Introduction
Apolipoprotein B mRNA-editing enzyme catalytic
polypeptides 3 (A3) are a class of intracellular innate immune
protein molecules with antiviral abilities in humans (1). The A3 family has seven members,
including APOBEC3A, B, C, D/E, F, G and H (1). The A3 family members can edit the
negative DNA strand generated during reverse transcription of a
virus, changing cytosine (C) into uracil (U), thereby transforming
guanine (G) into adenine (A) at corresponding sites on the positive
DNA strand and finally leading to the C/G-to-thymine (T)/A
hypermutation in the viral genome (2). Overall, the mutant viral genome is
degraded by U DNA transglucosylase and the hypermutation in the
negative DNA strand blocks the replication of the positive strand,
thereby inhibiting viral replication (2).
The members of the A3 family have been demonstrated
to induce the C/G-to-T/A hypermutation in the human
immunodeficiency virus (HIV) genome and to subsequently inhibit
viral replication (3–6). Luo et al (7) have demonstrated that APOBEC3F (A3F)
induces a G>A mutation in the genome of porcine endogenous
retrovirus (PERV) through a cytidine deamination mechanism, thereby
inhibiting PERV replication. Additionally, Noguchi et al
(8) have reported that high
expression levels of APOBEC3G (A3) protein in HepG2 cells
significantly reduce synthesis of the hepatitis B virus (HBV) and
induce the genomic hypermutation. Vieira et al (9) reported that the infection of high-risk
human papillomavirus (HPV) upregulates mRNA and protein expression
levels of APOBEC3B (A3B). After infection with inactivated HPV E6,
the HPV infection is no longer able to regulate expression levels
of A3B. It has been revealed that A3 can also target DNA viruses
requiring no reverse transcription when replicating, including the
TT virus, adeno-associated virus and herpes simplex virus 1
(10–13). A number of studies have demonstrated
that A3 serve an important role in mediating the clearance of
exogenous circular DNA from cells, which may clear the HPV genome
in persistently infected cells, and that the APOBEC protein may be
a driver for the HPV genomic mutations in cervical precancerous
lesions (14–16). Kondo et al (17) have reported that A3A is highly
expressed in patients with oropharyngeal cancer, which is involved
in the regulation of HPV16 infection and the integration in
oropharyngeal cancer.
HPV is a non-enveloped double-stranded DNA virus of
~8 kb. The genome can be divided into the following three regions:
The long control region (LCR); early region, containing 6 open
reading frames (ORFs), including E1, E2, E4, E5, E6 and E7; and
late regions, containing the L1 and L2 ORFs. Among these regions,
E6 and E7 are key oncogenes which serve important roles in
HPV-induced cervical cancer by interfering with the tumor
suppressor genes p53 and pRb, thus inhibiting normal cell
proliferation (18,19). The complete E2 protein regulates
viral mRNA transcription and DNA replication, negatively regulating
the expression of E6 and E7 oncogenes (18,19).
However, there are few systematic studies concerning
APOBEC3s-associated hypermutations in different regions of the HPV
genome or the effects of APOBEC3s on HPV protein expression levels,
particularly the E2 protein.
To elucidate whether the A3 family members function
in the regulation of HPV16 infection and the development of
cervical cancer in Uygur females from Xinjiang, China, samples of
cervical lesions were harvested from patients with high-risk HPV16
infections and analyzed. The mRNA and protein expressions of the A3
family members were detected. Additionally, the E2 region of the
HPV16 genome was amplified using the differential DNA denaturation
PCR (3D-PCR), followed by sequence analysis. In addition, to
investigate the effects of high expression levels of A3 on the
expression and editing of the E2 region of the HPV16 genome, a SiHa
cervical cancer cell model expressing high levels of A3 was
established in vitro.
Materials and methods
Patients
Cervical samples were obtained from 45 Uygur females
infected with high-risk HPV16 (age, 42.27±10.46 years), who were
admitted to the People's Hospital of Xinjiang Uygur Autonomous
Region (Urumqi, China), from January 2017 to December 2017.
According to the pathological diagnosis, there were 15 cases of
cervicitis, 15 cases of cervical intraepithelial neoplasia (CIN)
I–III and 15 cases of cervical cancer. The International Federation
of Gynecology and Obstetrics (FIGO, 2018) staging criteria were
used for CIN grading and cervical cancer staging (20). Informed consent was provided by every
patient and the study was approved by The Ethics Review Board of
the People's Hospital of Xinjiang Uygur Autonomous Region.
Study cell line
The human cervical carcinoma SiHa cell line was
purchased from the American Type Culture Collection (HTB-35). The
cells were verified by short tandem repeat (STR) testing. The cells
were cultured with the Dulbecco's modified eagle medium (11885092;
Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine
serum (CCS30013.01HI; EnmoAsai Biotechnology Co., Ltd., Changzhou,
China; http://www.mrcing.com/col.jsp?id=105), supplemented
with 1% penicillin-streptomycin (10,000 U/ml) (15140122; Gibco) and
incubated at 37°C with 5% CO2.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from cervical tissue samples
using a Total RNA extraction kit (DP431; Tiangen Biotech Co.,
Ltd.). The mRNAs of A3C (NM_014508.3), A3F (NM_001006666.2) and A3G
(NM_001349436.1) were obtained from the GenBank database
(uniprot.org/database/DB-0028) and the
ORFs were designed accordingly for the PCR primers (Table I). The extracted RNA was reverse
transcribed into cDNA using a PrimeScript™ RT reagent kit (cat. no.
RR037A; Takara Bio, Inc.). The reaction conditions were 37°C for 15
min and 85°C for 5 sec. RT-qPCR was performed using an AGS PCR
machine (AFD4800; Hangzhou AnYu Technologies Co., Ltd.). The
reaction conditions were set as follows: 95°C for 2 min; 95°C for 5
sec; 60°C for 10 sec; and 72 for 30 sec for a total of 40 cycles.
Target gene expression levels were determined with the
2−ΔΔCq method (21).
GADPH was used as an endogenous control.
 | Table I.PCR primer sequences. |
Table I.
PCR primer sequences.
| Gene | Primer sequences
(5′-3′) |
|---|
| APOBEC3C |
|
|
Forward |
GAGTGGGACAGGGACAAGCA |
|
Reverse |
GGCTCAGGATGACCAGGCAG |
| APOBEC3F |
|
|
Forward |
ATGAAGCCTCACTTCAGAAAC |
|
Reverse |
TCACTCGAGAATCTCCTGCA |
| APOBEC3G |
|
|
Forward |
TCTGGCTGTGCTACGAAGT |
|
Reverse |
GTGGAAGAATCTCATCTCTGG |
| GADPH |
|
|
Forward |
TCTCCTCTGACTTCAACAGCGAC |
|
Reverse |
CCCTGTTGCTGTAGCCAAATTC |
Western blot analysis
The tissue or SiHa cells were lysed using Biosharp
lysis (BL504A). The protein concentration was determined using the
bicinchoninic acid assay method. Protein samples (40 µg per lane)
were loaded onto a 4% gel, resolved SDS-PAGE and then subsequently
transferred onto a PVDF membrane. After blocking with 5% BSA at
room temperature for 1 h, the membrane was incubated with
anti-APOBEC3G (1:500; bs-15407R; BIOSS), anti-APOBEC3F (1:500;
ab227962; Abcam), anti-APOBEC3C (1:500; bs-12495R; BIOSS) and
anti-GAPDH (1:5,000; bs-50549R; BIOSS) primary antibodies at 4°C
for 16 h. Subsequently, the membranes were incubated with the
secondary antibody of goat anti-rabbit HRP conjugated IgG
(1:10,000; ZB-2301; OriGene Technologies, Inc.) at 37°C for 1 h in
the dark. The blot was developed using the ECL method (PE0010;
Beijing Solarbio Science & Technology Co., Ltd.). Protein bands
were imaged and analyzed using ImageJ software version 1.52s
(National Institutes of Health).
Lentiviral vector transfection
The A3C, A3F and A3G lentiviral expression vectors
were constructed by Shanghai GeneChem Co., Ltd. The lentiviral
expression vectors of PLVX-mCMV-ZsGreen-PGK-Puro-homo-APOBEC3C,
PLVX-mCMV- ZsGreen-PGK-Puro-Homo-APOBEC3F and PLVX-mCMV-
ZsGreen-PGK-Puro-homo-APOBEC3G were transfected into the SiHa cells
using Polybrene (Shanghai GeneChem Co., Ltd.) and Enhanced
Infection Solution (Shanghai GeneChem Co., Ltd.), at the
multiplicities of infection of 50, 10 and 20, respectively. The
blank and scrambled small interfering RNA (siRNA) negative control
groups were also set up. The siRNA was synthesized by Shanghai
GeneChem Co., Ltd; however, the sequences were not available.
Following addition of the lentiviral vectors, the cells were
incubated at 37°C in a 5% CO2 incubator for 16 h.
Subsequently, the cells were cultured with fresh DMEM for 72 h
prior to collection. The fluorescence intensity was observed using
an inverted fluorescence microscope.
3D-PCR amplification and hypermutation
analysis of the HPV16 E2 genome
HPV16 DNA was extracted from the cervical tissue
samples and SiHa cells using a Genomic DNA Extraction kit (DP304;
Tiangen Biotech Co., Ltd.). With the template of HPV16 DNA, the E2
region of the HPV16 genome was amplified with high-fidelity Taq
enzyme (14966005; Thermo Fisher Scientific, Inc.) using 3D-PCR at
different denaturation temperatures, at 89.3, 87.43, 85.2, 83.5 and
81.9°C. For the first round of PCR, the following primers were
used: Forward, 5′-GAGGACGAGGACAAGGAAA-3′ and reverse,
5′-AAGCACGCCAGTAATGTTG-3′. The reaction conditions were set as:
89.3°C for 1 min; 89.3°C for 45 sec, 52°C for 30 sec, 65°C for 50
sec, for a total of 40 cycles; followed by 65°C for 10 min. The
products were used as the templates for the second round of PCR,
with the following primers: Forward, 5′-ACGATGGAGACTCTTTGCC-3′ and
reverse, 5′-GCCAGTAATGTTGTGGATGC-3′. The reaction conditions were
set as follows: 87.4°C for 1 min; 87.4°C for 45 sec; 52°C for 30
sec, 65°C for 50 sec, for a total of 40 cycles; followed by 65°C
for 10 min. The obtained products were then used as the templates
for the next amplification with lowered denaturation temperatures,
ranging between 81.9°C and 89.3°C. The final products were
separated by 1.5% agarose gel electrophoresis. The target fragments
were recovered using a Gel Recovery kit (B518131-0100; Sangon
Biotech Co., Ltd.) and ligated into the pBS-T vector for
transformation. The positive clone was picked up and amplified by
liquid LB medium (B540111-0100; Sangon Biotech Co., Ltd.). DNA was
extracted from these amplified plasmids, which were subjected to
the Sanger sequencing on the ABI3100 sequencer (Xinjiang Dingju
Medical Laboratory) (http://dj-medlab.com/). The PCR product containing the
specified number of C-to-T mutations was cloned into the pBS-T
vector to prepare the reference plasmid [NC_001526.4 (1892..2989)]
(Xinjiang Dingju Medical Laboratory), which was verified by Sanger
sequencing. DNAMAN software version 6.0 (Lynnon Biosoft) was used
for sequence alignment.
Statistical analysis
The experiments were performed three times
independently. Data are presented as the mean ± SD. SPSS 16.0
software (SPSS, Inc.) was used for statistical analysis. One-way
ANOVA was performed for the comparison of mean values among groups
followed by Tukey's post hoc test. A χ2 test was used to
compare the hypermutation ratio. P<0.05 was considered to
indicate a statistically significant difference.
Results
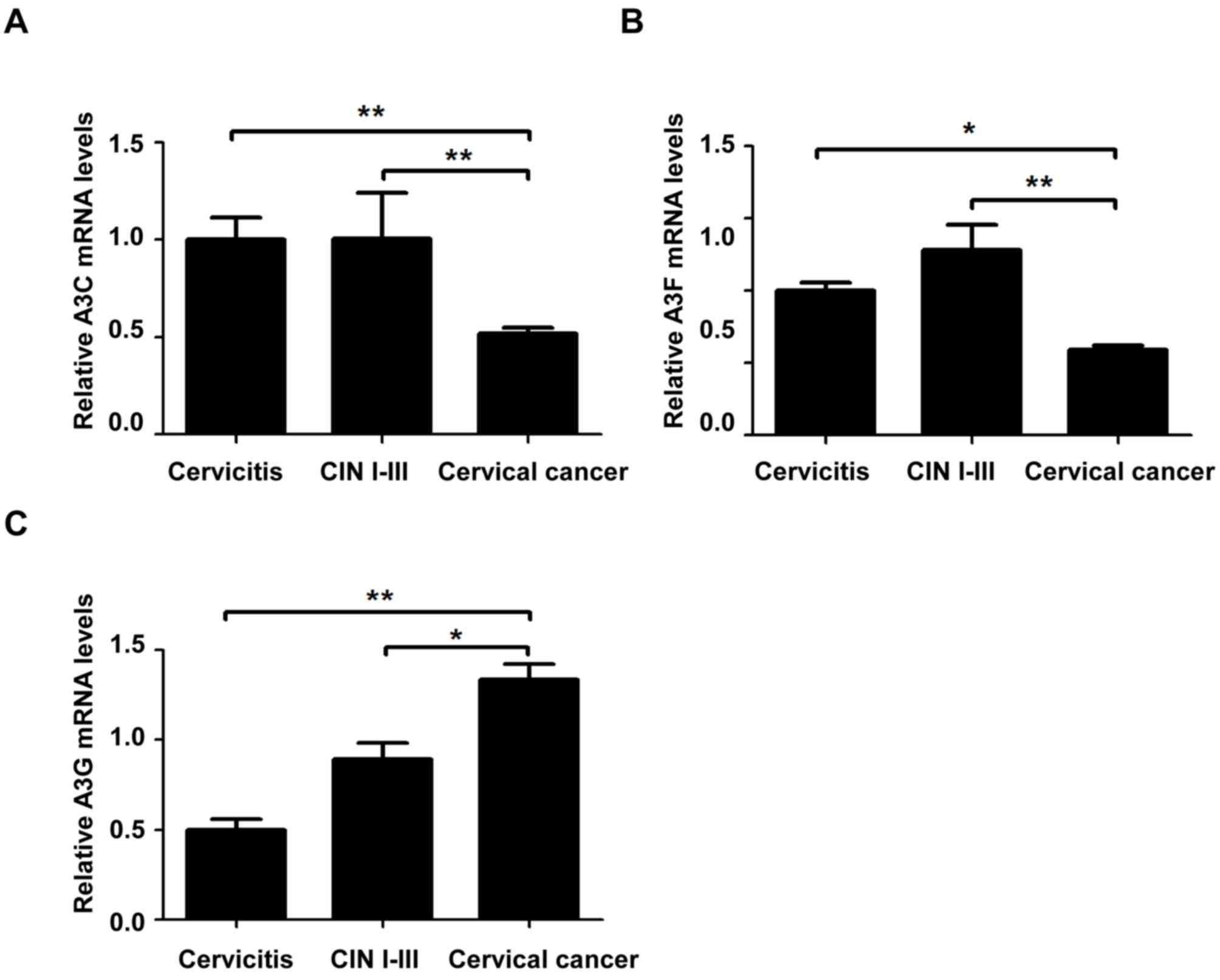
Relative mRNA expression levels of
A3C, A3F and A3G in cervical lesions of different grades
To investigate the mRNA expression levels of A3C,
A3F and A3G in cervical tissue samples, RT-qPCR was performed. The
mRNA expression levels of A3C in the cervical cancer group were
significantly lower compared with the CIN I–III and cervicitis
groups (P<0.01, Fig. 1A). The
mRNA expression levels of A3F in the cervical cancer group were
significantly lower compared with the cervicitis (P<0.05) and
CIN I–III (P<0.01) groups (Fig.
1B). The A3G mRNA expression in the cervical cancer group was
significantly higher compared with the CIN I–III (P<0.05) and
cervicitis groups (P<0.05, Fig.
1C). These results indicated that, with the development and
progression of cervical lesions, the relative mRNA expression
levels of A3C and A3F were significantly decreased, whereas the
relative A3G mRNA expression levels were significantly increased,
suggesting that A3 may be involved in the development of cervical
cancer.
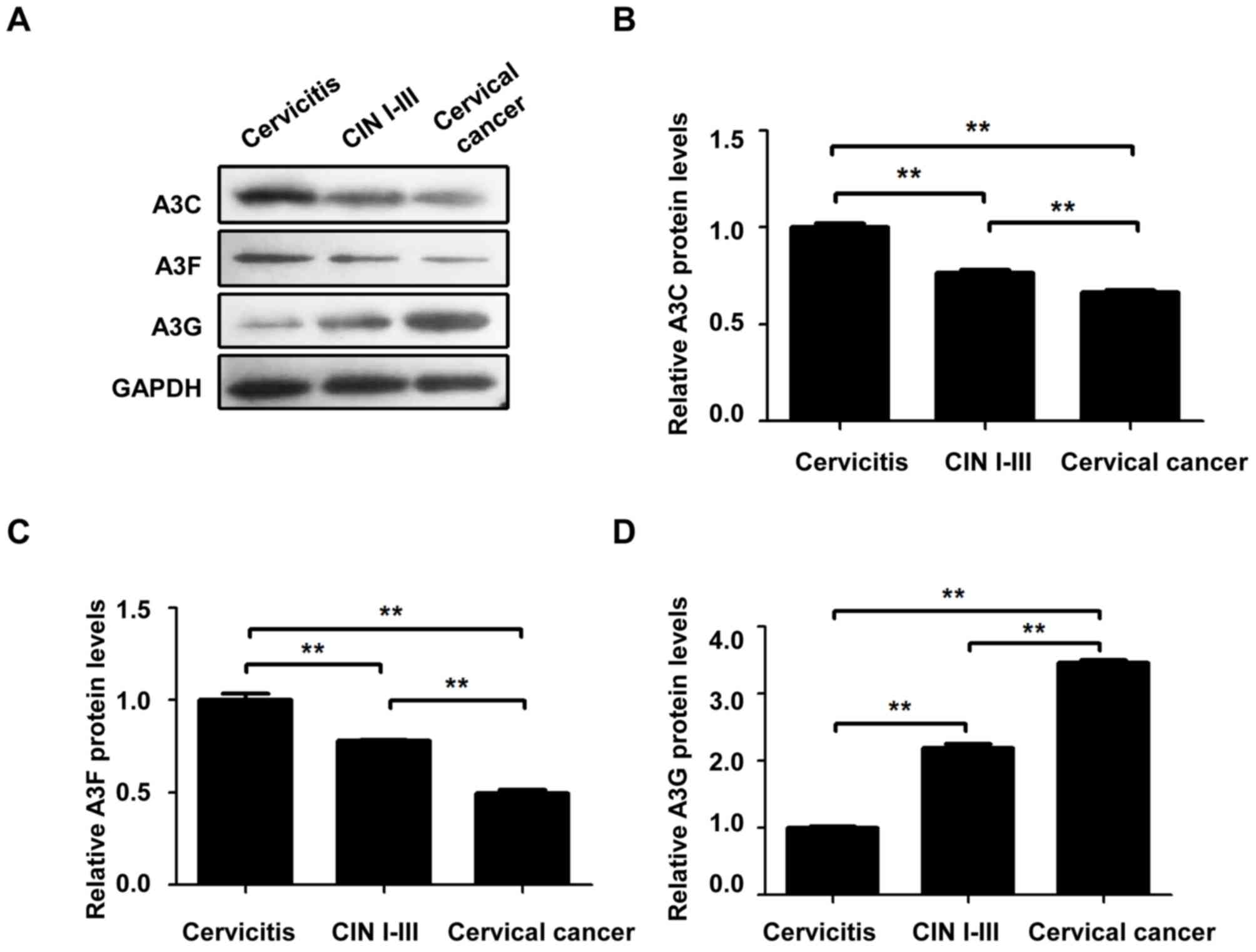
Relative protein expression levels of
A3C, A3F and A3G in cervical lesions of different grades
To investigate the protein expression levels of A3C,
A3F and A3G in these cervical tissue samples, western blot analysis
was performed. The protein expression levels of A3C (Fig. 2A and B) and A3F (Fig. 2A and C) in the cervical cancer group
were significantly lower compared with the cervicitis and CIN I–III
groups (P<0.01). The protein expression levels of A3C and A3F in
the CIN I–III group were significantly lower than the cervicitis
group (P<0.01). The A3G protein expression levels (Fig. 2A and C) in the cervical cancer group
were significantly higher compared with the cervicitis and CIN
I–III groups (P<0.01). The A3G protein expression levels in the
CIN I–III groups were significantly higher compared with the
cervicitis group (P<0.01). These results indicated that, with
the development and progression of cervical lesions, the relative
protein expression levels of A3C and A3F were significantly
decreased, whereas the relative A3G protein expression levels were
significantly increased, suggesting that A3 may be involved in the
cervical cancer pathogenesis and development.
3D-PCR detection of the HPV16 E2 gene
hypermutation in different grades of cervical lesions
DNA was extracted from the cervical tissues of the
patients with cervical cancer and 3D-PCR was performed to detect
the HPV16 E2 hypermutation. At the denaturation temperature of
≥87.4°C, the control DNA without mutations [0× base substitution
(b.s.)] was amplified and at the denaturation temperature of 85.2°C
the control DNA carrying 3 C>T mutants (3× b.s.) was amplified
(Fig. 3A), implying that at the
denaturation temperature of ≤85.2°C the hypermutation sample with
more than three base substitutions may be detected. Detection of
the cervicitis, CIN I–III and cervical cancer groups revealed that
in the 15 samples from the patients with cervicitis, hypermutation
was detected in one sample at the denaturation temperature of
85.2°C. For the 15 cases of CIN I–III, hypermutation was detected
in 5 cases at the denaturation temperature of 85.2°C and out of
these 5 cases hypermutation was detected in 2 cases at the
denaturation temperature of 83.5°C. For the 15 samples of cervical
cancer, hypermutation was detected in 5 cases at the denaturation
temperature of 85.2°C and out of these 5 cases, hypermutation was
detected in 3 cases at the denaturation temperature of 83.5°C
(Table II and Fig. 3A).
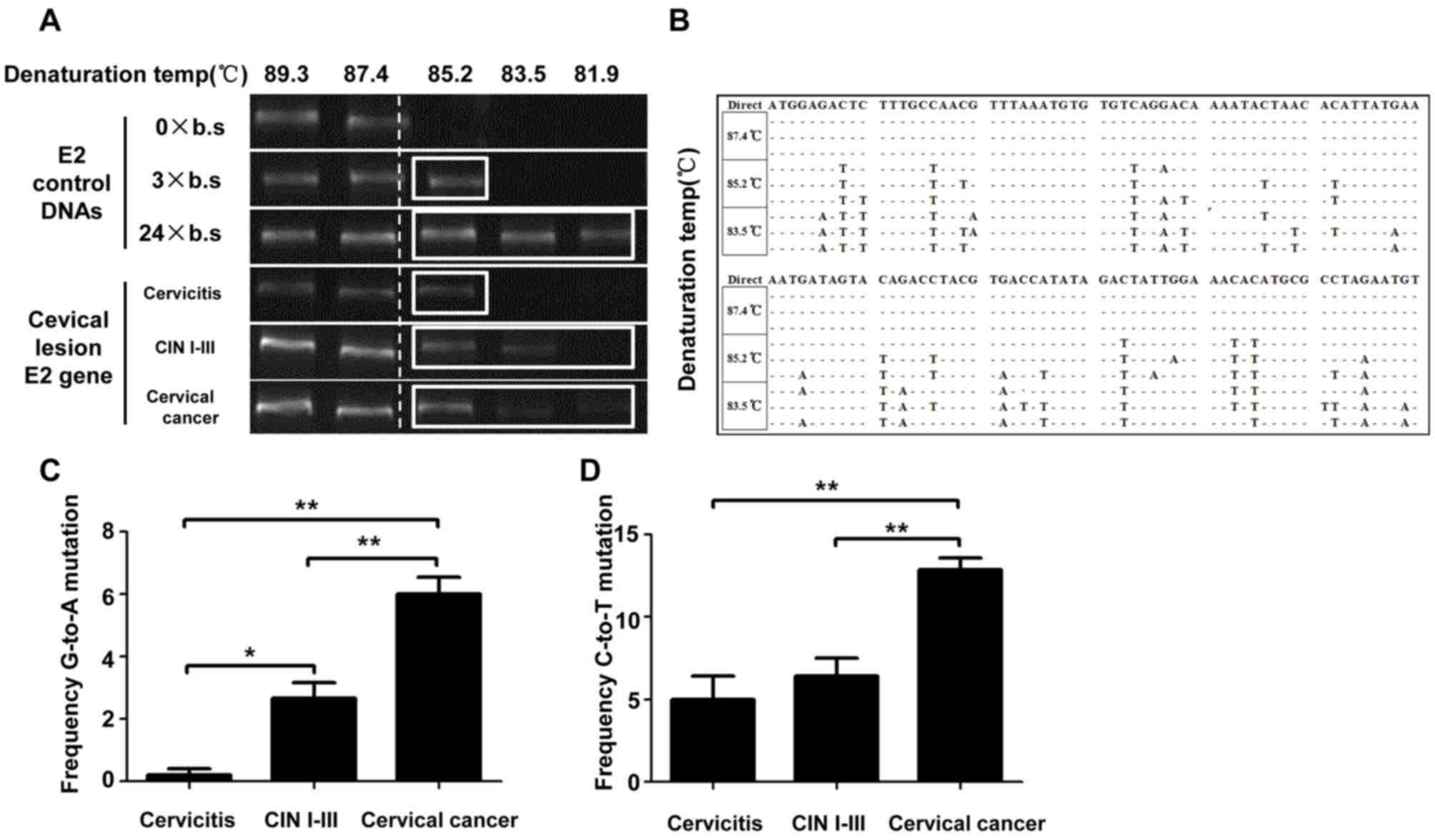 | Figure 3.Hypermutation of HPV16 E2 gene in
cervical lesions of different grades. The HPV16 E2 gene in cervical
tissues was amplified with 3D-PCR at different denaturing
temperatures of 89.3, 87.4, 85.2, 83.5 and 81.9°C. (A) 0× b.s., 3×
b.s. and 24× b.s. were used to construct the reference plasmids
with 0, 3 and 24 C-to-T base substitutions and the denaturation
temperatures for the hypermutation were detected in these samples.
The 3× b.s. sample exhibited bands at 85.2°C. Positive samples were
detected for the cervicitis, CIN I–III and cervical cancer groups
at the denaturing temperature of 85.2°C. (B) Base substitutions for
the hypermutation in single samples at different melting
temperatures of 87.4, 85.2 and 83.5°C. (C) Frequency of G-to-A
mutation. (D) Frequency of C-to-T mutation. *P<0.05,
**P<0.01. HPV, human papilloma virus; b.s., base substitution;
CIN I–III, cervical intraepithelial neoplasia I, II and III; C,
cytosine; G, guanine; A, adenine; T, thymine. |
 | Table II.Association between HPV16 E2
hypermutation and cervical lesions. |
Table II.
Association between HPV16 E2
hypermutation and cervical lesions.
|
| Second-round PCR
denaturing temperature, °C |
|
|
|---|
|
|
|
|
|
|---|
| Group | 87.4 | 85.2 | 83.5 | 81.9 | Hypermutation
ratio, % (85.2°C/87.4°C) | Hypermutation
ratio, % (83.5°C/87.4°C) |
|---|
| Cervicitis, n | 15 | 1 | 0 | 0 | 6.67 | 0.00 |
| CIN I–III, n | 15 | 5 | 2 | 2 | 33.33 | 13.33 |
| Cervical cancer,
n | 15 | 5 | 3 | 3 | 33.33 | 20.00 |
| χ2 |
|
|
|
| 3.85 | 3.15 |
| P-value |
|
|
|
| 0.146 | 0.207 |
For the samples with positive results in the
amplification at the melting temperature of 85.2°C in the 3D-PCR,
Sanger sequencing of the HPV16 E2 gene was performed. The results
revealed that the C>T and G>A hypermutations were observed in
the HPV16 E2 gene (Fig. 3B). The
rate of hypermutation in these cervical lesion samples of different
grades was further analyzed, revealing that there was no
significant difference in the rate of hypermutation among these
three groups at the denaturation temperature of 85.2°C
(χ2=3.85; P=0.146). When the temperature was dropped to
83.5°C, the rate of hypermutation was decreased and there was still
no significant difference in the ratio of hypermutation among these
three groups (χ2=3.15; P=0.207; Table II). Additionally, the number of base
substitutions in the single hypermutation sample was analyzed. The
number of G>A base substitutions in the hypermutation positive
cervical cancer cases (6.00±2.14) was significantly higher compared
with those in the CIN I–III (2.67±1.95) and cervicitis groups
(0.20±0.45; both P<0.05). Furthermore, the number of C>T base
substitutions in the hypermutation positive cervical cancer cases
(12.87±2.77) was significantly higher compared with those in the
CIN I–III (6.40±4.34) and cervicitis groups (5.00±3.16; both
P<0.05; Fig. 3C and D). These
results suggested that HPV16 E2 hypermutation may be increased with
the development and progression of cervical lesions.
Expression and hypermutation of HPV E2
in SiHa cells following APOBEC3s lentivirus transfection
The A3C, A3F and A3G overexpressing SiHa cell model
was established and the transcriptional level of HPV16 E2 and the
E2 hypermutation were investigated. Following transfection of A3F
and A3G, the mRNA and protein expression levels of HPV16 E2 in the
SiHa cells were significantly decreased compared with those in the
blank and the scrambled siRNA negative control groups (P<0.01;
Fig. 4A), whereas no significant
difference was observed in HPV16 E2 mRNA expression levels between
the A3C transfection and control groups (P>0.05). The HPV16 E2
gene in the SiHa cells was amplified by 3D-PCR. For the A3C
transfection groups, the hypermutations were observed at the
denaturation temperature of 85.2°C, while no hypermutations were
noted at other denaturation temperatures. Moreover, for the A3G
groups, the hypermutations were observed at the denaturation
temperatures of 85.2, 83.5 and 81.9°C. However, no hypermutations
were observed at the denaturation temperatures of 85.2, 83.5 or
81.9°C in the A3F groups. Then, the hypermutation positive samples
in the 3D-PCR were subjected to sequencing. After the transfection
of A3 proteins, a larger amount of G>A and C>T base
substitutions were detected in the HPV16 E2 gene in these
transfected SiHa cells (Fig. 4B).
There were a total of 126 C>T and 2 G>A base substitutions in
the A3G transfection group and 56 C>T and 34 G>A base
substitutions in the A3C transfection group (Table III). These results suggested that
the high expression levels of A3 proteins were associated with the
downregulated expression levels of HPV16 E2, and that high A3
expression induced these hypermutation in the HPV16 E2 gene.
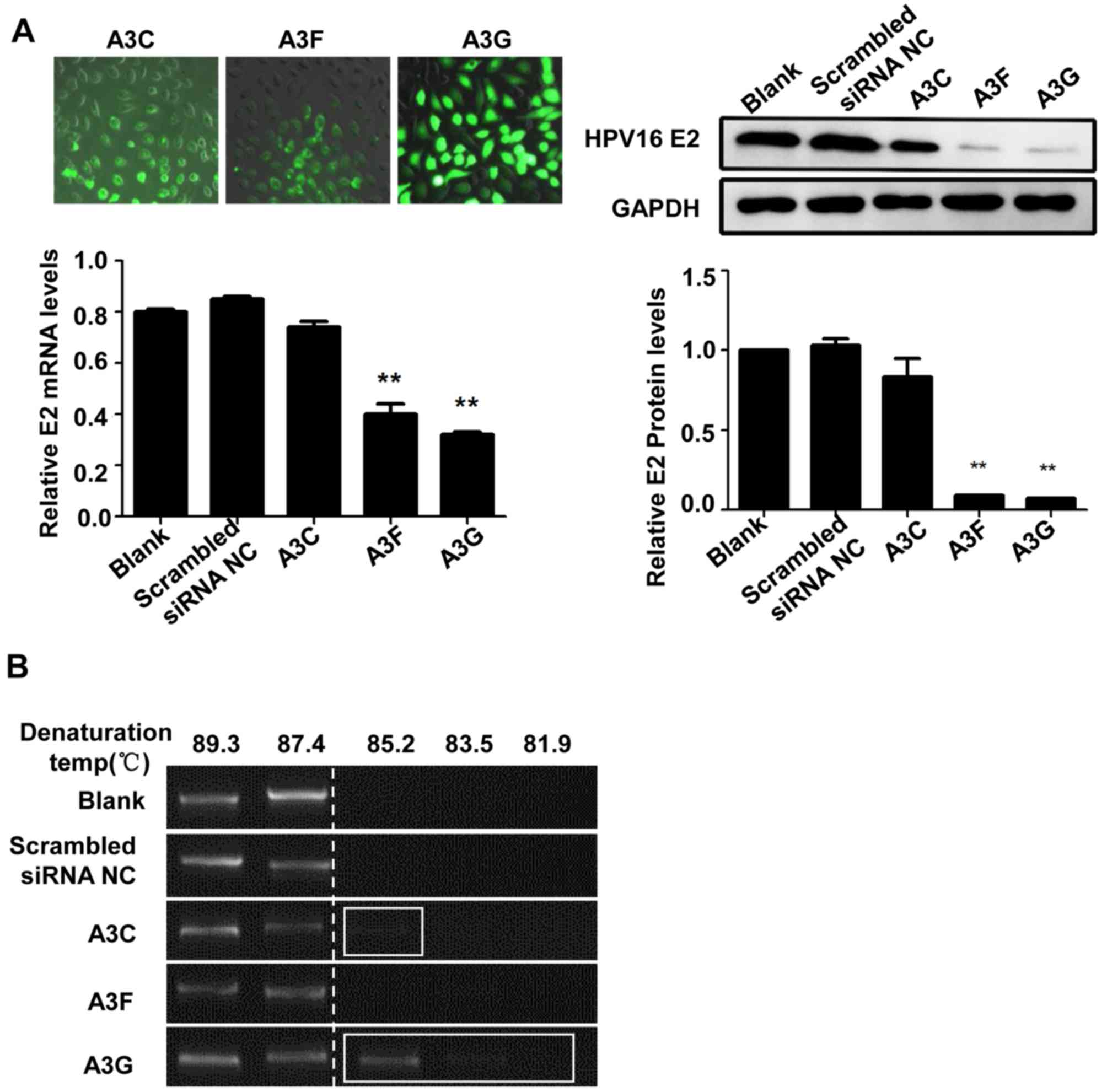 | Figure 4.HPV16 E2 gene expression and
hypermutation in SiHa cells transfected with APOBEC3s lentiviral
vectors. (A) GFP-labeled A3C, A3F and A3G lentiviruses were
transfected into SiHa cells and the fluorescence intensity was
observed using an inverted fluorescence microscope to determine the
transfection efficiency. Magnification, ×200. The mRNA and protein
expression levels of HPV16 E2 in A3 lentivirus-transfected SiHa
cells were confirmed using reverse transcription-quantitative PCR
and western blotting. (B) The HPV16 E2 genome hypermutation
following lentiviral transfection was detected using the
differential DNA Denaturation PCR. **P<0.01 vs. blank control or
scrambled siRNA control. HPV, human papilloma virus; APBOEC3s,
Apolipoprotein B mRNA-editing enzyme catalytic polypeptides; siRNA,
small interfering RNA; NC, negative control; A3C, APOBEC3C; A3F,
APOBEC3F; A3G, APOBEC3G; GFP, green fluorescent protein; C,
cytosine; G, guanine; A, adenine; T, thymine. |
 | Table III.Frequency of hypermutation base
substitutions in SiHa cells following lentiviral transfection as
detected by Sanger sequencing. |
Table III.
Frequency of hypermutation base
substitutions in SiHa cells following lentiviral transfection as
detected by Sanger sequencing.
|
| Frequency of base
substitutions |
|---|
|
|
|
|---|
| Variable | G→A | C→T | Other |
|---|
| A3C | 34 | 56 | 2 |
| A3F | 0 | 3 | 1 |
| A3G | 2 | 126 | 4 |
Discussion
High-risk HPV persistent infection is an important
factor for cervical cancer and precancerous lesions, which
integrate into the host genome after infection and thus induce
cervical lesions. The HPV16 is the most common genotype associated
with cervical cancer (22). The HPV
genome can be divided into the following three regions: The LCR,
early region (E1, E2, E4, E5, E6 and E7) and late region (L1 and
L2). The E2 gene regulates the processes of replication and
transcription of HPV DNA and is associated with the integration of
the HPV genome into the host genome (9,23,24).
Furthermore, the E2 gene regulates the function and transformation
of the E6 and E7 oncogenes (9). The
A3s family is a class of endogenous natural immune protein
molecules with antiviral capabilities (25). In this study, our results showed
that, during the progression of cervicitis to CIN I–III and then
cervical cancer, the mRNA and protein expression levels of A3C,
A3F, and A3G were significantly changed, suggesting that the
members of the A3s family may be involved in the development of
cervical cancer.
A potential role of A3 in viral infection is to edit
the negative DNA strand generated during viral reverse
transcription, deaminating the C into U and thereby causing the
C>T and G>A hypermutation in the viral genome (25–29).
Studies (30,31) have demonstrated that A3 induced this
hypermutation in viral DNA during HIV-1 replication, thereby
inhibiting the HIV-1 reverse transcription and DNA integration. It
has been shown that the A3G overexpression (both mRNA and protein
levels) induces HBV hypermutation and reduces HBV viral synthesis,
and the frequency of hypermutations is increased with elevated A3G
expression levels (both mRNA and protein) (8,32). A
large number of studies have confirmed that A3 induce mutations in
the HIV and HBV genomes, thereby inhibiting viral replication;
however, A3-induced mutation may also be a double-edged sword
(33–36). A3 induce the innate immune response
against the virus through targeted mutations, and meanwhile the A3
mutagenesis would induce the somatic cell mutation (31–38).
In the present study, 3D-PCR amplification and
sequence analysis was performed on the HPV16 genome from cervical
lesions of different grades. The results showed that there were
C>T and G>A hypermutations in the HPV16 E2 gene in Uygur
patients with cervicitis, CIN I–III and cervical cancer, which were
typical types of mutations caused by the APOBEC3 family (25–29).
However, in HPV16 E2 genomes isolated from clinical samples, there
were small numbers of base substitution hypermutation, and there
were no significant differences in the HPV16 E2 hypermutation
statistical data between the clinical samples of the cervicitis,
CIN I–III and cervical cancer groups. This may be because the
integrated HPV does not expose enough single-stranded DNA regions
required for the deaminase activity of A3 (30). Therefore, the poor ability of A3 to
integrate into HPV E2 gene may also lead to reduced frequency of
the A3-induced HPV16 hypermutation. Furthermore, in lesions
expressing high levels of A3 during the transcription and
replication, HPV16 may evolve into a mutant form possessing fewer
A3 target sites (39,40), and HPV16 may inhibit the antiviral
activity of A3 (39,40).
Analysis of persistent HPV infection in a cervical
keratinocyte model has demonstrated that A3 can induce
hypermutations in the HPV16 genome (37,40).
Additionally, A3 are involved in the editing of HPV E2
hypermutation (16,40,41). In
the present study, SiHa cell models overexpressing A3 were
established and these cells were subjected to HPV16 3D-PCR
amplification and sequence analysis. The results demonstrated that
A3G overexpression induced the downregulated expression of the
HPV16 E2 gene, and C>T hypermutations were observed in the E2
gene. Compared with previous studies showing that A3 induces HIV
hypermutation and inhibits viral replication (5,6), the
present study revealed that the frequency of the HPV E2
hypermutation in SiHa cells was lower, following overexpression of
A3G. The high expression levels of A3 may lead to E2
hypermutations, but low frequencies of these hypermutations may not
affect HPV16 replication. Studies have shown that A3 edits the HIV
hypermutation and thus inhibits the virus replication (4,5).
Compared with the frequency of A3-induced HIV hypermutation, the
frequency of HPV E2 region hypermutation in the SIHA cells
high-expressed with A3G is relatively lower. Based on these
findings, we speculate that the high A3s high expression leads to
the E2 hypermutation, rather than affects the HPV16 replication.
However, high A3G expression levels reduced the expression the E2
gene in SiHa cells. E2 is a key gene for the integration of HPV
into the host cells, which may enhance the expression levels of E6
and E7 oncogenes (9). Therefore, A3G
may regulate the integration of HPV into the host genome and
function in the development of cervical cancer. This may improve
the understanding of the molecular mechanisms underlying the
development of cervical cancer.
There are several limitations of the present study.
On one hand, the sample size was relatively small and it was
difficult to verify the clinical significance of HPV16
hypermutation in the pathogenesis of cervical cancer. On the other
hand, 3D-PCR technology could only detect the A/T-rich DNA sequence
compared with the reference sequence in the detection of HPV16
hypermutation. Therefore, HPV16 genome-wide sequencing would be
required to determine additional HPV16 mutations in clinical
samples. In the future, in-depth studies using high-throughput
sequencing technology, with an increased sequencing depth and
larger sample size are required to comprehensively evaluate the
biological significance of HPV16 hypermutations and the association
of HPV infection with the development of cervical cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China-Regional Science Fund (grant
no. 81460395).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
SS, HC and ZJ conducted the experiments, analyzed
the data and wrote the manuscript. MN and LW conceived and designed
the study and helped to revise the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Informed consent was provided by every patient and
the present study was approved by The Ethics Review Board of the
People's Hospital of Xinjiang Uygur Autonomous Region (Urumqi,
China; approval no. 2014048).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Warren CJ, Westrich JA, Doorslaer KV and
Pyeon D: Roles of APOBEC3A and APOBEC3B in human papillomavirus
infection and disease progression. Viruses. 9:E2332017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Salter JD, Bennett RP and Smith HC: The
APOBEC protein family: United by structure, divergent in function.
Trends Biochem Sci. 41:578–594. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ikeda T, Symeonides M, Albin JS, Li M,
Thali M and Harris RS: HIV-1 adaptation studies reveal a novel
Env-mediated homeostasis mechanism for evading lethal hypermutation
by APOBEC3G. PLoS Pathog. 14:e10070102018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ebrahimi D, Richards CM, Carpenter MA,
Wang J, Ikeda T, Becker JT, Cheng AZ, McCann JL, Shaban NM,
Salamango DJ, et al: Genetic and mechanistic basis for APOBEC3H
alternative splicing, retrovirus restriction, and counteraction by
HIV-1 protease. Nat Commun. 9:41372018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Anderson BD, Ikeda T, Moghadasi SA, Martin
AS, Brown WL and Harris RS: Natural APOBEC3C variants can elicit
differential HIV-1 restriction activity. Retrovirology. 15:782018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim EY, Lorenzo-Redondo R, Little SJ,
Chung YS, Phalora PK, Maljkovic Berry I, Archer J, Penugonda S,
Fischer W, Richman DD, et al: Human APOBEC3 induced mutation of
human immunodeficiency virus type-1 contributes to adaptation and
evolution in natural infection. PLoS Pathog. 10:e10042812014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Luo J: Stable expression of porcine
APOBEC3F in PK15 cells and its inhibitory effects on PERV mutation.
Guangxi University 2012.
|
|
8
|
Noguchi C, Hiraga N, Mori N, Tsuge M,
Imamura M, Takahashi S, Fujimoto Y, Ochi H, Abe H, Maekawa T, et
al: Dual effect of APOBEC3G on Hepatitis B virus. J Gen Virol.
88:432–440. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vieira VC, Leonard B, White EA, Starrett
GJ, Temiz NA, Lorenz LD, Lee D, Soares MA, Lambert PF, Howley PM
and Harris RS: Human papillomavirus E6 triggers upregulation of the
antiviral and cancer genomic DNA deaminase APOBEC3B. mBio.
5:e02234–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bulliard Y, Narvaiza I, Bertero A, Peddi
S, Röhrig UF, Ortiz M, Zoete V, Castro-Diaz N, Turelli P, Telenti
A, et al: Structure-function analyses point to a
polynucleotide-accommodating groove essential for APOBEC3A
restriction activities. J Virol. 85:1765–1776. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tsuge M, Noguchi C, Akiyama R, Matsushita
M, Kunihiro K, Tanaka S, Abe H, Mitsui F, Kitamura S, Hatakeyama T,
et al: G to A hypermutation of TT virus. Virus Res. 149:211–216.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gee P, Ando Y, Kitayama H, Yamamoto SP,
Kanemura Y, Ebina H, Kawaguchi Y and Koyanagi Y: APOBEC1-mediated
editing and attenuation of herpes simplex virus 1 DNA indicate that
neurons have an antiviral role during herpes simplex encephalitis.
J. Virol. 85:9726–9736. 2011. View Article : Google Scholar
|
|
13
|
Suspène R, Aynaud MM, Koch S, Pasdeloup D,
Labetoulle M, Gaertner B, Vartanian JP, Meyerhans A and Wain-Hobson
S: Genetic editing of herpes simplex virus 1 and Epstein-Barr
herpesvirus genomes by human APOBEC3 cytidine deaminases in culture
and in vivo. J Virol. 85:7594–7602. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lucifora J, Xia Y, Reisinger F, Zhang K,
Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz
T, et al: Specifific and nonhepatotoxic degradation of nuclear
hepatitis B virus cccDNA. Science. 343:1221–1228. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hirose Y, Onuki M, Tenjimbayashi Y, Mori
S, Ishii Y, Takeuchi T, Tasaka N, Satoh T, Morisada T, Iwata T, et
al: Within-host variations of human papillomavirus reveal APOBEC
signature mutagenesis in the viral genome. J Virol. 92:e00017–18.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stenglein MD, Burns MB, Li M, Lengyel J
and Harris RS: APOBEC3 proteins mediate the clearance of foreign
DNA from human cells. Nat Struct Mol Biol. 17:222–229. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kondo S, Wakae K, Wakisaka N, Nakanishi Y,
Ishikawa K, Komori T, Moriyama-Kita M, Endo K, Murono S, Wang Z, et
al: APOBEC3A associates with human papillomavirus genome
integration in oropharyngeal cancers. Oncogene. 36:1687–1697. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Graham SV: Human papillomavirus: Gene
expression, regulation and prospects for novel diagnostic methods
and antiviral therapies. Future Microbiol. 5:1493–1506. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xue Y, Bellanger S, Zhang W, Lim D, Low J,
Lunny D and Thierry F: HPV16 E2 is an immediate early marker of
viral infection, preceding E7 expression in precursor structures of
cervical carcinoma. Cancer Res. 70:5316–5325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bhatla N, Aoki D, Sharma DN and
Sankaranarayanan R: Cancer of the cervix uteri. Int J Gynaecol
Obstet. 143 (Suppl 2):S22–S36. 2018. View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
de Sanjose S, Quint WG, Alemany L, Geraets
DT, Klaustermeier JE, Lloveras B, Tous S, Felix A, Bravo LE, Shin
HR, et al: Human papillomavirus genotype attribution in invasive
cervical cancer: A retrospective cross-sectional worldwide study.
Lancet Oncol. 11:1048–1056. 2010. View Article : Google Scholar
|
|
23
|
Pande S, Jain N, Prusty BK, Bhambhani S,
Gupta S, Sharma R, Batra S and Das BC: Human papillomavirus type 16
variant analysis of E6,E7, and L1 gennes and long control regin in
biopsy samples from cervical cancer patients in North India. J Clin
Microbiol. 46:1060–1066. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Franceschi S: The IARC commitment to
cancer prevention: The example of papillomavirus and cervical
cancer. Recent Results Cancer Res. 166:277–297. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Suspène R, Guétard D, Henry M, Sommer P,
Wain-Hobson S and Vartanian JP: Extensive editing of both hepatitis
B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in
vivo. Proc Natl Acad Sci USA. 102:8321–8326. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Noguchi C, Ishino H, Tsuge M, Fujimoto Y,
Imamura M, Takahashi S and Chayama K: G to A hypermutation of
hepatitis B virus. Hepatology. 41:626–633. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Suspène R, Henry M, Guillot S, Wain-Hobson
S and Vartanian JP: Recovery of APOBEC3-edited human
immunodeficiency virus G->A hypermutants by differential DNA
denaturation PCR. J Gen Virol. 86:125–129. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nik-Zainal S, Alexandrov LB, Wedge DC, Van
Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J,
Stebbings LA, et al: Mutational processes molding the genomes of 21
breast cancers. Cell. 149:979–993. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Alexandrov LB, Nik-Zainal S, Wedge DC,
Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A,
Børresen-Dale AL, et al: Signatures of mutational processes in
human cancer. Nature. 500:415–421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Malim MH: APOBEC proteins and intrinsic
resistance to HIV-1 infection. Philos Trans R Soc Lond B Biol Sci.
364:675–687. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sheehy AM, Gaddis NC, Choi JD and Malim
MH: Isolation of a human gene that inhibits HIV-1 infection and is
suppressed by the viral Vif protein. Nature. 418:646–650. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kitamura K, Wang Z, Chowdhury S, Simadu M,
Koura M and Muramatsu M: Uracil DNA glycosylase counteracts
APOBEC3G-induced hypermutation of hepatitis B viral genomes:
Excision repair of covalently closed circular DNA. PLoS Pathog.
9:e10033612013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vieira VC and Soares MA: The role of
cytidine deaminases on innate immune responses against human viral
infections. Biomed Res Int. 2013:6830952013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nik-Zainal S, Wedge DC, Alexandrov LB,
Petljak M, Butler AP, Bolli N, Davies HR, Knappskog S, Martin S,
Papaemmanuil E, et al: Association of a germline copy number
polymorphism of APOBEC3A and APOBEC3B with burden of putative
APOBEC-dependent mutations in breast cancer. Nat Genet. 46:487–491.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Roberts SA, Sterling J, Thompson C, Harris
S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA,
et al: Clustered mutations in yeast and in human cancers can arise
from damaged long single-strand DNA regions. Mol Cell. 46:424–435.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kazanov MD, Roberts SA, Polak P,
Stamatoyannopoulos J, Klimczak LJ, Gordenin DA and Sunyaev SR:
APOBEC-induced cancer mutations are uniquely enriched in
early-replicating, gene-dense, and active chromatin regions. Cell
Rep. 13:1103–1109. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Roberts SA, Lawrence MS, Klimczak LJ,
Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL,
Saksena G, et al: An APOBEC cytidine deaminase mutagenesis pattern
is widespread in human cancers. Nat Genet. 45:970–976. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Burns MB, Lackey L, Carpenter MA, Rathore
A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N,
Nikas JB, et al: APOBEC3B is an enzymatic source of mutation in
breast cancer. Nature. 494:366–370. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mussil B, Suspène R, Aynaud MM, Gauvrit A,
Vartanian JP and Wain-Hobson S: Human APOBEC3A isoforms translocate
to the nucleus and induce DNA double strand breaks leading to cell
stress and death. PLoS One. 8:e736412013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang Z, Wakae K, Kitamura K, Aoyama S, Liu
G, Koura M, Monjurul AM, Kukimoto I and Muramatsu M: APOBEC3
deaminases induce hypermutation in human papillomavirus 16 DNA upon
beta interferon stimulation. J Virol. 88:1308–1317. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Burns MB, Temiz NA and Harris RS: Evidence
for APOBEC3B mutagenesis in multiple human cancers. Nat Genet.
45:977–983. 2013. View Article : Google Scholar : PubMed/NCBI
|