which demonstrated that miR-592 expression significantly decreased
following surgical excision of the tumors (Fig. 3C). miR-592 expression was also
analyzed in cultured media of CRC cells (HCT116 and SW480 cells).
Increased expression of miR-592 in media was dependent on time in
culture, and CRC cells released less miR-592 into media after
intracellular suppression with miR-592 inhibitor (P<0.05,
Fig. 3D and E).
Increasing evidence suggests that several
tumor-derived miRNAs are significantly dysregulated in the
peripheral blood of patients which can be used to differentiate
patients from healthy individuals, with a high diagnostic value
(21–23). A total of 30 serum samples collected
from 15 healthy individuals and 15 patients with CRC were analyzed
to determine whether serum miR-592 expression may be used to
diagnose patients with CRC. RT-qPCR analysis indicated that serum
miR-592 expression was significantly upregulated in patients with
CRC compared with HCs (P<0.001) (Fig.
3F). Furthermore, ROC analysis demonstrated that serum miR-592
expression may be used to differentiate patients with CRC from HCs,
with high sensitivity (86.6%) and specificity (73.4%), with an AUC
value of 0.88 (95% CI, 0.75–0.99; P<0.001) (Fig. 3G). Taken together, these results
suggest that elevated serum miR-592 expression may be a novel and
potential diagnostic biomarker for patients with CRC.
Serum miR-592 is a novel potential
biomarker for early diagnosis of CRC
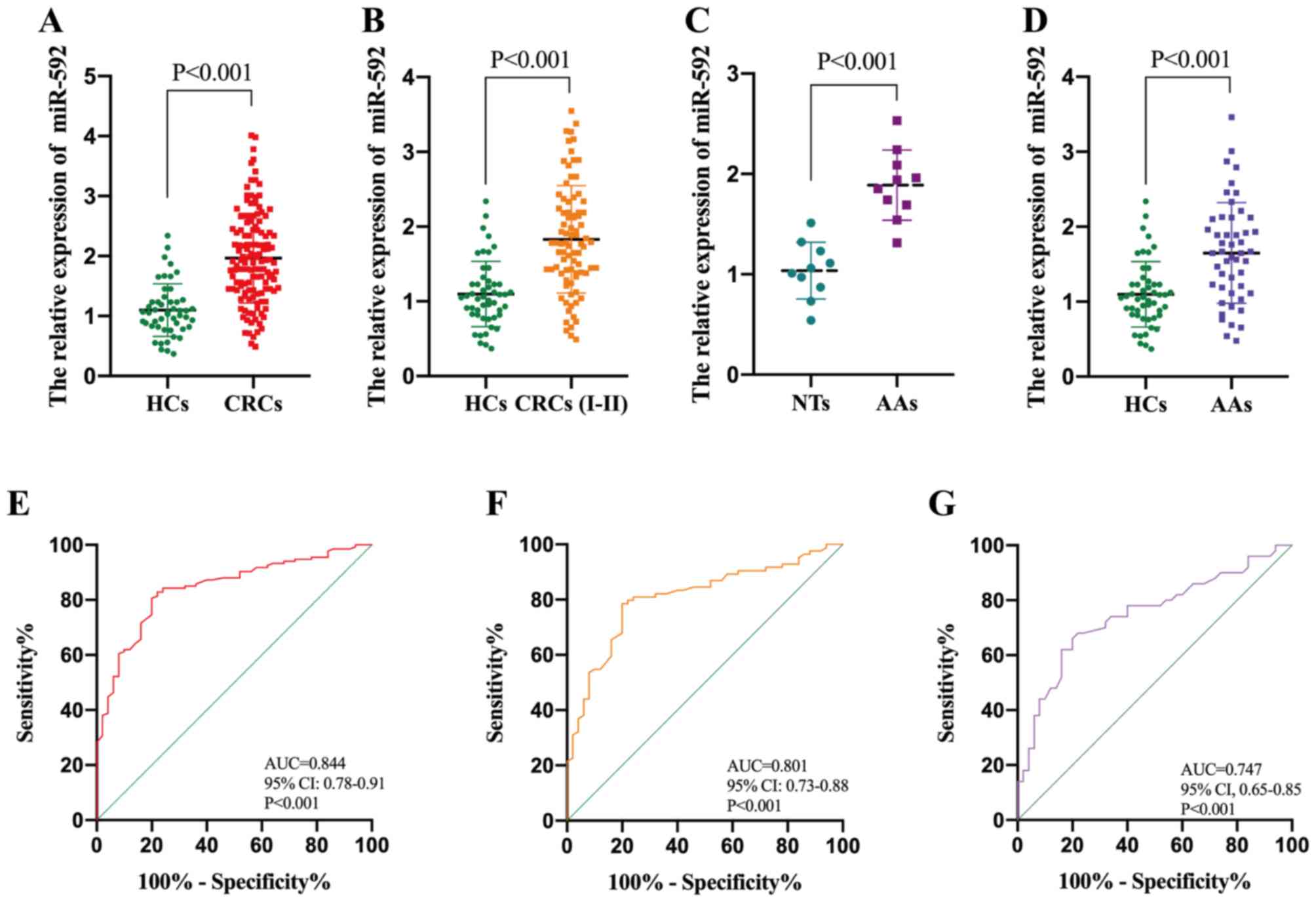
In order to validate the diagnostic value of serum
miR-592 in CRC, another independent cohort containing 134 patients
with CRC and 50 HCs was assessed. Consistently, serum miR-592
expression was significantly upregulated in patients with CRC
compared with HCs (Fig. 4A). ROC
analysis demonstrated that serum miR-592 expression may be used to
differentiate patients with CRC from HCs, with high sensitivity
(82.8%) and specificity (78.0%), and an AUC value of 0.844 (95% CI,
0.78–0.91; P<0.001) (Fig. 4E). In
addition, patients with CRC were classified into high group and low
groups, according to the median value of miR-592 expression (1.91).
The association between serum miR-592 expression and
clinicopathological characteristics of patients with CRC indicated
that elevated serum miR-592 expression was significantly associated
with large tumor size, advanced TNM stage and distant metastasis
(Table I). It has been reported that
~90% of CRC-associated mortalities are preventable if patients are
diagnosed at an early stage (24).
Thus, miR-592 expression levels in the serum of HCs and patients
with stages I–II of CRC were analyzed, in order to determine the
value of serum miR-592 as an early diagnostic biomarker for CRC.
The results demonstrated that miR-592 expression increased in the
peripheral blood of patients with stages I–II of CRC (Fig. 4B), which may be used to differentiate
patients at an early stage of CRC from HCs, with high sensitivity
(78.6%) and specificity (80.0%), and an AUC value of 0.801 (95% CI,
0.73–0.88; P<0.001) (Fig.
4F).
CRC typically develops in a progressive manner, from
normal colon epithelial cells, to adenomas and ultimately to
malignant cancer lesions (25). This
led to investigating the association between serum miR-592
expression and patients with AAs. RT-qPCR analysis demonstrated
that miR-592 expression was significantly upregulated in AA tissues
compared with normal tissues (NTs) (Fig.
4C). Furthermore, serum miR-592 expression was significantly
upregulated in patients with AA compared with HCs (Fig. 4D). ROC analysis indicated that serum
miR-592 expression may be used to differentiate patients with AA
from HCs, with high sensitivity (68.6%) and specificity (78.1%),
and an AUC value of 0.747 (95% CI, 0.65–0.85; P<0.001) (Fig. 4G). There was no difference in age,
sex and drinking status among the patients with AA and CRC
patients. Taken together, these results suggest that serum miR-592
is a potential biomarker for early diagnosis of CRC.
Discussion
The present study identified serum miR-592 as a
tumor-derived miRNA, which was significantly upregulated in
patients with CRC and AA. The results of the present study suggest
that circulating miR-592 may be used to differentiate patients with
CRC and AA from healthy individuals, with high value. Thus, serum
miR-592 is implicated as a novel potential biomarker for early
diagnosis of patients with CRC.
The biological impact of miR-592 has been reported
across several malignancies, including breast, gastric and
non-small cell lung cancer (26–28);
however, whether miR-592 takes on the role of an oncogene or tumor
suppressor is dependent on the tumor context. For example, miR-592
has been reported to be significantly downregulated in glioma,
suppressing the development of glioma by regulating Rho-associated
protein kinase (29). However, He
et al (27) demonstrated that
miR-592 is upregulated in gastric cancer (GC), promoting GC cell
proliferation, migration and invasion, while inducing
endothelial-to-mesenchymal transition via the phosphoinositide
3-kinase/AKT and mitogen-activated protein kinase/extracellular
signal-regulated kinase signaling pathways. miR-592 has been
reported to function as an oncogene in CRC (30), whereby upregulation of miR-592 is
associated with poor prognosis in patients with CRC (31). Consistent with the results of the
present study, Liu et al (31) also reported that miR-592 expression
is upregulated in clinical CRC serum samples. To the best of our
knowledge, the role of serum miR-592 as a novel diagnostic
biomarker for CRC has not been previously investigated. Using
independent cohorts, the present study demonstrated that serum
miR-592 may be used to differentiate patients at early stages of
CRC and patients with AA from HCs, with high diagnostic value.
Furthermore, the sensitivity and specificity of serum CEA (55 and
66%), CA19-9 (36 and 71%) and CA72-4 (25 and 66%) (32) are lower than those for serum miR-592,
respectively. Since dysregulated miR-592 in CRC tissues was
associated with poor prognosis of patients with CRC and elevated
serum miR-592 was demonstrated to be tumor-derived (30), it is hypothesized that serum miR-592
may have the ability to predict the prognosis of patients with CRC
in a non-invasive manner.
The present study posed several limitations. First,
the number of clinical samples was small. Prospective studies with
larger sample sizes are required to verify the function of
circulating miR-592 as a novel diagnostic biomarker for CRC.
Furthermore, the clinical data of patients with CRC, particularly
regarding the carcinoembryonic antigen, CA19-9 and CA72-4 were
limited. It is speculated that the combination of currently
available tumor biomarkers with miR-592 may improve the diagnostic
value or sensitivity and specificity for patients with CRC.
Previous studies have reported that tumor cells secrete miRNAs into
circulation via exosomes (33–35);
however, this phenomenon was not investigated in the present study.
Thus, future studies will aim to determine whether CRC cells have
the ability to release miR-592.
In conclusion, the results of the present study
suggest that serum miR-592 may be implicated as a novel potential
biomarker for the early and non-invasive detection of CRC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
present study are available in the GEO repository, [http://www.ncbi.nlm.nih.gov/geo].
Authors' contributions
LM designed the present study and drafted the
initial manuscript, while ZP acquired the clinical samples and
performed RT-qPCR. Both LM and ZP performed statistical analysis.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research and
Ethical Committee at Second Affiliated Hospital of Nanjing Medical
University (approval no. 2015-KY-040, Nanjing, China) and written
informed consent was provided by all patients prior to the study
start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Smith RA, von Eschenbach AC, Wender R,
Levin B, Byers T, Rothenberger D, Brooks D, Creasman W, Cohen C,
Runowicz C, et al: American Cancer Society guidelines for the early
detection of cancer: Update of early detection guidelines for
prostate, colorectal, and endometrial cancers. Also: Update
2001-testing for early lung cancer detection. CA Cancer J Clin.
51:38–75; quiz 77–80. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Carpelan-Holmstrom M, Louhimo J, Stenman
UH, Alfthan H, Jarvinen H and Haglund C: CEA, CA 242, CA 19-9, CA
72-4 and hCGbeta in the diagnosis of recurrent colorectal cancer.
Tumour Biol. 25:228–234. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hassan C, Pickhardt PJ, Laghi A, Kim DH,
Zullo A, Iafrate F, Di Giulio L and Morini S: Computed tomographic
colonography to screen for colorectal cancer, extracolonic cancer,
and aortic aneurysm: Model simulation with cost-effectiveness
analysis. Arch Intern Med. 168:696–705. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu X, Chen X, Zeng K, Xu M, He B, Pan Y,
Sun H, Pan B, Xu X and Xu T: DNA-methylation-mediated silencing of
miR-486-5p promotes colorectal cancer proliferation and migration
through activation of PLAGL2/IGF2/β-catenin signal pathways. Cell
Death Dis. 9:10372018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Han LC, Wang H, Niu FL, Yan JY and Cai HF:
Effect miR-214-3p on proliferation and apoptosis of breast cancer
cells by targeting survivin protein. Eur Rev Med Pharmacol Sci.
23:7469–7474. 2019.PubMed/NCBI
|
|
7
|
Wei YQ, Jiao XL, Zhang SY, Xu Y, Li S and
Kong BH: MiR-9-5p could promote angiogenesis and radiosensitivity
in cervical cancer by targeting SOCS5. Eur Rev Med Pharmacol Sci.
23:7314–7326. 2019.PubMed/NCBI
|
|
8
|
Wu HY, Wei Y and Pan SL: Down-regulation
and clinical significance of miR-7-2-3p in papillary thyroid
carcinoma with multiple detecting methods. IET Syst Biol.
13:225–233. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
An HJ, Park M, Kim J and Han YH: miR5191
functions as a tumor suppressor by targeting RPS6KB1 in colorectal
cancer. Int J Oncol. Aug 30–2019.(Epub ahead of print). View Article : Google Scholar
|
|
10
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tang Y, Zhao Y, Song X, Song X, Niu L and
Xie L: Tumor-derived exosomal miRNA-320d as a biomarker for
metastatic colorectal cancer. J Clin Lab Anal. 33:e230042019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Sui J, Shen X, Li C, Yao W, Hong
W, Peng H, Pu Y, Yin L and Liang G: Differential expression
profiles of microRNAs as potential biomarkers for the early
diagnosis of lung cancer. Oncol Rep. 37:3543–3553. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu X, Xu T, Hu X, Chen X, Zeng K, Sun L
and Wang S: Elevated circulating miR-182 acts as a diagnostic
biomarker for early colorectal cancer. Cancer Manag Res.
10:857–865. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Usuba W, Urabe F, Yamamoto Y, Matsuzaki J,
Sasaki H, Ichikawa M, Takizawa S, Aoki Y, Niida S, Kato K, et al:
Circulating miRNA panels for specific and early detection in
bladder cancer. Cancer Sci. 110:408–419. 2019.PubMed/NCBI
|
|
15
|
Abu-Duhier FM, Javid J, Sughayer MA, Mir
R, Albalawi T and Alauddin MS: Clinical significance of circulatory
miRNA-21 as an efficient non-invasive biomarker for the screening
of lung cancer patients. Asian Pac J Cancer Prev. 19:2607–2611.
2018.PubMed/NCBI
|
|
16
|
Imaoka H, Toiyama Y, Fujikawa H, Hiro J,
Saigusa S, Tanaka K, Inoue Y, Mohri Y, Mori T, Kato T, et al:
Circulating microRNA-1290 as a novel diagnostic and prognostic
biomarker in human colorectal cancer. Ann Oncol. 27:1879–1886.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lu JH, Zuo ZX, Wang W, Zhao Q, Qiu MZ, Luo
HY, Chen ZH, Mo HY, Wang F, Yang DD, et al: A two-microRNA-based
signature predicts first-line chemotherapy outcomes in advanced
colorectal cancer patients. Cell Death Discov. 4:1162018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nonaka R, Nishimura J, Kagawa Y, Osawa H,
Hasegawa J, Murata K, Okamura S, Ota H, Uemura M, Hata T, et al:
Circulating miR-199a-3p as a novel serum biomarker for colorectal
cancer. Oncol Rep. 32:2354–2358. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ho AS, Kim S, Tighiouart M, Gudino C, Mita
A, Scher KS, Laury A, Prasad R, Shiao SL, Ali N, et al: Association
of quantitative metastatic lymph node burden with survival in
hypopharyngeal and laryngeal cancer. JAMA Oncol. 4:985–989. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zou X, Li M, Huang Z, Zhou X, Liu Q, Xia T
and Zhu W: Circulating miR-532-502 cluster derived from chromosome
X as biomarkers for diagnosis of breast cancer. Gene.
722:1441042020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
D'Antona P, Cattoni M, Dominioni L, Poli
A, Moretti F, Cinquetti R, Gini E, Daffre E, Noonan DM, Imperatori
A, et al: Serum miR-223: A validated biomarker for detection of
early-stage non-small cell lung cancer. Cancer Epidemiol Biomarkers
Prev. 28:1926–1933. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cui Q: Significance of miR-27a and miR-31
in early diagnosis and prognosis of colorectal cancer. Oncol Lett.
18:3092–3096. 2019.PubMed/NCBI
|
|
24
|
Smith RA, Andrews KS, Brooks D, Fedewa SA,
Manassaram-Baptiste D, Saslow D, Brawley OW and Wender RC: Cancer
screening in the United States, 2017: A review of current American
Cancer Society guidelines and current issues in cancer screening.
CA Cancer J Clin. 67:100–121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang J, Raju GS, Chang DW, Lin SH, Chen Z
and Wu X: Global and targeted circulating microRNA profiling of
colorectal adenoma and colorectal cancer. Cancer. 124:785–796.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hou W, Zhang H, Bai X, Liu X, Yu Y, Song L
and Du Y: Suppressive role of miR-592 in breast cancer by
repressing TGF-β2. Oncol Rep. 38:3447–3454. 2017.PubMed/NCBI
|
|
27
|
He Y, Ge Y, Jiang M, Zhou J, Luo D, Fan H,
Shi L, Lin L and Yang L: MiR-592 promotes gastric cancer
proliferation, migration, and invasion through the PI3K/AKT and
MAPK/ERK signaling pathways by targeting spry2. Cell Physiol
Biochem. 47:1465–1481. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Z, Li B, Niu L and Ge L: miR-592
functions as a tumor suppressor in human non-small cell lung cancer
by targeting SOX9. Oncol Rep. 37:297–304. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gao S, Chen J, Wang Y, Zhong Y, Dai Q,
Wang Q and Tu J: MiR-592 suppresses the development of glioma by
regulating Rho-associated protein kinase. Neuroreport.
29:1391–1399. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fu Q, Du Y, Yang C, Zhang D, Zhang N, Liu
X, Cho WC and Yang Y: An oncogenic role of miR-592 in tumorigenesis
of human colorectal cancer by targeting Forkhead Box O3A (FoxO3A).
Expert Opin Ther Targets. 20:771–782. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu M, Zhi Q, Wang W, Zhang Q, Fang T and
Ma Q: Up-regulation of miR-592 correlates with tumor progression
and poor prognosis in patients with colorectal cancer. Biomed
Pharmacother. 69:214–220. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Carpelan-Holmstrom M, Louhimo J, Stenman
UH, Alfthan H and Haglund C: CEA, CA 19-9 and CA 72-4 improve the
diagnostic accuracy in gastrointestinal cancers. Anticancer Res.
22:2311–2316. 2002.PubMed/NCBI
|
|
33
|
Sun L, Liu X, Pan B, Hu X, Zhu Y, Su Y,
Guo Z, Zhang G, Xu M, Xu X, et al: Serum exosomal miR-122 as a
potential diagnostic and prognostic biomarker of colorectal cancer
with liver metastasis. J Cancer. 11:630–637. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J,
Zhou K, Liu X, Ren X, Wang F, et al: Cancer-derived exosomal
miR-25-3p promotes pre-metastatic niche formation by inducing
vascular permeability and angiogenesis. Nat Commun. 9:53952018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu B, Su S, Patil DP, Liu H, Gan J,
Jaffrey SR and Ma J: Molecular basis for the specific and
multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun. 9:4202018. View Article : Google Scholar : PubMed/NCBI
|