Introduction
Gastric cancer (GC) is one of the most severe tumor
types with a high mortality rate (1,2) and poor
prognosis (3,4). The global pattern of histone
modifications may serve as a predictor of the risk of recurrence of
human cancer (5,6). Histone modification, as a notable
component of epigenetics, occurs in a diverse range of biological
processes. Aberrant post-translational modification of histone
tails by methylation is closely associated with tumor development,
progression, prognosis and recurrence (7). For example, di-methylation of lysine 9
of histone H3 (H3K9me2) is correlated with gene repression and
serves a well-established function in heterochromatin formation and
gene transcription regulation in human cancer (8). Among well-studied histone methylations,
the methylation pattern of H3K9 is associated with gene regulation
including repression (9).
Euchromatic histone lysine methyltransferase 2 (EHMT2; also known
as G9a), which is a lysine methyltransferase that contributes to
the epigenetic silencing of tumor suppressor genes, is required for
H3K9me2 (10). EHMT2 may catalyze a
modification at histone 3 lysine 9 including H3K9me1 and H3K9me2;
H3K9me1 is associated with gene activation, whereas H3K9me2 is
predominant in silenced genes (11).
EHMT2-dependent H3K9me2 is associated with gene silencing and
functions primarily through the recruitment of H3K9me2-binding
proteins that prevent transcriptional activation (12). EHMT2 has been reported to be
overexpressed in pancreatic (13),
breast (14,15), lung (16,17),
hepatocellular (18), colorectal
carcinoma (19) and GC (20).
The abnormal expression level of EHMT2 and H3K9me2
has been identified in multiple types of cancer, including
hematologic malignancies (21).
However, the clinical significance of EHMT2, H3K9me2 and their
interactions in solid tumor types, including in GC, remains
unclear. A previous study has revealed that H3K9me2 may contribute
to DNA methylation via DNA (cytosine-5-) methyltransferase 3 αb to
repress E-cadherin in the epithelial-mesenchymal
transition-associated metastasis of GC (22). Additionally, the hypoxic silencing of
tumor suppressor Runt-related transcription factor 3 may also be
mediated by upregulated EHMT2 and histone deacetylase 1 in GC cells
(20). Increased EHMT2 levels in GC
tissues may also promote tumor invasion and metastasis, and are
associated with an with advanced stage and shorter overall survival
time in a SET domain-independent manner (23). Previously, accumulating evidence has
indicated that investigation into the clinical importance of EHMT2
levels and H3K9me2 methylation patterns may be of help for the
diagnosis and treatment of GC (24–26).
The aim of the present study was to evaluate the
methylation pattern of H3K9me2 and EHMT2 expression levels in GC
and adjacent healthy tissues, and to reveal the association between
the increased EHMT2 expression and H3K9me2 methylation levels.
Materials and methods
Clinical cases with GC
A total of 118 archived paraffin-embedded GC
specimen blocks were selected retrospectively from the Department
of Pathology of Yancheng Hospital (Jiangsu, China). The specimens
were collected from patients (82 men and 36 women) with GC who
underwent surgery between March 2010 and December 2011. Medical
records, including clinicopathological parameters and follow-up
data, were also obtained. The inclusion criteria were as follows:
i) No other serious or fatal diseases and ii) if the patients died
during the follow-up period, the cause of mortality should be the
secondary change, including tumor progression, cachexia, recurrence
and metastasis, and follow-up data were complete. The American
Joint Committee on Cancer staging system (8th edition) (27) was used for pathological
tumor-node-metastasis (pTNM) staging. The mean and median follow-up
period was 38.1 months and 41.0 months (range, 1–60 months),
respectively. The present study was ethically approved and
supervised by the Committee for Ethical Review of Research of
Yancheng Hospital.
Immunohistochemical staining
The selected paraffin blocks were sliced into 4-µm
tissue sections, heated for 60 min at 65°C and cooled down. The
sections were subsequently deparaffinized, rehydrated and placed
with ethylene diamine tetraacetic acid antigen retrieval solution
(pH 8.0) to retrieve the antigens, followed by incubation with 3%
H2O2 for 25 min at room temperature (RT).
Following blocking with 3% BSA for 30 min at RT, the sections were
treated with primary antibodies against H3K9me2 (1:200; cat. no.
ab1220; Abcam) and EHMT2 (1:800; cat. no. ab185050; Abcam) in a wet
box at 4°C overnight. The sections were further incubated with
horseradish peroxidase-conjugated secondary antibodies (cat. no.
K5007; Dako; Agilent Technologies GmbH) for 50 min at RT. The
staining was visualized using freshly prepared
3,3′-diaminobenzidine reagent (cat. no. G1211; Wuhan Servicebio
Technology Co., Ltd.). The chromogenic reaction was stopped when
the nuclei appeared brown/yellow under the microscope;
subsequently, the nuclei were counterstained with hematoxylin
staining solution at RT. Following dehydration and washing, the
slides were mounted with coverslips, and the staining was evaluated
under an Axiocam 105 light microscope (magnification, ×100 and
×400; Carl Zeiss AG). Cells treated with PBS instead of the primary
antibody were used as a negative control.
Evaluation of immunostained epithelial
tissues
The staining was blindly examined by two associate
professors from the Department of Pathology, Medical School,
Southeast University (Nanjing, China). The analyzed areas were
tumor cells in the cancerous tissues and epithelial cells in the
adjacent healthy epithelial tissues. The scoring system was
described in previous studies (28,29). In
brief, the final score of immunostaining was calculated as the sum
product of the scale of staining intensity and the scale of
staining area. The scale of intensity was as follows: 0=negative;
1=weak staining; 2=moderate staining and 3=intensive staining; and
the scale of staining area was as follows: 0=0-5% positive cells;
1=6-25% positive cells; 2=26-50% positive cells; 3=51-75% positive
cells and 4=76-100% positive cells. The expression patterns were
classified into two groups: Low (score >8) and high (score
>8) scoring group. The patients were divided into three groups
according to H3K9me2 and EHMT2 expression: i) Low expression group
for patients with low expression levels of H3K9me2 and EHMT2; ii)
high expression group for patients with high expression levels of
H3K9me2 and EHMT2 and iii) other group for patients with high
H3K9me2 expression and low EHMT2 expression, and vice versa.
Statistical analysis
Statistical analysis was performed using SPSS
software (v.19; IBM Corp.). Data are presented as the mean ±
standard error of the mean. The histone modification levels of
cancerous and adjacent healthy tissues were assessed using a paired
Student's t-test. The associations between clinicopathological
variables and the immunostaining scores of histone methylation were
analyzed using a χ2 test or Fischer's exact test.
Kaplan-Meier (K-M) analysis with a log-rank test was used to
determine the contribution of the clinicopathological features and
the immunostaining expression patterns to the patients' survival
time. Multivariate analysis using Cox's proportional hazard
regression was used to examine the clinical value of the levels of
the studied protein and the clinicopathological parameters of the
patients. Statistical analyses were performed using SPSS Statistics
v.17 (SPSS Inc.). P<0.05 was considered to indicate a
statistically significant difference.
Results
Clinicopathological profiles of the
patients
In the present study, the tissue samples from 82
male and 36 female patients with GC were studied; the median age
was 59.7 years (age range, 33–83 years), and the
clinicopathological variables are summarized in Table I. The samples comprised 16
well-differentiated cases, 62 poorly differentiated cases and 40
moderately differentiated cases. The pTNM staging was as follows:
30 cases were stage I (25.4%), 33 were stage II (28%), 47 were
stage III (39.8%) and 8 were IV (6.8%). Among the 118 patients, the
5-year survival rate following gastrectomy was 36.4%.
 | Table I.Clinicopathological characteristics
of patients with gastric carcinoma (n=118). |
Table I.
Clinicopathological characteristics
of patients with gastric carcinoma (n=118).
|
Characteristics | Patient, n | Percentage, % |
|---|
| Sex |
|
Male | 82 | 69.5 |
|
Female | 36 | 30.5 |
| Age, years |
|
≤60 | 59 | 50.0 |
|
>60 | 59 | 50.0 |
| Differentiation
degree |
|
Well-differentiated | 16 | 13.6 |
|
Moderately differentiated | 40 | 33.9 |
| Poorly
differentiated | 62 | 52.5 |
| Lymph node
metastasis |
|
Negative | 51 | 43.2 |
|
Positive | 67 | 56.8 |
| Distal
metastasis |
|
Negative | 110 | 93.2 |
|
Positive | 8 | 6.8 |
| pTNM |
| I | 30 | 25.4 |
| II | 33 | 28.0 |
|
III | 47 | 39.8 |
| IV | 8 | 6.8 |
| Survival time,
months following operation |
| ≤5 | 75 | 63.6 |
|
>5 | 43 | 36.4 |
H3K9me2 and EHMT2 expression patterns
are associated with clinicopathological characteristics in patients
with GC
The expression of histone methylation marker H3K9me2
and histone methyltransferase EHMT2 were evaluated in the surgical
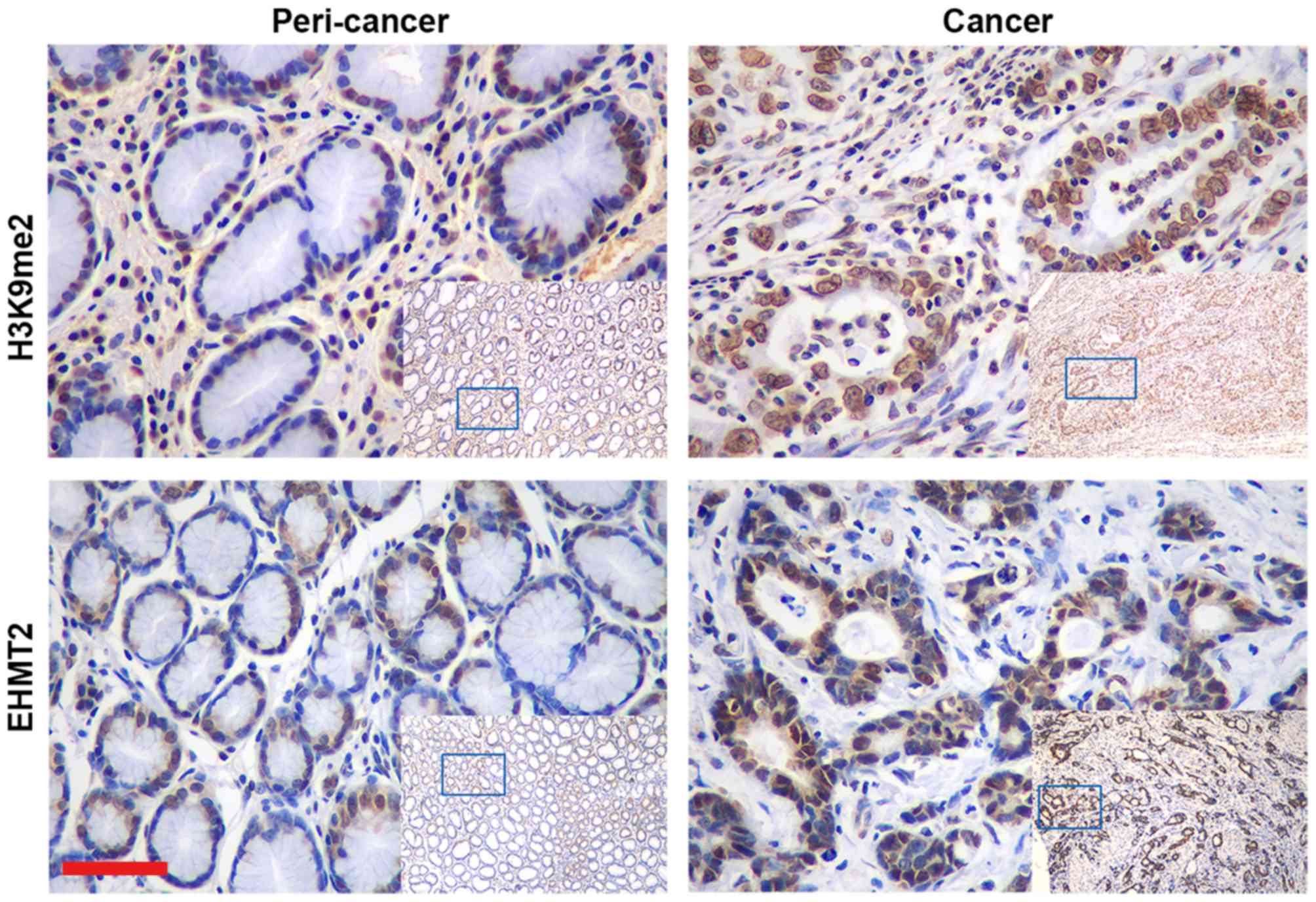
samples from 118 patients with GC. H3K9me2 was localized in the
nuclei of epithelial cells, whereas EHMT2 mainly labelled the
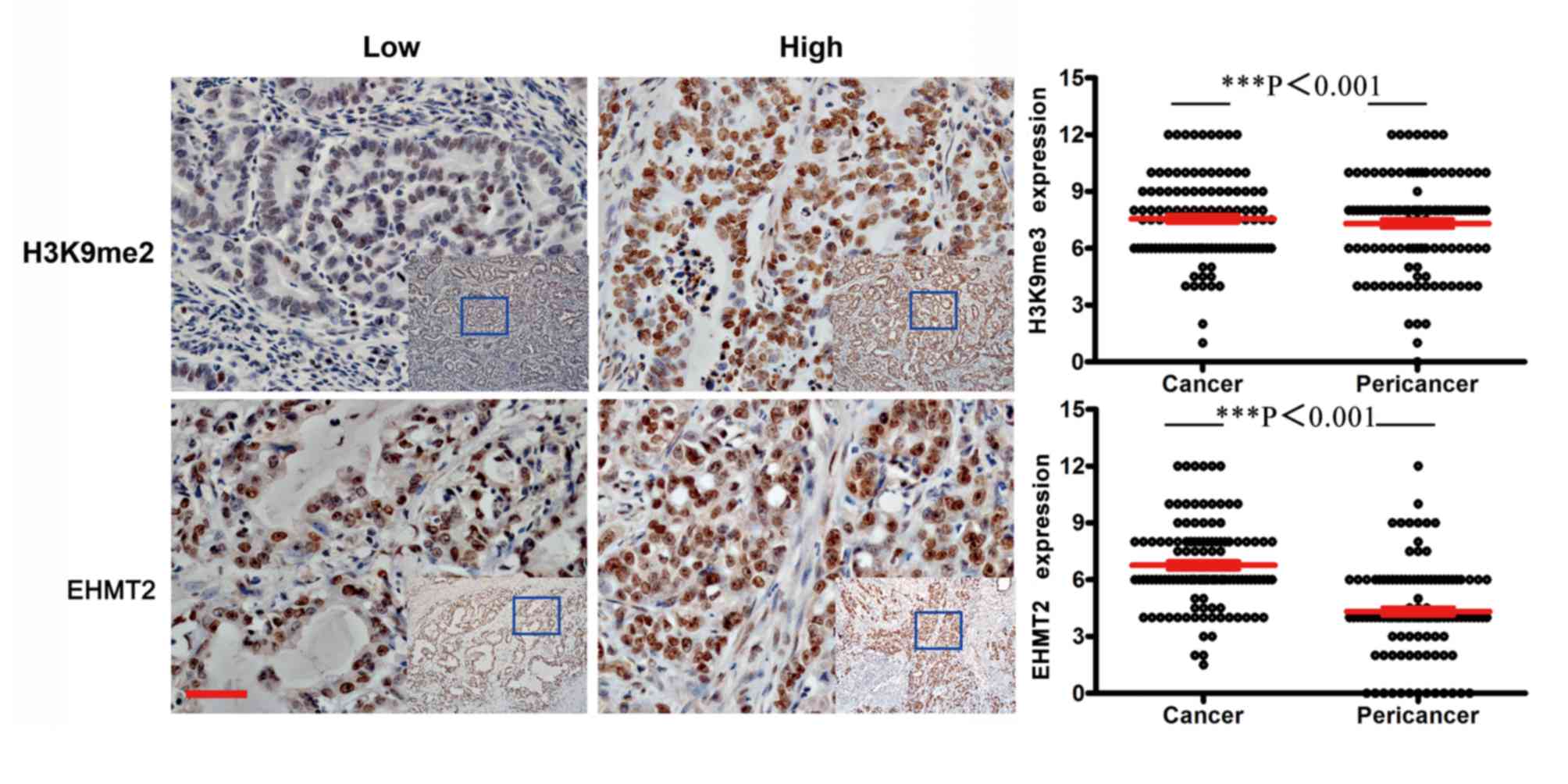
nucleus with partial staining in the cytoplasm (Fig. 1). Example tissues exhibiting low and
high expression areas are presented in Fig. 2. The assessment of the positive
immunoreaction in the epithelial cells of cancerous and
non-cancerous tissues demonstrated that cancerous tissues exhibited
significantly stronger immunostaining compared with adjacent
healthy tissues (P<0.001; Table
II).
 | Table II.Differential expression of histone
methylation between gastric cancer and peri-cancer tissues. |
Table II.
Differential expression of histone
methylation between gastric cancer and peri-cancer tissues.
| Histone
methylation | Cancer | Peri-cancer | P-value |
|---|
| H3K9me2 |
7.557±0.206 |
7.311±0.236 |
<0.001a |
| EHMT2 | 6.765±0.216 | 4.319±0.228 |
<0.001a |
To clarify the association between patient
clinicopathological characteristics, EHMT2 expression and H3K9me2
methylation, the data were analyzed using a χ2 test; the
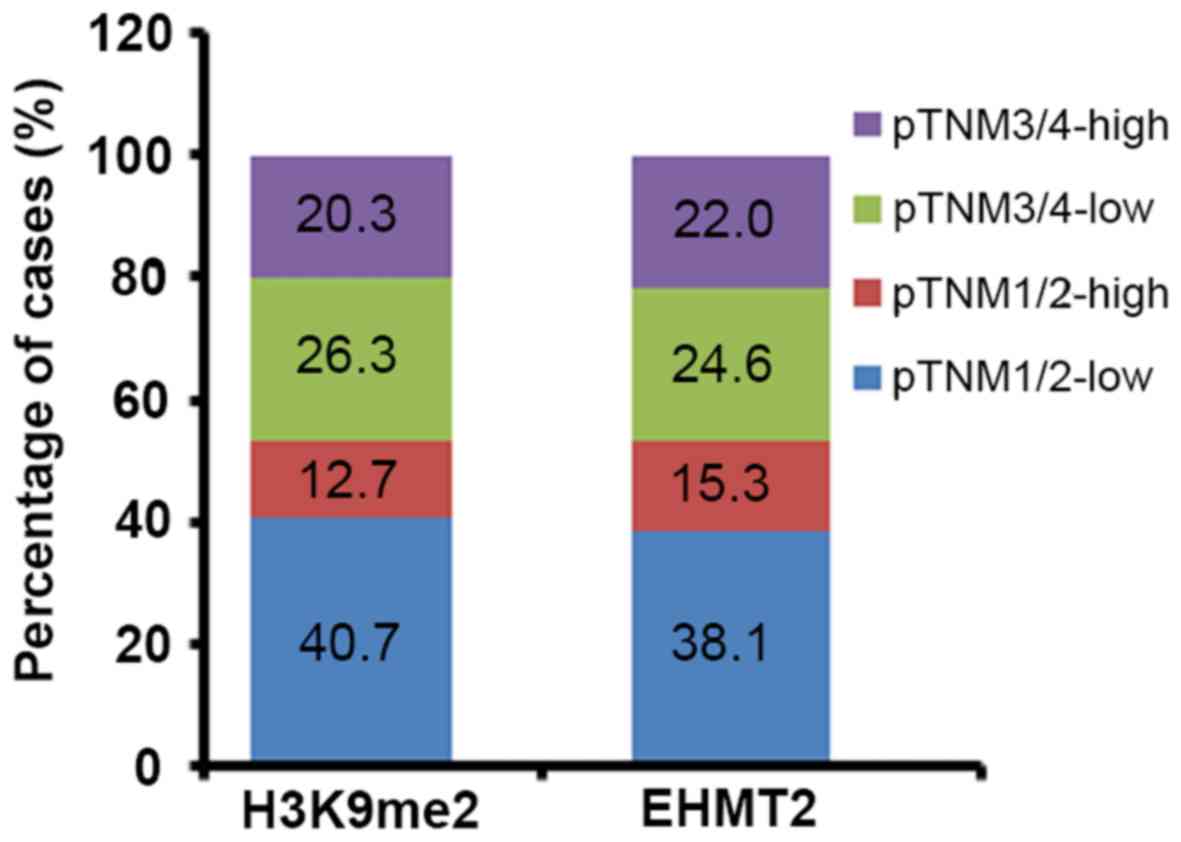
results demonstrated that EHMT2 overexpression and H3K9me2
methylation levels were significantly associated with the degree of
differentiation (P=0.025 and P=0.031, respectively), lymph node
involvement (P=0.021 and P=0.021, respectively) and pTNM stage
(P=0.036 and P=0.022, respectively), but not sex, age or distal
metastasis (Table III; Fig. 3). The results of distal metastasis
may have been affected by the low ratio of positive cases (6.8%) in
the studied samples.
 | Table III.Association between H3K9me2 or EHMT2
expression patterns and clinicopathological characteristics of
patients with gastric carcinoma (n=118). |
Table III.
Association between H3K9me2 or EHMT2
expression patterns and clinicopathological characteristics of
patients with gastric carcinoma (n=118).
|
| H3K9me2
expression | EHMT2
expression |
|---|
|
|
|
|
|---|
|
Characteristics | Low | High | χ2 | P-value | Low | High | χ2 | P-value |
|---|
| Sex |
|
| 0.798 | 0.372 |
|
| 0.031 | 0.861 |
|
Male | 57 | 25 |
|
| 51 | 31 |
|
|
|
Female | 22 | 14 |
|
| 23 | 13 |
|
|
| Age, years |
|
| 0.957 | 0.328 |
|
| 0.145 | 0.703 |
|
≤60 | 37 | 22 |
|
| 38 | 21 |
|
|
|
>60 | 42 | 17 |
|
| 36 | 23 |
|
|
| Differentiation
degree |
|
| 4.661 | 0.031a |
|
| 5.027 | 0.025a |
| Well +
moderately differentiated | 43 | 13 |
|
| 41 | 15 |
|
|
| Poorly
differentiated | 36 | 26 |
|
| 33 | 29 |
|
|
| Lymph node
metastasis |
|
| 5.352 | 0.021a |
|
| 5.346 | 0.021a |
|
#x00A0; Negative | 40 | 11 |
|
| 38 | 13 |
|
|
|
#x00A0; Positive | 39 | 28 |
|
| 36 | 31 |
|
|
| Distal
metastasis |
|
| 1.114 | 0.291 |
|
| 0.554 | 0.457 |
|
#x00A0; Negative | 75 | 35 |
|
| 68 | 42 |
|
|
|
#x00A0; Positive | 4 | 4 |
|
| 6 | 2 |
|
|
| pTNM |
|
| 5.217 | 0.022a |
|
| 4.392 | 0.036a |
|
#x00A0; I+II | 48 | 15 |
|
| 45 | 18 |
|
|
|
#x00A0; III+IV | 31 | 24 |
|
| 29 | 26 |
|
|
EHMT2 overexpression was significantly associated
with H3K9me2 methylation level (P=0.040; data not shown). The
results indicated that EHMT2 and H3K9me2 expression patterns
exhibited notable consistency. As presented in Fig. 3, H3K9me2 exhibited similar ratios of
cases of each stage of pTNM to EHMT2 when analyzed based on the
expression level, which further confirmed their positive
association.
Effect of H3K9me2 and EHMT2 on the
survival of patients with GC
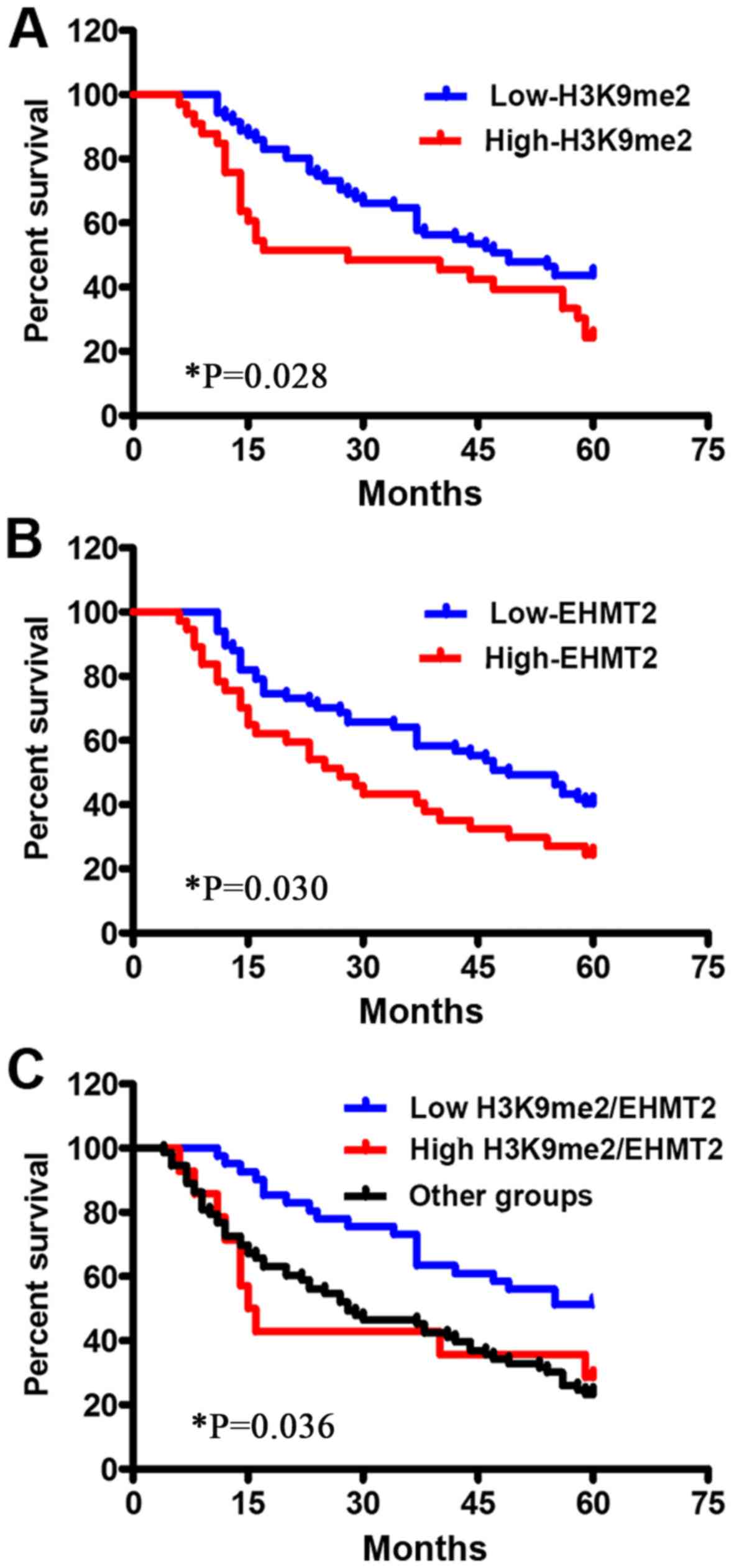
To understand the impact of H3K9me2 and EHMT2
expression patterns on the survival of patients with GC, overall
survival time based on H3K9me2 and EHMT expression levels was
further analyzed by K-M analysis with a log-rank test. Patients
with low expression levels of H3K9me2 exhibited a significantly
longer survival time compared with those in the high expression
group (P<0.05). Overexpression of EHMT2 presented the same trend
(P<0.05; Fig. 4; Table S1). Mean progression-free survival
time of patients in the low and high H3K9me2 expression groups were
45.521±2.195 and 34.061±3.861 months, respectively, and in the low
and high EHMT2 expression level groups were 41.761±2.399 and
32.243±3.364 months, respectively (Table S1). The median survival period
exhibited similar tendencies. Univariate K-M survival curves
demonstrated that tumor differentiation degree, lymph node
involvement, distal metastasis and pTNM stage were significantly
associated with a shorter survival time of patients with GC
(P<0.001; Table S2). However,
the sex and age of patients exhibited no association with patient
survival time in the present study.
Combination of EHMT2 overexpression
and H3K9me2 level is an effective prognostic marker for patients
with GC
To identify the independent risk factors for the
prognosis of patients with GC, multivariate Cox's regression
analysis was conducted to evaluate the clinicopathological features
of patients with GC and the expression levels of H3K9me2 and EHMT2.
The results demonstrated that the expression patterns of H3K9me2
and EHMT2, differentiation degree, lymph node involvement, distal
metastasis and pTNM stage exhibited independent significant
prognostic effects on patient survival time (P<0.05; Table IV). To present the significant
factors more accurately, the patients were regrouped by combining
the expression patterns of H3K9me2 and EHMT2. The expression
patterns were classified into two groups: Low (score ≤8) and high
(score >8) scoring groups. The patients were allocated into
three groups: i) Low expression group for patients with low
expression levels of H3K9me2 and EHMT2; ii) high expression group
for patients with high expression levels of H3K9me2 and EHMT2 and
iii) other group for patients with high H3K9me2 expression and low
EHMT2 expression, and vice versa. The survival analysis revealed
that the high expression group exhibited the shortest survival time
(31.467±5.671 months), the low expression group exhibited the
longest survival period (46.000±2.711 months) and the other group
exhibited an intermediate survival duration (37.943±2.830 months;
Table S1). Therefore, the
combination of H3K9me2 and EHMT2 expression patterns may be used as
a more accurate indicator for the overall survival time of patients
with GC compared with either H3K9me2 or EHMT2 alone (P=0.036;
Table S1). In addition,
multivariate Cox analysis revealed that the combined expression
patterns of H3K9me2 and EHMT2, differentiation degree, lymph node
involvement, distal metastasis and pTNM stage significantly
predicted the survival time of patients with GC (P<0.05;
Table IV).
 | Table IV.Multivariate Cox analysis of overall
survival based on individual and combined groups of the histone
signatures. |
Table IV.
Multivariate Cox analysis of overall
survival based on individual and combined groups of the histone
signatures.
| A, Individual
groups of histone signatures |
|---|
|
|---|
|
Characteristics | HR | 95% CI | P-value |
|---|
| Sex | 1.088 | 0.633–1.868 | 0.939 |
| Age | 0.773 | 0.462–1.293 | 0.539 |
| Differentiation
degree | 0.514 | 0.299–0.882 | 0.016a |
| Lymph node
metastasis | 0.230 | 0.113–0.467 |
<0.001c |
| Distal
metastasis | 0.402 | 0.185–0.872 | 0.021a |
| pTNM | 0.110 | 0.044–0.271 |
<0.001c |
| H3K9me2 | 1.050 | 0.637–1.729 | 0.042a |
| EHMT2 | 1.004 | 0.599–1.683 | 0.045a |
|
| B, Combined
groups of histone signatures |
|
|
Characteristics | HR | 95% CI | P-value |
|
| Differentiation
degree | 0.531 | 0.313–0.901 | 0.019a |
| Lymph node
metastasis | 0.234 | 0.116–0.474 |
<0.001c |
| Distal
metastasis | 0.388 | 0.183–0.822 | 0.013a |
| pTNM | 0.106 | 0.043–0.258 |
<0.001c |
| Low expression
group | 0.009b |
|
|
| High expression
group | 1.178 | 0.709–1.957 | 0.018a |
| Other groups | 1.434 | 0.715–2.876 | 0.020a |
Discussion
Alterations of epigenetic regulation genes,
including DNA methylation, histone modifications, chromatin
remodeling and non-coding RNA regulation, have been detected in
early carcinogenesis and cancer progression (30–34). A
number of them have been proposed as biomarkers for cancer
detection and tumor prognosis (35–37). A
number of diverse factors, including DNA damage, DNA and histone
methylation, in addition to environmental influence, are associated
with GC-associated mortality in China (17). Previously, increasing evidence has
revealed that aberrant epigenetic regulation serves an important
function during tumorigenesis. Histone methylations and their
corresponding catalytic enzymes were the focus of a previous study;
among various best-studied histone methylations, H3K9 methylation
is associated with gene repression (9). Epigenetic alterations of tumor genes
are present during carcinogenesis and may provide novel biomarkers
for diagnosis (38–40).
EHMT2 was first identified as a gene located in the
major histocompatibility complex locus in mice and human leukocyte
antigen locus in humans (41).
Previous studies have indicated that EHMT2 serves an important
function during the carcinogenesis and development of a tumor
(13,14,17,42,43).
EHMT2 is also a crucial factor in a variety of biological
progresses, including behavior plasticity, lymphocyte development,
stem cell differentiation and tumor cell growth (44). The clinical importance of EHMT2
expression in numerous cancer tissues has been studied (16); however, the expression pattern of
EHMT2 and its significance in GC is largely unclear. Although the
crucial functions of H3K9me2 and EHMT2 have been elucidated in
several tumor types (45,46), the associations between H3K9me2
methylation pattern and EHMT2 expression level in GC are unknown.
EHMT2 is significantly upregulated in numerous different tumor
types compared with matched normal controls, and knocking down
EHMT2 or the pharmacological inhibition of its activity suppresses
tumor cell growth and invasion, indicating that EHMT2 may be an
oncogenic and metastatic factor (47,48). In
addition, high expression levels of EHMT2 are correlated with a
poor overall survival in patients with lung adenocarcinoma
(16).
EHMT2 is a main histone lysine methylation enzyme
which catalyzes the modification at histone 3 lysine 9, including
H3K9me1 and H3K9me2. The patterns of H3K9me1 or H3K9me2 are
different during development, as there are either more mono- or
dimethylations at H3K9 during the maturation of the auditory system
(49). During the development of the
zebrafish retina, EHMT2 expression and H3K9me2 markers have been
noted to be closely associated (50).
H3K9 methylation is a crucial event in reprogramming
to pluripotency (51). Numerous
studies have demonstrated that the global level of H3K9me2 is
associated with the prognosis of prostate and kidney cancer
(52,53). These results indicate that there is
an association between EHMT2 expression and H3K9me2 markers.
The focus of the present study was the function of
EHMT2 and H3K9me2 in the processes associated with a poor prognosis
of GC. H3K9me2 and EHMT2 expression exhibited strong immunostaining
in GC tumor tissues compared with adjacent healthy tissues. There
was an association between the levels of H3K9me2 and EHMT2. The
results also demonstrated that EHMT2 expression was associated with
the differentiation degree and lymph node metastasis. These results
were consistent with a previous study (23), which demonstrated that increased
EHMT2 expression in GC tissues correlated with an advanced stage
and promoted tumor invasion and metastasis. Depletion of EHMT2 may
be of therapeutic value by inhibiting cell proliferation and
inducing apoptosis in GC (54). High
expression levels of H3K9me2 or EHMT2 were significantly associated
with a worse overall survival time in patients with GC. Patients
with combined lower levels of H3K9me2 and EHMT2 exhibited a better
survival rate and prognosis compared with those with combined
higher levels of H3K9me2 and EHMT2, in addition to the other group.
Furthermore, the results of the present study suggested that the
development of GC is associated with pathological grade, lymph node
metastasis and TNM stage.
In conclusion, EHMT2 expression level and H3K9me2
methylation level may be associated with the development risk and
prognosis of GC. Patients with increased EHMT2 and H3K9me2 levels
exhibited worse overall survival and a poorer prognosis compared
with other patients. Overexpression of EHMT2 and H3K9me2 levels may
be predictor markers of progression and prognosis in patients with
GC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural
Science Foundation of China (grant nos. 81672414 and 81472548).
Availability of data and materials
The analyzed data sets generated during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
PC conceived the study and was a major contributor
in writing the manuscript. QQ and ZZ contributed to the analysis
and interpretation of the data. XS and SY were involved in drafting
the manuscript and the re-analysis of the immunohistochemistry
scores. ZY acquired the data and performed statistical analysis. RS
also acquired the data. YL performed the immunostaining. DG made
contributions to the experimental studies. HF designed the research
protocols and gave final approval of the version to be published.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was reviewed and ethically
approved by the Committee for Ethical Review of Research of
Yancheng Hospital (Yancheng, China), and written informed consent
was obtained from the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Norollahi SE, Alipour M, Rashidy-Pour A,
Samadani AA and Larijani LV: Regulatory fluctuation of WNT16 gene
expression is associated with human gastric adenocarcinoma. J
Gastrointest Cancer. 50:42–47. 2017. View Article : Google Scholar
|
|
3
|
Nashimoto A, Akazawa K, Isobe Y, Miyashiro
I, Katai H, Kodera Y, Tsujitani S, Seto Y, Furukawa H, Oda I, et
al: Gastric cancer treated in 2002 in Japan: 2009 annual report of
the JGCA nationwide registry. Gastric Cancer. 16:1–27. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Theuer CP, Kurosaki T, Ziogas A, Butler J
and Anton-Culver H: Asian patients with gastric carcinoma in the
United States exhibit unique clinical features and superior overall
and cancer specific survival rates. Cancer. 89:1883–1892. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fraga MF, Ballestar E, Villar-Garea A,
Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S,
Petrie K, et al: Loss of acetylation at Lys16 and trimethylation at
Lys20 of histone H4 is a common hallmark of human cancer. Nat
Genet. 37:391–400. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wood LD, Parsons DW, Jones S, Lin J,
Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al: The
genomic landscapes of human breast and colorectal cancers. Science.
318:1108–1113. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Benard A, Goossens-Beumer IJ, van Hoesel
AQ, de Graaf W, Horati H, Putter H, Zeestraten EC, van de Velde CJ
and Kuppen PJ: Histone trimethylation at H3K4, H3K9 and H4K20
correlates with patient survival and tumor recurrence in
early-stage colon cancer. BMC Cancer. 14:5312014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tachibana M, Ueda J, Fukuda M, Takeda N,
Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T and Shinkai Y:
Histone methyltransferases G9a and GLP form heteromeric complexes
and are both crucial for methylation of euchromatin at H3-K9. Genes
Dev. 19:815–826. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dawson MA and Kouzarides T: Cancer
epigenetics: From mechanism to therapy. Cell. 150:12–27. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tachibana M, Sugimoto K, Nozaki M, Ueda J,
Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, et al: G9a
histone methyltransferase plays a dominant role in euchromatic
histone H3 lysine 9 methylation and is essential for early
embryogenesis. Genes Dev. 16:1779–1791. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Barski A, Cuddapah S, Cui K, Roh TY,
Schones DE, Wang Z, Wei G, Chepelev I and Zhao K: High-resolution
profiling of histone methylations in the human genome. Cell.
129:823–837. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Scheer S and Zaph C: The Lysine
Methyltransferase G9a in Immune Cell Differentiation and Function.
Front Immunol. 8:4292017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tian YF, Wang HC, Luo CW, Hung WC, Lin YH,
Chen TY, Li CF, Lin CY and Pan MR: Preprogramming therapeutic
response of PI3K/mTOR dual inhibitor via the regulation of EHMT2
and p27 in pancreatic cancer. Am J Cancer Res. 8:1812–1822.
2018.PubMed/NCBI
|
|
14
|
Casciello F, Al-Ejeh F, Kelly G, Brennan
DJ, Ngiow SF, Young A, Stoll T, Windloch K, Hill MM, Smyth MJ,
Gannon F and Lee JS: G9a drives hypoxia-mediated gene repression
for breast cancer cell survival and tumorigenesis. Proc Natl Acad
Sci U S A. 114:7077–7082. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim K, Son MY, Jung CR, Kim DS and Cho HS:
EHMT2 is a metastasis regulator in breast cancer. Biochem Biophys
Res Commun. 496:758–762. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang T, Zhang P, Li W, Zhao T, Zhang Z,
Chen S, Yang Y, Feng Y, Li F, Shirley Liu X, Zhang L, Jiang G and
Zhang F: G9A promotes tumor cell growth and invasion by silencing
CASP1 in non-small-cell lung cancer cells. Cell Death Dis.
8:e27262017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang L, Dong X, Ren Y, Luo J, Liu P, Su D
and Yang X: Targeting EHMT2 reverses EGFR-TKI resistance in NSCLC
by epigenetically regulating the PTEN/AKT signaling pathway. Cell
Death Dis. 9:1292018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qin J, Li Q, Zeng Z, Wu P, Jiang Y, Luo T,
Ji X, Zhang Q, Hao Y and Chen L: Increased expression of G9A
contributes to carcinogenesis and indicates poor prognosis in
hepatocellular carcinoma. Oncol Lett. 15:9757–9765. 2018.PubMed/NCBI
|
|
19
|
Qin J, Zeng Z, Luo T, Li Q, Hao Y and Chen
L: Clinicopathological significance of G9A expression in colorectal
carcinoma. Oncol Lett. 15:8611–8619. 2018.PubMed/NCBI
|
|
20
|
Lee SH, Kim J, Kim WH and Lee YM: Hypoxic
silencing of tumor suppressor RUNX3 by histone modification in
gastric cancer cells. Oncogene. 28:184–194. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Renneville A, Van Galen P, Canver MC,
McConkey M, Krill-Burger JM, Dorfman DM, Holson EB, Bernstein BE,
Orkin SH, Bauer DE, et al: EHMT1 and EHMT2 inhibition induces fetal
hemoglobin expression. Blood. 126:1930–1939. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cui H, Hu Y, Guo D, Zhang A, Gu Y, Zhang
S, Zhao C, Gong P, Shen X, Li Y, et al: DNA methyltransferase 3A
isoform b contributes to repressing E-cadherin through cooperation
of DNA methylation and H3K27/H3K9 methylation in EMT-related
metastasis of gastric cancer. Oncogene. 37:4358–4371. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hu L, Zang MD, Wang HX, Zhang BG, Wang ZQ,
Fan ZY, Wu H, Li JF, Su LP, Yan M, et al: G9A promotes gastric
cancer metastasis by upregulating ITGB3 in a SET domain-independent
manner. Cell Death Dis. 9:2782018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang C, Wei S, Hu J and Xiong Z:
Upregulated expression of G9a is correlated with poor prognosis of
gastric cancer patients. Medicine (Baltimore). 98:e182122019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Y, Guo D, Sun R, Chen P, Qian Q and Fan
H: Methylation patterns of Lys9 and Lys27 on histone H3 correlate
with patient outcome in gastric cancer. Dig Dis Sci. 64:439–446.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee KH, Park JW, Sung HS, Choi YJ, Kim WH,
Lee HS, Chung HJ, Shin HW, Cho CH, Kim TY, et al: PHF2 histone
demethylase acts as a tumor suppressor in association with p53 in
cancer. Oncogene. 34:2897–2909. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ji X, Bu ZD, Yan Y, Li ZY, Wu AW, Zhang
LH, Zhang J, Wu XJ, Zong XL, Li SX, Shan F, et al: The 8th edition
of the American Joint Committee on Cancer tumor-node-metastasis
staging system for gastric cancer is superior to the 7th edition:
results from a Chinese mono-institutional study of 1663 patients.
Gastric Cancer. 21:643–652. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Axiotis CA, Monteagudo C, Merino MJ,
LaPorte N and Neumann RD: Immunohistochemical detection of
P-glycoprotein in endometrial adenocarcinoma. Am J Pathol.
138:799–806. 1991.PubMed/NCBI
|
|
29
|
Fisher KE, Cohen C, Siddiqui MT, Palma JF,
Lipford EH III and Longshore JW: Accurate detection of BRAF p.V600E
mutations in challenging melanoma specimens requires stringent
immunohistochemistry scoring criteria or sensitive molecular
assays. Hum Pathol. 45:2281–2293. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sina AA, Carrascosa LG, Liang Z, Grewal
YS, Wardiana A, Shiddiky MJA, Gardiner RA, Samaratunga H, Gandhi
MK, Scott RJ, et al: Epigenetically reprogrammed methylation
landscape drives the DNA self-assembly and serves as a universal
cancer biomarker. Nat Commun. 9:49152018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Noberini R, Osti D, Miccolo C, Richichi C,
Lupia M, Corleone G, Hong SP, Colombo P, Pollo B, Fornasari L, et
al: Extensive and systematic rewiring of histone post-translational
modifications in cancer model systems. Nucleic Acids Res.
46:3817–3832. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu Q, Lian JB, Stein JL, Stein GS,
Nickerson JA and Imbalzano AN: The BRG1 ATPase of human SWI/SNF
chromatin remodeling enzymes as a driver of cancer. Epigenomics.
9:919–931. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kumar R, Li DQ, Müller S and Knapp S:
Epigenomic regulation of oncogenesis by chromatin remodeling.
Oncogene. 35:4423–4436. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ma Y, Yang Y, Wang F, Moyer MP, Wei Q,
Zhang P, Yang Z, Liu W, Zhang H, Chen N, et al: Long non-coding RNA
CCAL regulates colorectal cancer progression by activating
Wnt/β-catenin signalling pathway via suppression of activator
protein 2α. Gut. 65:1494–1504. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tahara T and Arisawa T: DNA methylation as
a molecular biomarker in gastric cancer. Epigenomics. 7:475–486.
2015. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lomberk G, Blum Y, Nicolle R, Nair A,
Gaonkar KS, Marisa L, Mathison A, Sun Z, Yan H, Elarouci N, et al:
Distinct epigenetic landscapes underlie the pathobiology of
pancreatic cancer subtypes. Nat Commun. 9:19782018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tvardovskiy A, Schwämmle V, Kempf SJ,
Rogowska-Wrzesinska A and Jensen ON: Accumulation of histone
variant H3.3 with age is associated with profound changes in the
histone methylation landscape. Nucleic Acids Res. 45:9272–9289.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Okugawa Y, Grady WM and Goel A: Epigenetic
alterations in colorectal cancer: Emerging biomarkers.
Gastroenterology. 149:1204–1225 e12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Costa-Pinheiro P, Montezuma D, Henrique R
and Jerónimo C: Diagnostic and prognostic epigenetic biomarkers in
cancer. Epigenomics. 7:1003–1015. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gu L, Frommel SC, Oakes CC, Simon R, Grupp
K, Gerig CY, Bär D, Robinson MD, Baer C, Weiss M, et al ICGC
Project on Early Onset Prostate Cancer, : BAZ2A (TIP5) is involved
in epigenetic alterations in prostate cancer and its overexpression
predicts disease recurrence. Nat Genet. 47:22–30. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Brown SE, Campbell RD and Sanderson CM:
Novel NG36/G9a gene products encoded within the human and mouse MHC
class III regions. Mamm Genome. 12:916–924. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang YF, Zhang J, Su Y, Shen YY, Jiang DX,
Hou YY, Geng MY, Ding J and Chen Y: G9a regulates breast cancer
growth by modulating iron homeostasis through the repression of
ferroxidase hephaestin. Nat Commun. 8:2742017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wei L, Chiu DK, Tsang FH, Law CT, Cheng
CL, Au SL, Lee JM, Wong CC, Ng IO and Wong CM: Histone
methyltransferase G9a promotes liver cancer development by
epigenetic silencing of tumor suppressor gene RARRES3. J Hepatol.
67:758–769. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mayr C, Helm K, Jakab M, Ritter M,
Shrestha R, Makaju R, Wagner A, Pichler M, Beyreis M, Staettner S,
et al: The histone methyltransferase G9a: A new therapeutic target
in biliary tract cancer. Hum Pathol. 72:117–126. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Casciello F, Al-Ejeh F, Kelly G, Brennan
DJ, Ngiow SF, Young A, Stoll T, Windloch K, Hill MM, Smyth MJ, et
al: G9a drives hypoxia-mediated gene repression for breast cancer
cell survival and tumorigenesis. Proc Natl Acad Sci USA.
114:7077–7082. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Salzberg AC, Harris-Becker A, Popova EY,
Keasey N, Loughran TP, Claxton DF and Grigoryev SA: Genome-wide
mapping of histone H3K9me2 in acute myeloid leukemia reveals large
chromosomal domains associated with massive gene silencing and
sites of genome instability. PLoS One. 12:e01737232017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dong C, Wu Y, Yao J, Wang Y, Yu Y,
Rychahou PG, Evers BM and Zhou BP: G9a interacts with Snail and is
critical for Snail-mediated E-cadherin repression in human breast
cancer. J Clin Invest. 122:1469–1486. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liu S, Ye D, Guo W, Yu W, He Y, Hu J, Wang
Y, Zhang L, Liao Y, Song H, et al: G9a is essential for
EMT-mediated metastasis and maintenance of cancer stem cell-like
characters in head and neck squamous cell carcinoma. Oncotarget.
6:6887–6901. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ebbers L, Runge K and Nothwang HG:
Differential patterns of histone methylase EHMT2 and its catalyzed
histone modifications H3K9me1 and H3K9me2 during maturation of
central auditory system. Cell Tissue Res. 365:247–264. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Olsen JB, Wong L, Deimling S, Miles A, Guo
H, Li Y, Zhang Z, Greenblatt JF, Emili A and Tropepe V: G9a and
ZNF644 physically associate to suppress progenitor gene expression
during neurogenesis. Stem Cell Reports. 7:454–470. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sridharan R, Gonzales-Cope M, Chronis C,
Bonora G, McKee R, Huang C, Patel S, Lopez D, Mishra N, Pellegrini
M, et al: Proteomic and genomic approaches reveal critical
functions of H3K9 methylation and heterochromatin protein-1γ in
reprogramming to pluripotency. Nat Cell Biol. 15:872–882. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Seligson DB, Horvath S, McBrian MA, Mah V,
Yu H, Tze S, Wang Q, Chia D, Goodglick L and Kurdistani SK: Global
levels of histone modifications predict prognosis in different
cancers. Am J Pathol. 174:1619–1628. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Mosashvilli D, Kahl P, Mertens C,
Holzapfel S, Rogenhofer S, Hauser S, Büttner R, Von Ruecker A,
Müller SC and Ellinger J: Global histone acetylation levels:
Prognostic relevance in patients with renal cell carcinoma. Cancer
Sci. 101:2664–2669. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lin X, Huang Y, Zou Y, Chen X and Ma X:
Depletion of G9a gene induces cell apoptosis in human gastric
carcinoma. Oncol Rep. 35:3041–3049. 2016. View Article : Google Scholar : PubMed/NCBI
|