Introduction
Cervical cancer (CC) is a prevalent gynecological
malignancy with poor prognosis globally (1). In recent years, despite improvements in
the diagnosis and therapeutic strategies for CC, patients suffering
from CC still presented metastasis and relapse, which remain the
main reasons for tumor-related deaths (2). However, the underlying mechanisms of CC
development have not yet been sufficiently elucidated. Therefore,
further understanding of the mechanism on CC occurrence and
development is urgently imperative to identify novel diagnostic and
therapeutic biomarkers for CC patients. Emerging literature has
demonstrated that microRNAs (miRNAs) are emerging as tumor
regulators of multiple tumors (3,4).
miRNAs may serve as key regulators of gene
expression via targeting 3-untranslated regions (3′-UTRs) of target
genes, leading to the inhibition of protein translation or mRNA
degradation. Moreover, recently, miRNAs have been verified to play
pivotal functions in various events which are involved in
oncogenesis, such as cell apoptosis, proliferation, survival, and
metastasis, through repressing expression of their targets. For
example, Fang et al (5)
proposed that miR-21 enhanced triple-negative breast cancer cell
proliferation and invasion through regulating PTEN; Xiao et
al (6) reported that miR-144
suppressed colorectal cancer proliferation and migration through
GSPT1; Yang et al (7) claimed
that miR-203 inhibited gastric carcinoma cell proliferation,
migration and invasion via targeting Slug. However, the precise
functions of miR-100 in CC require further investigation.
Therefore, the present study was performed to confirm the roles of
miR-100 in CC carcinogenesis.
Epithelial-to-mesenchymal transition (EMT) is a
notable process involved in tumor-associated metastases and
invasion (8). In the progress of
EMT, obvious changes on cell adhesion, polarities, and motile
property have been confirmed. In general, EMT is typically featured
by the upregulation of mesenchymal marker and downregulation of
epithelial marker (9). Moreover, EMT
is considered to be one of the critical steps in the metastatic
cascades of multiple malignant tumors, including hepatocellular
carcinoma (10), prostate carcinoma
(11) and renal cell carcinoma
(12). In addition, the AKT/mTOR
signalling pathway has essential roles in basic cellular processes,
including cell apoptosis, differentiation and growth, exerting
oncogenic functions in tumorigenesis of different malignancies
(13,14). Thus, in the present study, it was
investigated whether miR-100 regulated CC progression through
regulation of EMT and AKT/mTOR pathway.
Special AT-rich sequence-binding protein 1 (SATB1)
is a nuclear matrix-associated protein and implicated in regulating
tissue-specific gene expression, having emerged as a novel
modulator of oncogenic pathways (15). SATB1 has been reported to be involved
in the metastasis and growth of numerous malignant tumors. For
example, Qi et al (16)
indicated that SATB1 promoted EMT and metastases in prostate
cancer; Li et al (17) found
that SATB1 facilitated tumor oral squamous cell carcinoma
metastases and invasiveness; studies by Pan et al (18) demonstrated that SATB1 was correlated
with metastasis and progression of breast carcinoma. These studies
suggested that SATB1 exerted oncogenic roles in tumor progression.
However, the detailed roles of SATB1 in CC still need to be further
elucidated.
Patients and methods
CC tissue specimens
Fifty-eight pairs of CC tissues and matched adjacent
normal tissues were collected from CC patients who underwent
surgical resection in the Second Hospital of Shandong University
(Jinan, China) between April 2015 and October 2017. The CC patients
received no treatments before tissue collection. All patients
involved in the present study provided written informed consent.
The fresh tissue sample was frozen in liquid nitrogen immediately,
then stored at −80°C for further assays. The present study was
approved by the Ethics Committee of the Second Hospital of Shandong
University.
CC cell lines and cell cultures
Human CC cells (C-33A, HeLa, SiHa and Ca-Ski) and
normal cervical epithelial cell line (Ect1-E6E7) were purchased
from the Committee on Type Culture Collection of the Chinese
Academy of Sciences (Shanghai, China). All the cells were
maintained in Dulbeccos modified Eagles medium (DMEM) with 10%
fetal bovine serum (FBS) in a humidified incubator containing 5%
CO2 at 37°C.
Cell transfections
miR-100 mimics, inhibitor or negative controls (NC)
were obtained from GenePharma. Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) was utilized to transfect them into
CC cells according to the manufacturer's proposal.
qRT-PCR
Total RNAs were isolated from the cultured cells and
tissue specimens using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), followed by reverse transcription into cDNA by
PrimeScript RT reagent kit (Takara Biotechnology, Co., Ltd.).
qRT-PCR was conducted with SYBR®-Green PCR Master Mix
(Takara Biotechnology, Co., Ltd.) on an ABI 7500 system (Applied
Biosystems). The relative expression levels of genes were detected
with the 2−ΔΔCt method. Expression of candidate genes
was normalized to that of glyceraldehyde-3-phosphate dehydrogenase
(GAPDH) whereas U6 was an endogenous control for miR-100. The
sequences of the primers are shown in Table I.
 | Table I.Primer sequences for qRT-PCR. |
Table I.
Primer sequences for qRT-PCR.
| Primer | Sequence |
|---|
| miR-100 | F:
5′-ACACTCCAGCTGGGAACCCGTAGATCCGAAC-3′ |
| miR-100 | R:
5′-TGGTGTCGTGGAGTCG-3′ |
| U6 | F:
5′-CTCGCTTCGGCAGCACA-3′ |
| U6 | R:
5′-AACGCTTCACGAATTTGCGT-3′ |
| SATB1 | F:
5′-GTGGAAGCCTTGGGAATCC-3′ |
| SATB1 | R:
5′-CTGACAGCTCTTCTTCTAGTT-3′ |
| GAPDH | F:
5′-GGCCAAGGTCATCCATGACAA-3′ |
| GAPDH | R:
5′-TCTTCTGACACCTACCGGGGA-3′ |
Immunohistochemistry (IHC)
For IHC assays, CC tissues and adjacent normal
tissues were formalin-fixed, paraffin-embedded and cut into 4 µm
sections. Then, the slides were deparaffinized in xylene and
rehydrated through a descending series of ethanol. Antigen
retrieval was performed with citrate buffer in microwave oven.
Subsequently, slides were blocked with 3% hydrogen peroxidase in
methanol and 10% goat serum. Slides were incubated with SATB1
antibody (1:200; ab109122; Abcam) overnight at 4°C and then with
HRP-labeled goat anti-rabbit IgG (1:2,000, ab205718; Abcam) for 30
min at room temperature. The slides were stained with DAB as the
chromogen and counterstained with hematoxylin. Then, a bright-field
microscope (Olympus BX50; Olympus Corporation) was used to image
and analyze the slides. Five visual fields were randomly chosen
from each section by double-blind method. The double-blind method
was utilized to randomly select 5 visual fields from each section.
The expression levels were determined following the ratio of
positive cells:stained cells. The cells <25% were considered as
negative (−) while the ratio >25% was positive (+) (19,20).
Cell proliferation assay
MTT assays were carried out to assess the
proliferation abilities of CC cells with different transfections.
Briefly, transfected CC cells were plated into 96-well plate and
incubated for 0, 24, 48 or 72 h at 37°C. Then, each well was added
with MTT solution (5 mg/ml, 10 µl) and incubated for 4 h.
Subsequently, dimethyl sulfoxide (DMSO) was added to each well to
dissolve the formazan crystals. Finally, the OD490 was
measured using the Multiskan Spectrum equipment (Thermo Fisher
Scientifc, Inc.).
5-Bromo-2-deoxyuridine (BrDU)
assay
Cell proliferation rate was also assess by measuring
the BrdU immunofluorescence incorporation of DNA during mitosis
using the BrdU assay (Sigma-Aldrich; Merck KGaA) in accordance with
the manufacturer's instructions. Cells (1×104) were
cultured in 24-well plates for 8 h and treated with DHC for another
24 h, and then incubated with 10 µg-ml BrdU (Sigma-Aldrich; Merck
KGaA) for 0.5 h. Each experiment was independently performed three
times, and a two-tailed unpaired Student's t-test was performed to
analyze the significance.
Transwell assays
Transwell chamber (8.0 µm pore size; Corning Co.)
precoated with or without Matrigel (BD Biosciences) was utilized to
perform invasion and migration assays. CC cells with different
transfections in serum-free medium were seeded into the top
chamber. Medium containing 10% FBS, as a chemoattractant, was added
into the bottom chambers. Followed by incubation at 37°C with 5%
CO2 for 48 h, cells remaining on the top surface were
scraped away while cells attached to the bottom surface were fixed,
stained and detected under an inverted microscope (Olympus
Corporation) from five randomly selected visual fields.
Western blot analysis
Ice-cold Protein Lysis Buffer (Beyotime)
supplemented with protease inhibitor cocktail (Sigma-Aldrich; Merck
KGaA) was used to lyse the CC cells. Protein concentrations were
determined using a BCA Protein assay kit (Beyotime) following the
manufacturer's instructions. Proteins were separated with 10%
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE), followed by transferred onto PVDF membrane (Millipore)
which was blocked in TBST with 5% skim milk for 2 h at room
temperature. Then, the membrane was incubated with the following
primary antibodies overnight at 4°C: SATB1 (1:1,000; ab109122),
PI3K (1:1,000, ab191606) (both from Abcam), p-PI3K (1:1,000,
BS4605; Xinle Biotechnology), Akt (1:1,000, sc-56878), p-Akt
(1:1,000, sc-81433) (both from Santa Cruz Biotechnology, Inc.),
mTOR (1:1000, ab32028), p-mTOR (1:1,000, ab109268), E-cadherin
(1:1,000, ab133597), Vimentin (1:1,000, ab137321) and GAPDH
(1:10,000, ab128915) (all from Abcam). The membranes were washed
three times in TBST incubated with HRP-conjugated secondary
antibody (1:2,000, ab205718; Abcam) at room temperature for 2 h and
then washed with TBST. ECL reagents (Thermo Fisher Scientifc, Inc.)
were utilized to visualize the proteins.
Dual-luciferase reporter assays
The wild-type (WT) and mutant (MUT) SATB1 3′-UTR
were designed and prepared by GenePharma Co., Ltd., and were
inserted into pGL3 plasmids (Promega). Subsequently, CC cells were
cotransfected with either the SATB1-3′-UTR-MUT or SATB1-3′-UTR-WT
reporter vector, along with miR-100 mimics by Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). After incubation for
48 h, the dual-luciferase reporter assay system (Promega) was used
to determine the luciferase activity in transfected cells according
to the manufacturer's recommendations.
Xenograft tumor formation assay
This study was approved by the Ethics Committees on
Animal Research of the Second Hospital of Shandong University.
Female nude mice (4–6 week-old) were randomly divided into two
groups. The HeLa cells infected with lentivirus containing
lentiviral miR-100 (lenti-miR-100) or the negative control
(lenti-control) were injected subcutaneously into right flank of
mice. The size of tumors was measured every 3 days. The tumor
volumes were calculated as follows: Volume = (length ×
width2)-2. The mice were sacrificed at the end of the
observation periods to excise the tumors.
Statistical analysis
All assays were repeated at least 3 times. All
statistical analysis was carried out using SPSS software version
17.0 (SPSS Inc.). Student's t-test and one-way ANOVA followed by
Tukey's post hoc test were utilized to analyze 2 or multiple
groups. Kaplan-Meier curve and log-rank test were utilized to
analyze the overall survival of CC patients. P<0.05 indicates
statistically significant difference.
Results
Low miR-100 expression level predicts
poor prognosis of CC patients
To elucidate the functions of miR-100 in CC
progression, we detected the miR-100 expression in CC tissue
specimens and matched normal tissue samples by performing qRT-PCR.
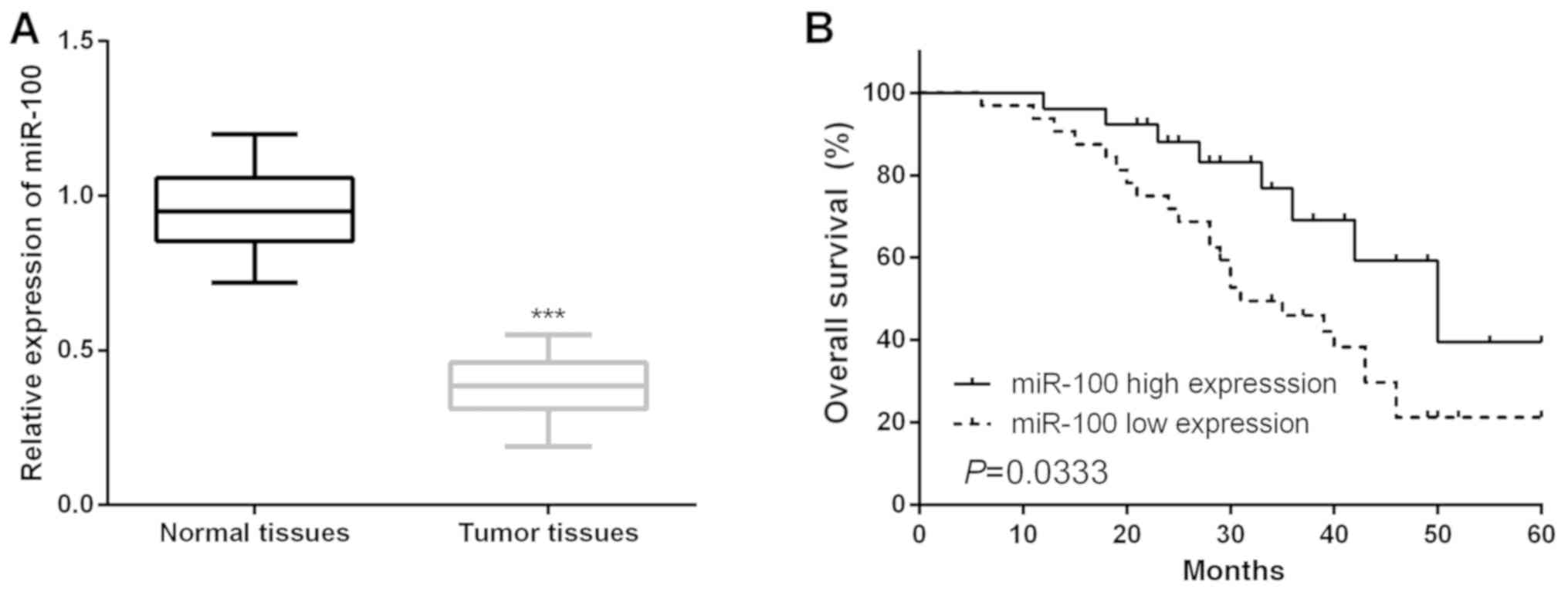
It was clear that the expression of miR-100 in CC tissues was
prominently decreased in comparison with that in paired healthy
tissue samples (Fig. 1A). Moreover,
the association between miR-100 expression and the
clinicopathological features of CC patients was explored by
classifying the CC patients into two groups on the basis of mean
expression of miR-100. As presented in Table II, lower miR-100 expression was
notably associated with poorer clinicopathologic features of CC
patients. The association between miR-100 and the OS of CC patients
was determined via Kaplan-Meier analysis and the log-rank test.
Results showed that CC patients who had lower miR-100 expression
presented poorer OS compared with patients with higher miR-100
expression (Fig. 1B).
 | Table II.Correlation of miR-100 expression
with the clinicopathological characteristics of the cervical cancer
patients. |
Table II.
Correlation of miR-100 expression
with the clinicopathological characteristics of the cervical cancer
patients.
|
|
|
miRNA-100a expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | Cases (n=58) | High (n=21) | Low (n=37) | P-value |
|---|
| Age (years) |
|
|
| 0.4128 |
|
>60 | 30 | 9 | 21 |
|
|
≤60 | 28 | 12 | 16 |
|
| Family history of
cancer |
|
|
| 0.4651 |
|
Yes | 28 | 8 | 20 |
|
| No | 30 | 13 | 17 |
|
| Tumor size
(cm) |
|
|
| 0.0921 |
|
≥5.0 | 26 | 7 | 19 |
|
|
<5.0 | 32 | 14 | 18 |
|
| TNM stage |
|
|
| 0.0018b |
|
I–II | 27 | 17 | 10 |
|
|
III | 31 | 4 | 27 |
|
| Lymph node
metastasis |
|
|
| 0.0025b |
|
Yes | 16 | 5 | 11 |
|
| No | 42 | 16 | 26 |
|
| Menopause |
|
|
| 0.2468 |
|
Yes | 27 | 10 | 17 |
|
| No | 32 | 11 | 20 |
|
| FIGO stage |
|
|
| 0.0031b |
|
I–II | 26 | 17 | 9 |
|
|
III–IV | 32 | 4 | 28 |
|
| Distant
metastasis |
|
|
| 0.4396 |
|
Yes | 29 | 7 | 22 |
|
| No | 29 | 14 | 15 |
|
| HPV infection |
|
|
| 0.4952 |
|
Negative | 30 | 11 | 20 |
|
|
Positive | 28 | 10 | 17 |
|
Restoration of miR-100 significantly
repressed CC cell proliferation
For a better understanding of the functions of
miR-100 in CC development, the effects of miR-100 on CC cell
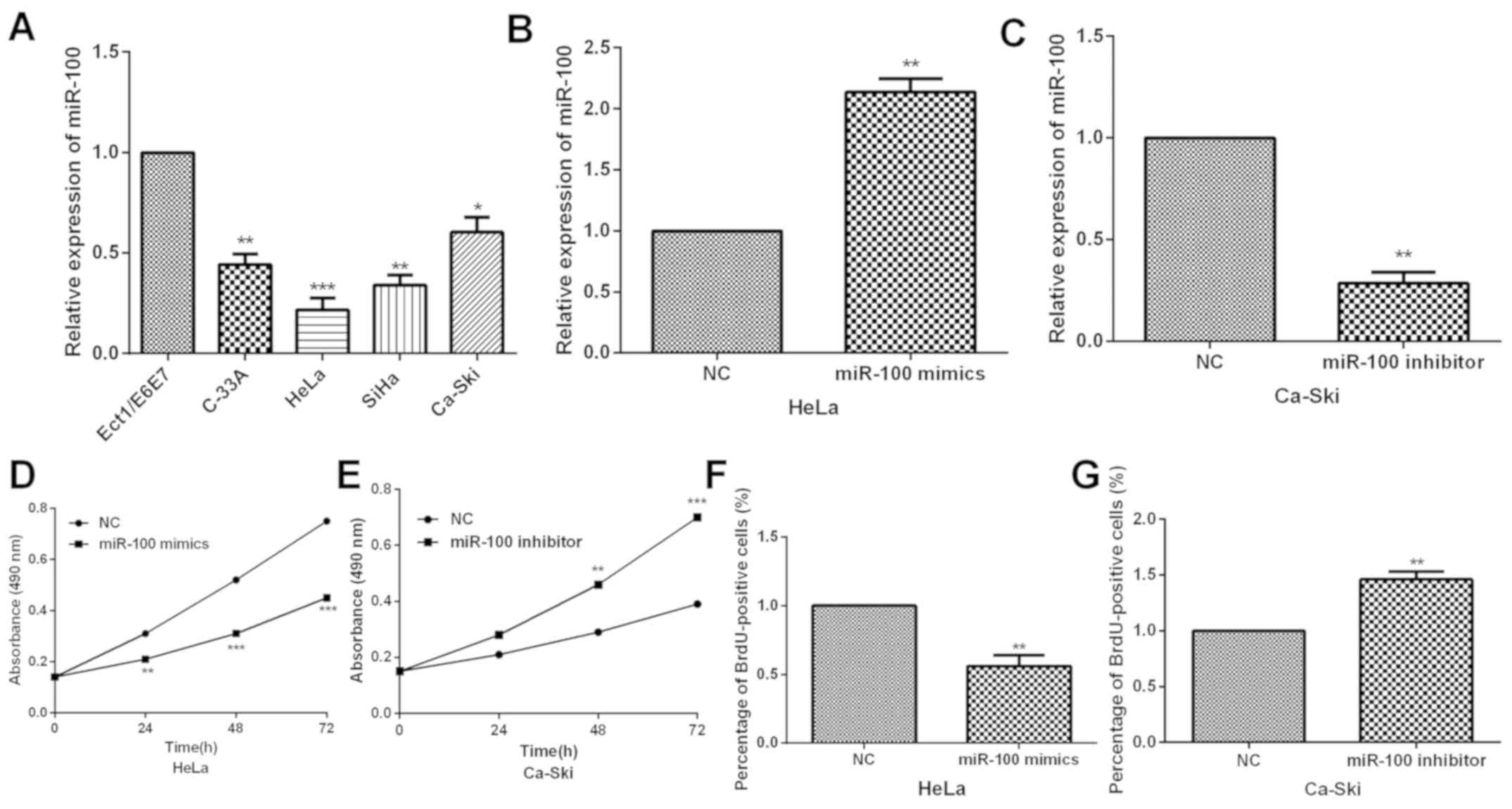
proliferation was investigated. qRT-PCR was carried out to detect
the miR-100 expression in CC cells and normal cervical epithelial
cells. The results demonstrated significant decrease of miR-100
expression in CC cells compared with the Ect1-E6E7 cells (Fig. 2A). Subsequently, due to the
relatively low or high endogenous miR-100 expression, HeLa and
Ca-Ski cells were selected to study the potential functions and
mechanisms of miR-100. The miR-100 expression was altered by
transfecting miR-100 mimics or inhibitor into HeLa and Ca-Ski cells
and qRT-PCR was carried out to examine the efficiency. miR-100 was
dramatically overexpressed by miR-100 mimics in HeLa cells,
whereas, obviously downregulated by miR-100 inhibitor in Ca-Ski
cells (Fig. 2B and C). Then, MTT
assay was conducted to investigate the influence of miR-100 on CC
cell proliferation ability. Data indicated that miR-100
overexpression prominently inhibited CC cell proliferation while
miR-100 inhibition markedly enhanced the proliferation capacity
(Fig. 2D and E). To further explore
the potential function of miR-100 in cervical cancer cells, BrdU
assay was performed to assess the effects of miR-100 on HeLa and
Ca-Ski cells. Compared to controls, miR-100 mimics resulted in a
decrease in BrdU positive cells, whereas, miR-100 inhibitor
increased BrdU positive cells (Fig. 2F
and G). This observation indicates that miR-100 has a
stimulating effect on the proliferation of HeLa and Ca-Ski
cells.
miR-100 overexpression notably
suppresses CC cell invasion and migration
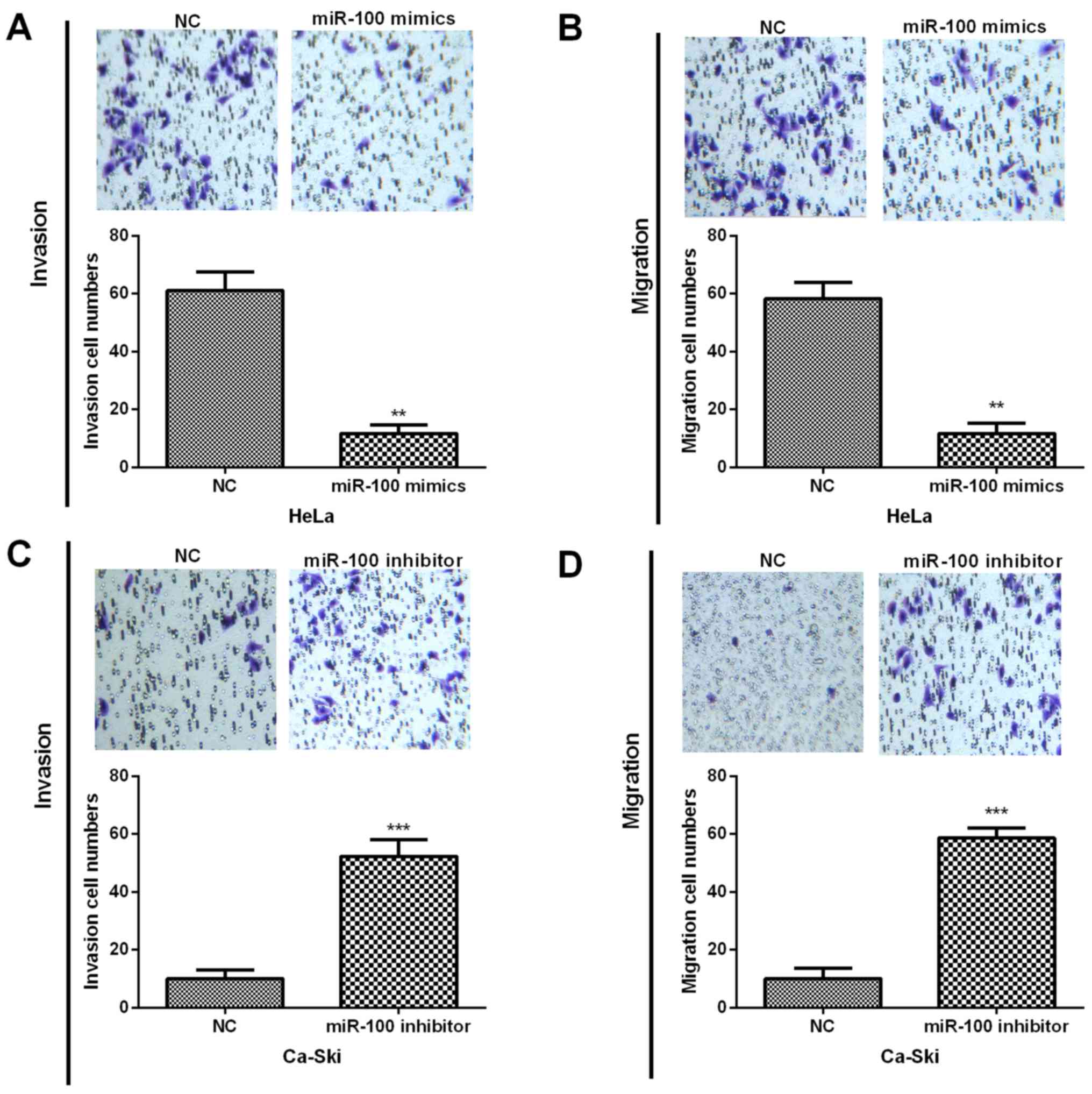
To further confirm whether miR-100 could regulate
the CC progression and metastasis, Transwell assays were performed
and results indicated that miR-100 overexpression dramatically
repressed HeLa cell invasion and migration abilities (Fig. 3A and B). In contrast, the inhibition
of miR-100 in Ca-Ski cells prominently promoted the invasion and
migration capacities (Fig. 3C and
D). These results revealed that miR-100 played inhibitory roles
in CC progression.
SATB1 is a functional target of
miR-100 in CC cells
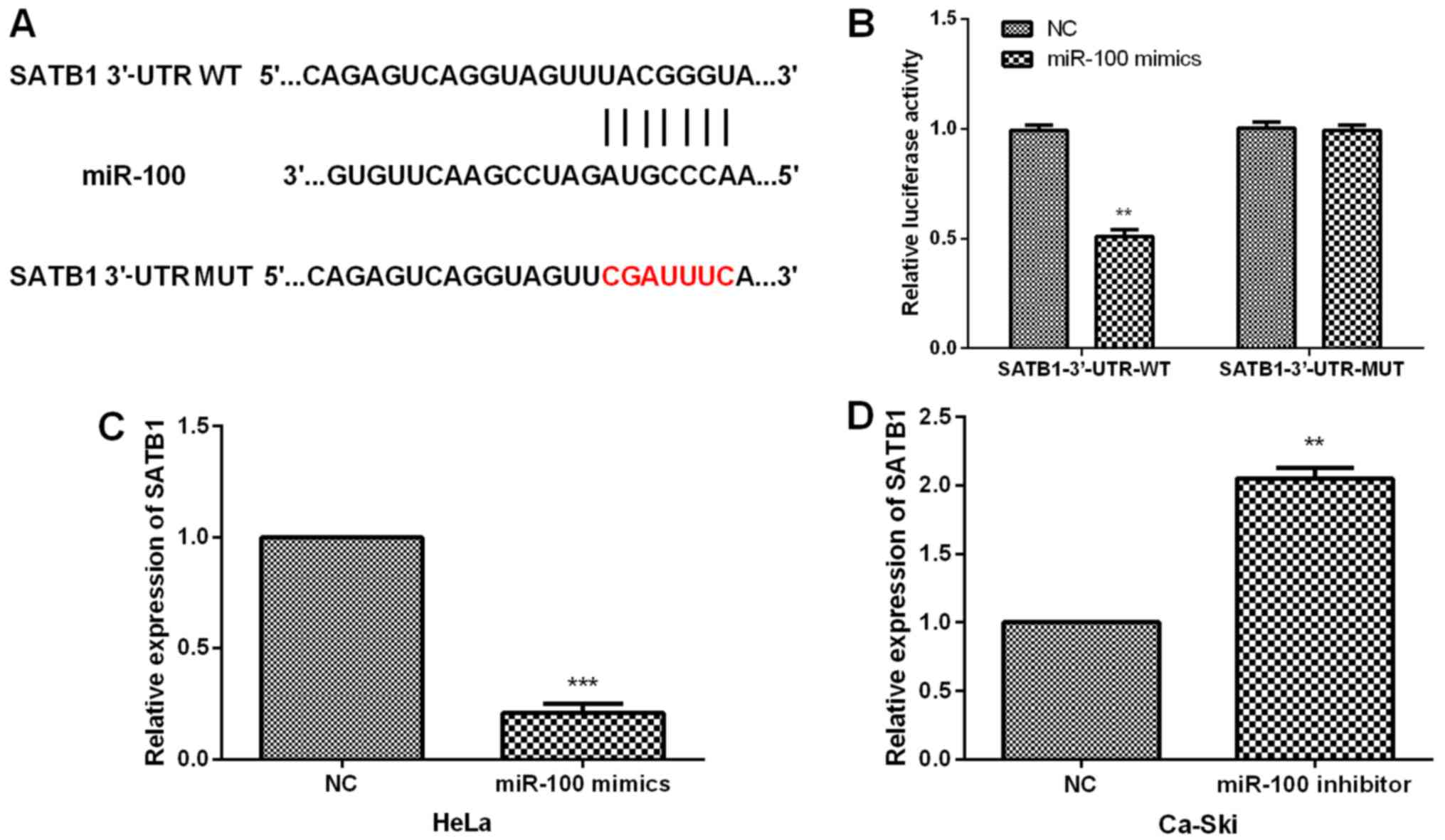
To explore the underlying mechanism of the
functional roles of miR-100 in CC, the putative targets of miR-100
were analyzed and target scan analysis suggested that SATB1 was a
potential target for miR-100 (Fig.
4A). Luciferase reporter assay was carried out to determine the
interaction between miR-100 and SATB1. Results demonstrated that
miR-100 overexpression prominently declined the luciferase activity
of SATB1-3′-UTR-WT; on the other hand, miR-100 mimics did not
change the luciferase activity of CC cells with SATB1-3′-UTR-MUT
(Fig. 4B). In addition, to
investigate whether miR-100 had effects on SATB1 expression,
qRT-PCR analysis was performed to measure the SATB1 expression in
CC cells with transfections of miR-100 mimics or inhibitor. Data
indicated that resumption of miR-100 evidently suppressed the SATB1
expression in HeLa cells whereas miR-100 inhibition obviously
enhanced the SATB1 expression in Ca-Ski cells (Fig. 4C and D).
miR-100 regulates AKT/mTOR signaling
pathway and EMT in CC cells
As we verified that SATB1 was a target for miR-100,
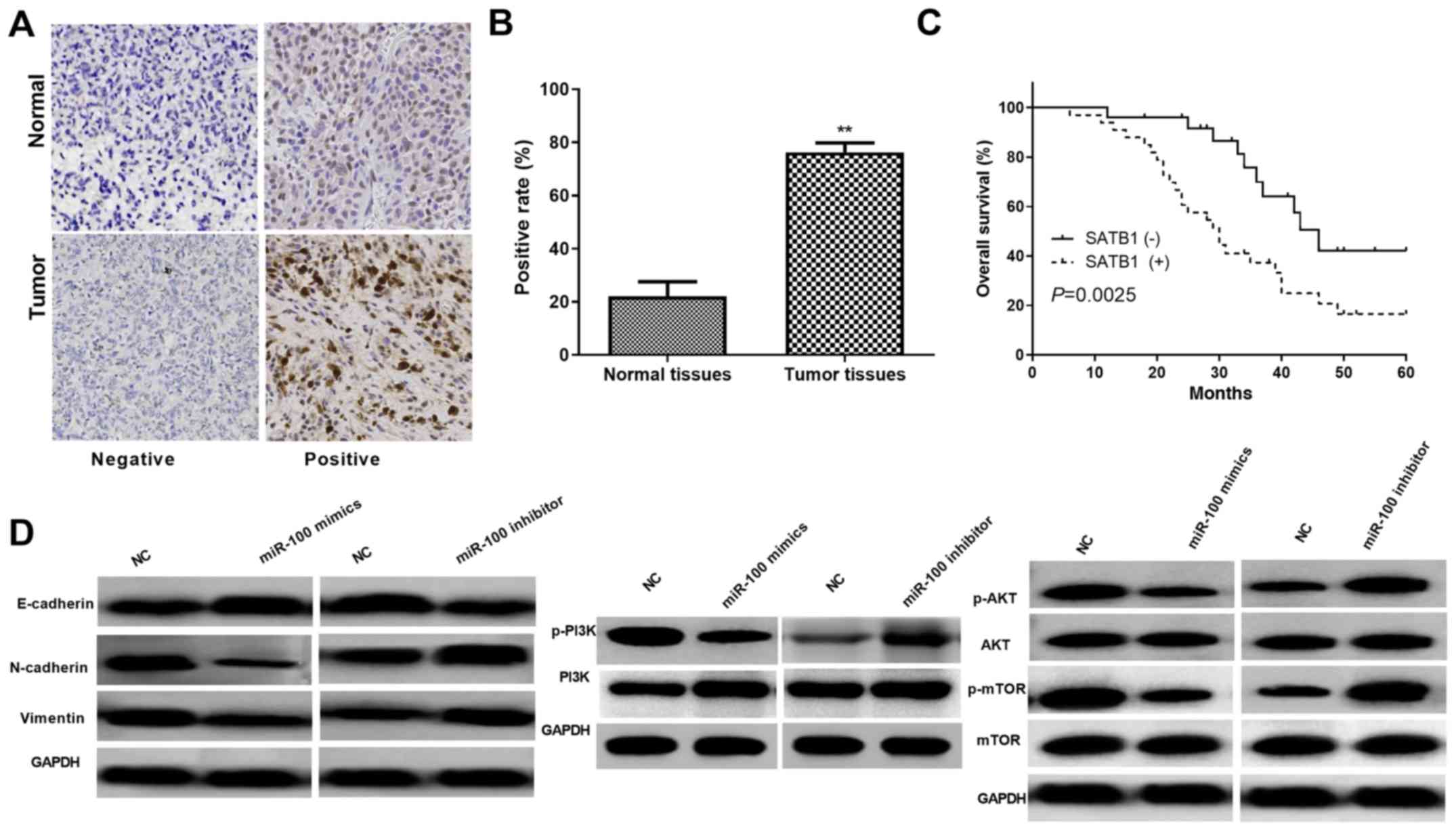
the expression of SATB1 in CC tissues was measured by IHC assays.
As shown in Fig. 5A and B, the
findings revealed that SATB1 mainly localized at the nucleus and
was dramatically upregulated in CC tissue samples. In addition,
Kaplan-Meier analysis further indicated that CC patients with
relatively higher SATB1 level had significantly decreased OS
(Fig. 5C). Moreover, to clarify the
specific molecular mechanisms under miR-100 suppressing CC cell
progression, our studies were extended on AKT/mTOR signaling
pathway and EMT. Western blot analysis demonstrated that expression
of p-AKT and p-mTOR was obviously inhibited by miR-100
overexpression in HeLa cells while there was no prominent variation
in AKT and mTOR expression; whereas, miR-100 inhibition notably
increased the p-AKT and p-mTOR expression in Ca-Ski cells (Fig. 5D). As EMT is a pivotal progress in
tumor cell metastasis and invasion, the functions of miR-100 in CC
cell EMT was analyzed by detecting the protein levels of EMT
markers. Results demonstrated that the E-cadherin expression was
obviously increased while the N-cadherin and vimentin expression
was significantly decreased in miR-100 overexpressed HeLa cells.
Whereas, suppression of miR-100 reversed these results (Fig. 5D). Overall, these results implied
that miR-100 inhibited CC progression by regulation of AKT/mTOR
signaling pathway and EMT.
miR-100 inhibits CC cell tumor growth
in vivo
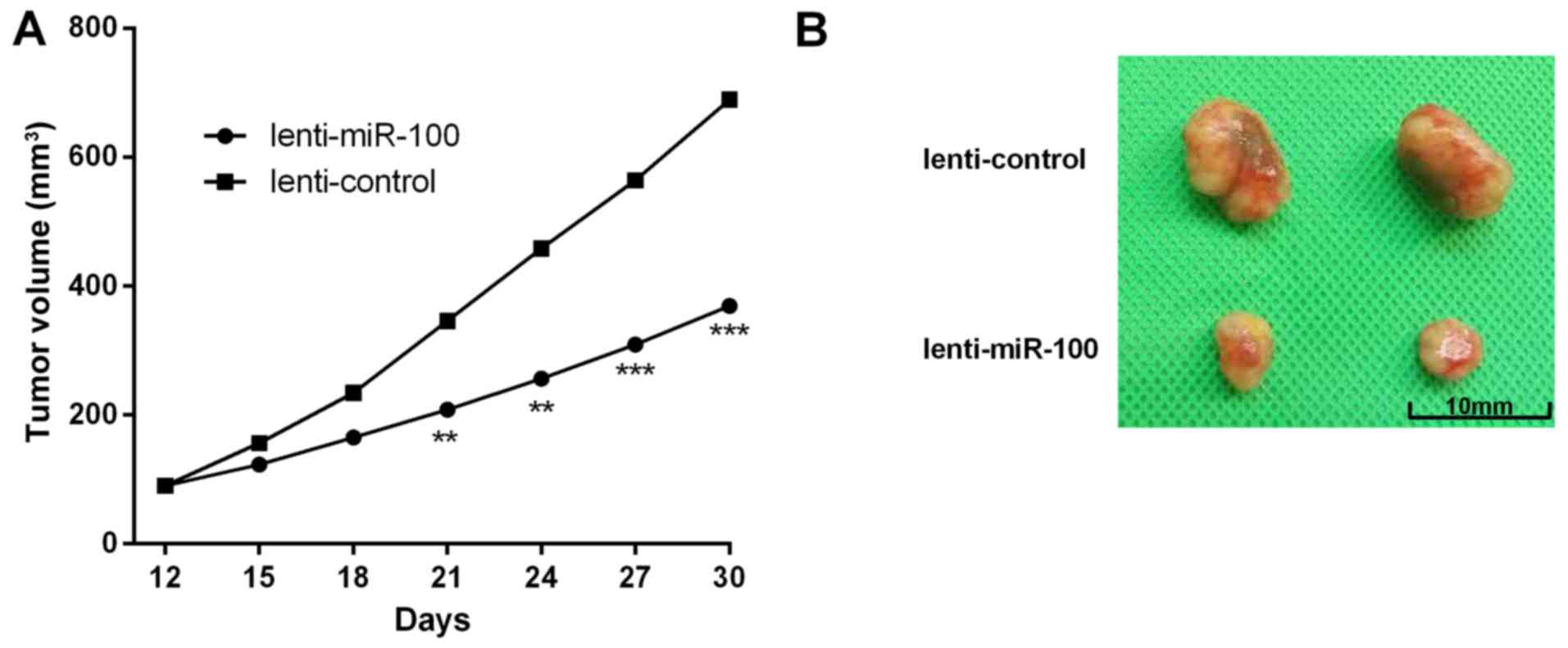
In the present study, we further investigated the
effects of miR-100 on CC tumor growth in vivo. HeLa cells
were stably transfected with lenti-miR-100 or the negative control.
Tumor volume analysis indicated that the volume of mice in
lenti-miR-23b group were obviously smaller than the controls. In
addition, the growth rate was significantly slower than the control
groups (Fig. 6A and B).
Discussion
CC is currently the leading factor for deaths in
women of childbearing ages, and existing conventional therapeutic
screening approaches remain ineffective for advanced stage CC
(21). Therefore, elucidating the
mechanisms underlying CC progression and identifying biomarkers for
prevention, early diagnosis and treatment of CC patients have
become the hot spot of CC research (22). Emerging evidence has indicated that
certain aberrantly expressed miRNAs are involved in tumorigenesis
through regulating the expression of their targets and serve as
novel biomarkers for the diagnosis and prognosis of different human
malignancies, including CC (23).
For example, Chen and Liu (24)
reported that miR-744 inhibited CC growth and progression through
inducing apoptosis via regulation of Bcl-2. Chen et al
(25) confirmed that miR-206 and
miR-34a functioned as novel prognostic and therapeutic biomarkers
in CC. Hence, identification of miRNAs and their targets implicated
in tumorigenesis may provide key clues to develop novel diagnostic
methods and therapies for CC patients.
Previous studies demonstrated that miR-100 exerted
tumor suppressive functions in numerous cancers by modulating
different targets. For instance, Liu et al (26) proposed that miR-100 repressed cell
proliferation, invasion and migration and promoted chemosensitivity
in osteosarcoma via regulating IGFIR; Luan et al (27) proposed that miR-100 upregulation
suppressed glioblastoma cell chemosensitivity, proliferation and
migration through FGFR3; studies by Qureshi et al (28) demonstrated that miR-100 was a novel
non-invasive biomarker for earlier diagnosis of bladder cancer.
Thus, we assumed that miR-100 might serve as a cancer repressor in
CC. To test the hypothesis, series of experiments was performed.
Data revealed that miR-100 expression was obviously decreased in CC
and the low miR-100 expression was related to its malignant
progression and poor prognosis. In addition, functional experiments
indicated that miR-100 restoration repressed the proliferation,
invasion and migration capacities of CC cells via regulation of
AKT/mTOR signaling pathway and EMT progress. Whereas, miR-100
overexpression was also able to inhibit CC tumorigenesis in
vivo. These results demonstrated the potential tumor inhibitory
roles of miR-100 in CC progression.
SATB1 has been confirmed to be implicated in various
cellular processes, including invasion, migration, apoptosis and
proliferation (29,30). In the present study, the functional
effects of SATB1 wewe further investigated on CC progression.
Results in the present study successfully revealed that SATB1 was
upregulated in CC, which demonstrated poor OS of CC patients.
Moreover, SATB1 was confirmed to be a target for miR-100 in CC
cells. Our findings illustrated that functions of miR-100 were
regulated by SATB1. Thus, our results provided new insight into the
miR-100 biological functions in CC cells.
Collectively, evidence was provided that miR-100
exerted an inhibitory role in CC progression both in vitro
and in vivo. It was confirmed that miR-23b expression was
notably declined in CC tissues and cell lines, whereas, the low
miR-23b expression was related to poor OS and worse prognosis. The
functional assays indicated that miR-23b overexpression prominently
decreased the proliferation, invasion and migration abilities of CC
cells through regulation of the AKT/mTOR signaling pathway and EMT.
Moreover, SATB1 was identified as one functional target for miR-23b
in CC cells. Hence, the findings of the present study provide a new
clue to the functions of miR-23b/SATB1 in CC progression as novel
diagnostic and prognosis markers.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CH, XQ and NZ participated in the conception and
design of the study, and performed the experiments. HJ performed
the analysis and interpretation of the data. SZ wrote the
manuscript, assisted with the statistical analysis of the data and
contributed with constructive discussions. HY was involved in the
conception of the study and provided the patients’ clinical data as
well as crucial experimental materials. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Second Hospital of Shandong University (Jinan,
China), and the Ethics Committee on Animal Research of Shandong
University. All patients involved in the present study provided
written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Carr KR, Ioffe YJ, Filippova M,
Duerksen-Hughes P and Chan PJ: Combined ultrasound-curcumin
treatment of human cervical cancer cells. Eur J Obstet Gynecol
Reprod Biol. 193:96–101. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yu Q, Yang X, Duan W, Li C, Luo Y and Lu
S: miRNA-346 promotes proliferation, migration and invasion in
liver cancer. Oncol Lett. 14:3255–3260. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Puik JR, Meijer LL, Le Large TY, Prado MM,
Frampton AE, Kazemier G and Giovannetti E: miRNA profiling for
diagnosis, prognosis and stratification of cancer treatment in
cholangiocarcinoma. Pharmacogenomics. 18:1343–1358. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fang H, Xie J, Zhang M, Zhao Z, Wan Y and
Yao Y: miRNA-21 promotes proliferation and invasion of
triple-negative breast cancer cells through targeting PTEN. Am J
Transl Res. 9:953–961. 2017.PubMed/NCBI
|
|
6
|
Xiao R, Li C and Chai B: miRNA-144
suppresses proliferation and migration of colorectal cancer cells
through GSPT1. Biomed Pharmacother. 74:138–144. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang L, Liang H, Wang Y, Gao S, Yin K, Liu
Z, Zheng X, Lv Y, Wang L, Zhang CY, et al: MiRNA-203 suppresses
tumor cell proliferation, migration and invasion by targeting Slug
in gastric cancer. Protein Cell. 7:383–387. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Deng B, Zhang S, Zhang Y, Miao Y, Meng X
and Guo K: Knockdown of Tripartite Motif Containing 28 suppresses
the migration, invasion and epithelial-mesenchymal transition in
ovarian carcinoma cells through downregulation of Wnt/β-catenin
signaling pathway. Neoplasma. 64:893–900. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu HY and Cai XP: miR-338-3p suppresses
epithelial-mesenchymal transition and metastasis in human nonsmall
cell lung cancer. Indian J Cancer. 52 (Suppl 3):E168–E171. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen H, Jia W and Li J: ECM1 promotes
migration and invasion of hepatocellular carcinoma by inducing
epithelial-mesenchymal transition. World J Surg Oncol. 14:1952016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Colditz J, Rupf B, Maiwald C and Baniahmad
A: Androgens induce a distinct response of epithelial-mesenchymal
transition factors in human prostate cancer cells. Mol Cell
Biochem. 421:139–147. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
He H, Dai J, Xu Z, He W, Wang X, Zhu Y and
Wang H: Fbxw7 regulates renal cell carcinoma migration and invasion
via suppression of the epithelial-mesenchymal transition. Oncol
Lett. 15:3694–3702. 2018.PubMed/NCBI
|
|
13
|
Chen X, Liao Y, Yu Y, Zhu P, Li J, Qin L,
Liao W and Huang Z: Elevation of MAP17 enhances the malignant
behavior of cells via the Akt/mTOR pathway in hepatocellular
carcinoma. Oncotarget. 8:92589–92603. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xue L, Wang Y, Yue S and Zhang J: Low
MiR-149 expression is associated with unfavorable prognosis and
enhanced Akt/mTOR signaling in glioma. Int J Clin Exp Pathol.
8:11178–11184. 2015.PubMed/NCBI
|
|
15
|
Cai S, Han HJ and Kohwi-Shigematsu T:
Tissue-specific nuclear architecture and gene expression regulated
by SATB1. Nat Genet. 34:42–51. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qi H, Fu X, Li Y, Pang X, Chen S, Zhu X,
Li F and Tan W: SATB1 promotes epithelial-mesenchymal transition
and metastasis in prostate cancer. Oncol Lett. 13:2577–2582. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li YC, Bu LL, Mao L, Ma SR, Liu JF, Yu GT,
Deng WW, Zhang WF and Sun ZJ: SATB1 promotes tumor metastasis and
invasiveness in oral squamous cell carcinoma. Oral Dis. 23:247–254.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pan Z, Jing W, He K, Zhang L and Long X:
SATB1 is correlated with progression and metastasis of breast
cancers: A meta-analysis. Cell Physiol Biochem. 38:1975–1983. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hu XT, Chen W, Zhang FB, Shi QL, Hu JB,
Geng SM and He C: Depletion of the proteasome subunit PSMA7
inhibits colorectal cancer cell tumorigenicity and migration. Oncol
Rep. 22:1247–1252. 2009.PubMed/NCBI
|
|
20
|
Hu XT, Chen W, Wang D, Shi QL, Zhang FB,
Liao YQ, Jin M and He C: The proteasome subunit PSMA7 located on
the 20q13 amplicon is overexpressed and associated with liver
metastasis in colorectal cancer. Oncol Rep. 19:441–446.
2008.PubMed/NCBI
|
|
21
|
Kessler TA: Cervical cancer: Prevention
and early detection. Semin Oncol Nurs. 33:172–183. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cordeiro MN, De Lima RC, Paolini F, Melo
AR, Campos AP, Venuti A and De Freitas AC: Current research into
novel therapeutic vaccines against cervical cancer. Expert Rev
Anticancer Ther. 18:365–376. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Y, Zhang D, Wang F, Xu D, Guo Y and
Cui W: Serum miRNAs panel (miR-16-2*, miR-195, miR-2861, miR-497)
as novel non-invasive biomarkers for detection of cervical cancer.
Sci Rep. 5:179422015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen XF and Liu Y: MicroRNA-744 inhibited
cervical cancer growth and progression through apoptosis induction
by regulating Bcl-2. Biomed Pharmacother. 81:379–387. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen AH, Qin YE, Tang WF, Tao J, Song HM
and Zuo M: MiR-34a and miR-206 act as novel prognostic and therapy
biomarkers in cervical cancer. Cancer Cell Int. 17:632017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu Y, Zhu ST, Wang X, Deng J, Li WH,
Zhang P and Liu BS: MiR-100 inhibits osteosarcoma cell
proliferation, migration, and invasion and enhances
chemosensitivity by targeting IGFIR. Technol Cancer Res Treat.
15:NP40–NP48. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Luan Y, Zhang S, Zuo L and Zhou L:
Overexpression of miR-100 inhibits cell proliferation, migration,
and chemosensitivity in human glioblastoma through FGFR3. Onco
Targets Ther. 8:3391–3400. 2015.PubMed/NCBI
|
|
28
|
Qureshi A, Fahim A, Kazi N, Farsi Kazi SA
and Nadeem F: Expression of miR-100 as a novel ancillary
non-invasive biomarker for early detection of bladder carcinoma. J
Pak Med Assoc. 68:759–763. 2018.PubMed/NCBI
|
|
29
|
Xiao T, Fu L and Jie Z: SATB1
overexpression correlates with gastrointestinal neoplasms invasion
and metastasis: A meta-analysis for Chinese population. Oncotarget.
8:48282–48290. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang B, Xiong F, Wang S, Lang X, Wang X
and Zhou H: Effect of SATB1 silencing on the proliferation,
invasion and apoptosis of TE-1 esophageal cancer cells. Oncol Lett.
13:2915–2920. 2017. View Article : Google Scholar : PubMed/NCBI
|