Introduction
Renal cell carcinoma (RCC) is one of the most common
cancers, accounting for >140,000 deaths worldwide each year
(1). RCC has been considered to be
an immunogenic tumor (2).
Interleukin-2, interferon α and immune checkpoint inhibitors have
been used clinically in RCC therapy (3–5). Novel
immunotherapies, such as the combination of anti-programmed death
ligand-1 antibody and anti-cytotoxic T-lymphocyte-associated
protein 4 antibody, have demonstrated clinical benefit (6). However, only a proportion of patients
with RCC benefit from the novel immunotherapies (7). The immune suppression microenvironment
of RCC remains to be investigated.
B7 family member H4 (B7-H4), also known as B7× or
B7S1, is one of the members of the B7 superfamily of co-stimulatory
molecules and serves as an inhibitory modulator of the T-cell
response (8). B7-H4 mRNA is widely
expressed in human peripheral tissues, but its protein expression
in normal tissues seems to be limited (9). To date, B7-H4 has been detected in
several types of human cancer tissue. In ovarian cancer, B7-H4
expression is associated with tumor-infiltrated antigen-presenting
cells (APCs) (10). In colorectal
carcinoma, B7-H4 facilitates tumor proliferation and metastasis
(11). The expression of B7-H4 in
lung cancer is associated with decreased PFS under nivolumab
treatment (12). Elevated B7-H4 in
breast cancer is associated with an ‘immune-cold’ microenvironment
(13). In renal cell carcinoma,
intrahepatic cholangiocarcinoma and thyroid cancer, the expression
level of B7-H4 is positively associated with tumor progression
(14–16). Previous studies have reported that
B7-H4, which is a type I transmembrane glycoprotein, binds to its
corresponding receptor on lymphocytes, thus negatively regulating
the immune response (8,17,18).
However, certain tumors express B7-H4 protein in intracellular
compartments (19–22). Unlike tumor-associated macrophages,
B-cells and dendritic cells (DCs) with membrane expression of
B7-H4, tumor cells expressing intracellular B7-H4 do not inhibit
T-cell immunity (21). Therefore, it
appears that intracellular B7-H4 has a distinct biological function
to membrane-located B7-H4 (22,23). In
order to investigate the biological function of B7-H4 in kidney
cancer cells, the present study constructed negative control
(NC)/786-O and B7-H4/786-O cell lines, and screened the
differentially expressed genes (DEGs) in B7-H4/786-O cells by
microarray analysis. The identified DEGs were subsequently
validated by western blotting.
Materials and methods
Cell culture
The 786-O cell line (catalogue TCHu186) was
purchased from the Shanghai Cell Bank of the Shanghai Institute for
Biological Sciences. The cells were cultured in RPMI 1640 medium
(Gibco, Thermo Fisher Scientific, Inc.) containing 10% fetal bovine
serum (Gibco, Thermo Fisher Scientific, Inc.) at 37°C, 5%
CO2. A total of 1×106 786-O cells/well in
24-well plates were transfected with 50 µl/well LV5-B7-H4
lentivirus (Genepharma Co., Ltd) or empty vector (Genepharma Co.,
Ltd) with 5 µg/ml polybrene (Genepharma Co., Ltd) according the
manufacturer's instructions. The stable expression clones were
selected in medium containing 0.5 mg/ml G418 (Thermo Fisher
Scientific, Inc.) 24 h post-transfection. After 2 weeks, NC/786-O
and B7-H4/786-O stable transgenic cells were obtained and
subsequent experiments were performed.
To knock down B7-H4 expression in breast cancer cell
line SK-BR-3, 1×106 cells were seeded in 6-well plates
24 h prior to transfection. When the cells reached 70–80%
confluence in each well, they were treated with Opti-MEM Reduced
Serum Medium (Gibco, Thermo Fisher Scientific, Inc.) containing 50
nM B7-H4 small interfering (si)RNA (Genepharma Co., Ltd) and 10 µl
Lipofectamine® 2000 (Invitrogen, Thermo Fisher
Scientific, Inc.) for 6 h. The cell supernatant was replaced with
fresh medium and cells were cultured for 48 h prior to harvesting
and subsequent experiments. The B7-H4 siRNA sequences were as
follows: Forward, 5′-GCUGGAGCAAUUGCACUCAUCAUUG(dTdT)-3′ and
reverse, 5′-CAAUGAUGAGUGCAAUUGCUCCAGC(dTdT)-3′.
Microarray processing
For total RNA extraction, NC/786-O and B7-H4/786-O
cells were harvested. Triplicate samples were prepared for each
cell line. Total RNA was extracted using TRIzol reagent
(Invitrogen, Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Total RNA was quantified using a NanoDrop
ND-2000 spectrophotometer (NanoDrop Technologies; Thermo Fisher
Scientific, Inc.) and the RNA integrity was assessed using an
Agilent Bioanalyzer 2100 (Agilent Technologies, Inc.). Total RNA
was transcribed into double-stranded cDNA and labeled with
cyanine-3-cytidine triphosphate. The labeled cDNAs were hybridized
onto Agilent Human Gene Expression microarrays (Agilent
Technologies, Inc.). The arrays were scanned by the Agilent Scanner
G2505C (Agilent Technologies, Inc.). The heatmap of the gene
expression in the two cell lines (triplicates of each cell line)
was produced by Multiexperimental Viewer software version 4.8.1
(MeV development team).
DEG analysis
Feature Extraction software version 10.7.1.1
(Agilent Technologies, Inc.) was used to analyze array images to
obtain raw data. GeneSpringGX version 11.0 (Agilent Technologies)
was employed for the basic analysis of the raw data, which were
normalized with the quantile algorithm. Probes that had ≥10% values
in any one of all conditions and had flags in ‘Detected’ were
selected for further data analysis. Genes with fold-change ≥2 and
P<0.05 calculated by unpaired Student's t-test between
B7-H4/786-O and NC/786-O cells were identified as DEGs.
Functional enrichment analysis
To explore the functions in which DEGs were enriched
in the B7-H4/786-O cell line, the identified DEGs were subjected to
functional enrichment analysis with the Datasets for Annotation,
Visualization and Integrated Discovery version 6.7 (david.abcc.Nicfcrf.gov). Gene Ontology (GO)
(http://geneontology.org/) terms and Kyoto
Encyclopedia of Genes and Genome (KEGG) pathways (https://www.kegg.jp/) with P<0.05 were screened
out.
Western blot analysis
NC/786-O and B7-H4/786-O cells were harvested and
lysed in RIPA buffer. RIPA cell lysis buffer (cat. no. P0013B) was
purchased from Beyotime Institute of Biotechnology. Total proteins
were extracted, concentrations were determined by bicinchoninic
acid assay, separated using 12% SDS-PAGE (100 µg/lane), and the
proteins were transferred onto PVDF membranes. Upon blocking in 5%
BSA/PBS buffer at room temperature for 30 min, the membranes were
blotted with the primary antibodies at 4°C overnight and
subsequently washed with PBS-0.05% (v/v) Tween 20 (PBST) three
times. The primary antibodies used were as follows: NME1 (1:500;
cat. no. 11086-2-AP, Proteintech Group, Inc.); membrane
metalloendopeptidase (MME; 1:500; cat. no 18008-1-AP; Proteintech
Group, Inc.); vanin 1 (VNN1; 1:300; cat. no. 21745-1-AP;
Proteintech Group, Inc.); matrix metalloproteinase 7 (MMP7; 1:300;
cat. no. 10374-2-AP; Proteintech Group, Inc.); tumor necrosis
factor α (TNF-α; 1:2,000; cat. no. 60291-1-Ig; Proteintech Group,
Inc.); human C-X-C motif chemokine ligand (CXCL) 1/2/3 (1 µg/ml;
cat. no. MAB276; R&D Systems, Inc.); CXCL8 (1:500; cat. no.
27095-1-AP; Proteintech Group, Inc.); human C-C motif chemokine
ligand 20 (CCL20; 1 µg/ml); cat. no. MAB360; R&D Systems,
Inc.); B7-H4 (1:500; cat. no. 12080-1-AP; Proteintech Group, Inc.);
β-actin (1: 5,000; cat. no. 66009-1-Ig; Proteintech Group, Inc.)
and α-tubulin (1:2,000; cat. no. 11224-1-AP; Proteintech Group,
Inc.) were used. Then, the membranes were incubated with the
secondary antibodies at room temperature for 1 h and subsequently
washed three times with PBST. The secondary antibodies horse
anti-mouse IgG (H&L)-horseradish peroxidase (HRP; 1:1,000; cat.
no. 7076;) and goat anti-rabbit IgG (H&L)-HRP (1:1,000; cat.
no. 7074) were purchased from Cell Signaling Technology, Inc. The
membranes were immersed in ECL detection reagent and imaged with a
Gel Doc™ EZ system (Bio-Rad Laboratories, Inc.). β-actin or
α-tubulin were used as loading controls. The intensities of the
bands were determined by Image Lab software version 4.1 (Bio-Rad
Laboratories, Inc.). Three independent experiments were
performed.
The Cancer Genome Atlas (TCGA)
analysis
RNA-sequencing TCGA data were analysed using the
Gene Expression Profiling Interactive Analysis (GEPIA) website
(gepia.cancer-pku.cn/index.html), which is an
interactive web server for analysing RNA-sequencing expression data
from TCGA and Genotype-Tissue Expression databases (24). The correlation between B7-H4 and the
expression of chemokine genes CXCL1, CXCL2, CXCL3, CXCL8 and CCL20
was evaluated by the Pearson's correlation test. P<0.05 was
considered to indicate a statistically significant difference.
Immunohistochemistry (IHC)
The use of clinical samples was approved by the
Soochow University Ethical Review Board, and written informed
consent was obtained from the patients. A total of 59 specimens
fixed with 10% formalin at room temperature for 24 h and embedded
in paraffin were collected from the Second Affiliated Hospital of
Soochow University were used for IHC analysis. The patients
included 36 males and 23 females (mean age, 59 years; age range
39–78 years). All patients had a pathological diagnosis of renal
cell carcinoma and underwent radical nephrectomy. Sections (4-µm)
were deparaffinized in serial grades of xylene followed by
rehydration in sequentially increasing dilutions of ethanol. Upon
antigen retrieval was performed by saline sodium citrate (microwave
heating for 30 min), endogenous peroxidase was blocked by 3.0%
hydrogen peroxide at room temperature for 1 h. Non-specific
interactions were blocked using 1.5% blocking serum (Dako, Agilent
Technologies, Inc.) in PBS for 10 min at room temperature. Then the
sections were incubated with primary antibodies against B7-H4
(1:100; cat. no. 12080-1-AP; Proteintech Group, Inc.); CCL20
(1:200; cat. no. 26572-1-AP; Proteintech Group, Inc.) and CXCL8
(1:200; cat. no. 27095-1-AP; Proteintech Group, Inc.) at 4°C
overnight. The sections were then incubated with a HRP-conjugated
goat anti-rabbit secondary antibody (1:50; cat. no. GK600705; Gene
Tech Co., Ltd.) at room temperature for 1 h. Then the sections were
stained by the avidin-biotin immunoperoxidase method. For negative
control staining, the primary antibodies were omitted in the
procedure. The sections were observed under a light microscope
(CX43; Olympus Corporation) at ×400 magnification and evaluated and
graded independently by two investigators. The Allred scoring
system was used (25). The signal
intensity was graded from 0 to 3 (0, none; 1, weak; 2,
intermediate; and 3, strong), and the percentage of positive tumor
cells was scored according to a scale from 1 to 5 (1, <1; 2,
1–10; 3, 10–30; 4, 30–60; and 5, >60%). An Allred score of B7-H4
<80 was designated as B7-H4low, while >80 was
designated as B7-H4high.
Mouse and tumor models
Non-obese diabetic/severe combined immunodeficiency
mice (age, 4 weeks; male) were purchased from Joinn Laboratories
(China), Co., Ltd. Mice were kept in pathogen-free facilities under
conventional conditions with controlled temperature (22±2°C) and
humidity (55±10%) and 12-h dark/light cycles. The mice were allowed
free access to food and water. All animal experiments were approved
and conducted in compliance with institutional guidelines
established by the Ethics Committee of Soochow University (Suzhou,
China). Mice were subcutaneously inoculated into the left axilla
with 5×106 786-O transfected cells suspended in
serum-free PBS. Mice were intraperitoneally injected with isotype
IgG or anti-CXCL8 antibody once every 3 days at a dose of 1 mg/kg
(n=6 mice/group). Tumor growth was assessed twice a week. The
orthogonal tumor dimensions were measured with a calliper. The
tumor volume was calculated according to the formula V=a ×
b2/2, where V is the volume, a is the maximum diameter
and b is the smallest diameter. When the tumor diameter reached 20
mm or tumor volume reached 1,500 mm3, the mice were
preemptively euthanized by cervical dislocation and the tumor
tissues were isolated and weighed. During the present study, the
maximum body weight loss is in a mouse due to cachexia was 9.8%,
and the longest diameter exhibited by a single subcutaneous tumor
was 1.8 cm. No mice exhibited multiple tumors.
Flow cytometry
Flow cytometry analysis of tumor infiltrated
neutrophils in tumor tissues was performed as described previously
(26). Tumor tissues were
mechanically dissected into small pieces and further digested with
a mixture of Liberase™ TL (Roche Applied Science) and DNase. Cells
digested from the tumor tissues were filtered through a 70-µm
filter, fixed with 4% paraformaldehyde at room temperature for 15
min, and incubated in PBS containing 1% FBS (cat. no. 10099141;
Gibco, Thermo Fisher Scientific, Inc.) and 3 mM EDTA at room
temperature for 20 min to block non-specific binding. The cells
were incubated with an anti-mouse cluster of differentiation (CD)
16/32 antibody (2.5 µg/106 cells; cat. no. MAB1460;
R&D Systems, Inc.) at 4°C for 30 min. Cells were stained with
fluorescein isothiocyanate-conjugated CD45 (0.25 µg/106
cells; cat. no. 147709, Biolegend, Inc.);
P-phycoerythrin-conjugated CD11b (0.25 µg/106 cells;
cat. no. 101207; Biolegend, Inc.) and APC-conjugated lymphocyte
antigen 6 complex, locus G antibodies (0.06 µg/106
cells; cat. no. 127613; Biolegend, Inc.) at 4°C for 30 min. Flow
cytometry data acquisition was performed on a BD FACSCalibur (BD
Biosciences) and analysed by FlowJo version 7.6 (BD
Biosciences).
Statistical analysis
All data are expressed as the mean ± SD from at
least triplicate experiments. For the correlation analysis between
B7-H4 and upregulated DEGs in TCGA data, the Pearson's correlation
test was used. For the chemokine IHC score, the unpaired Student's
t-test was used to evaluate the difference in two groups
(dichotomized according B7-H4 level). For multiple groups in the
mouse model, quantitative data was analysed by the one-way ANOVA.
Multiple comparisons among the groups were performed using the
Tukey's post hoc test. SPSS 19.0 software (IBM Corp.) was used for
all statistical analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification of DEGs in microarray
datasets and functional enrichment analysis
To identify the effects of B7-H4 overexpression on
the function of renal cell carcinoma cells, the total mRNA
extracted from NC/786-O and B7-H4/786-O cells was subjected to
microarray analysis. The heatmap of the gene expression in the two
cell lines (triplicates of each cell line) is presented in Fig. 1A. In total, 724 upregulated and 804
downregulated DEGs were obtained. The significantly enriched KEGG
pathways of upregulated genes in the B7-H4/786-O cell line were
analyzed. The KEGG pathways of upregulated genes with P≤0.01 are
shown in Fig. 1B. The results
revealed that there was a significant association with ‘rheumatoid
arthritis’, ‘legionellosis’, ‘TNF signaling pathway’,
‘nucleotide-binding oligomerization domain (NOD)-like receptor
signaling pathway’, ‘NF-κB signaling pathway’ and ‘chemokine
signaling pathway’ (Fig. 1B). GO
terms enrichment analysis for upregulated DEGs was performed. The
top 20 biological processes (BPs) are presented in Fig. 1C. The results revealed that
‘inflammatory response’, ‘immune response’, ‘cell chemotaxis’ and
‘cellular response to lipopolysaccharide’ were associated with the
upregulated DEGs (Fig. 1C).
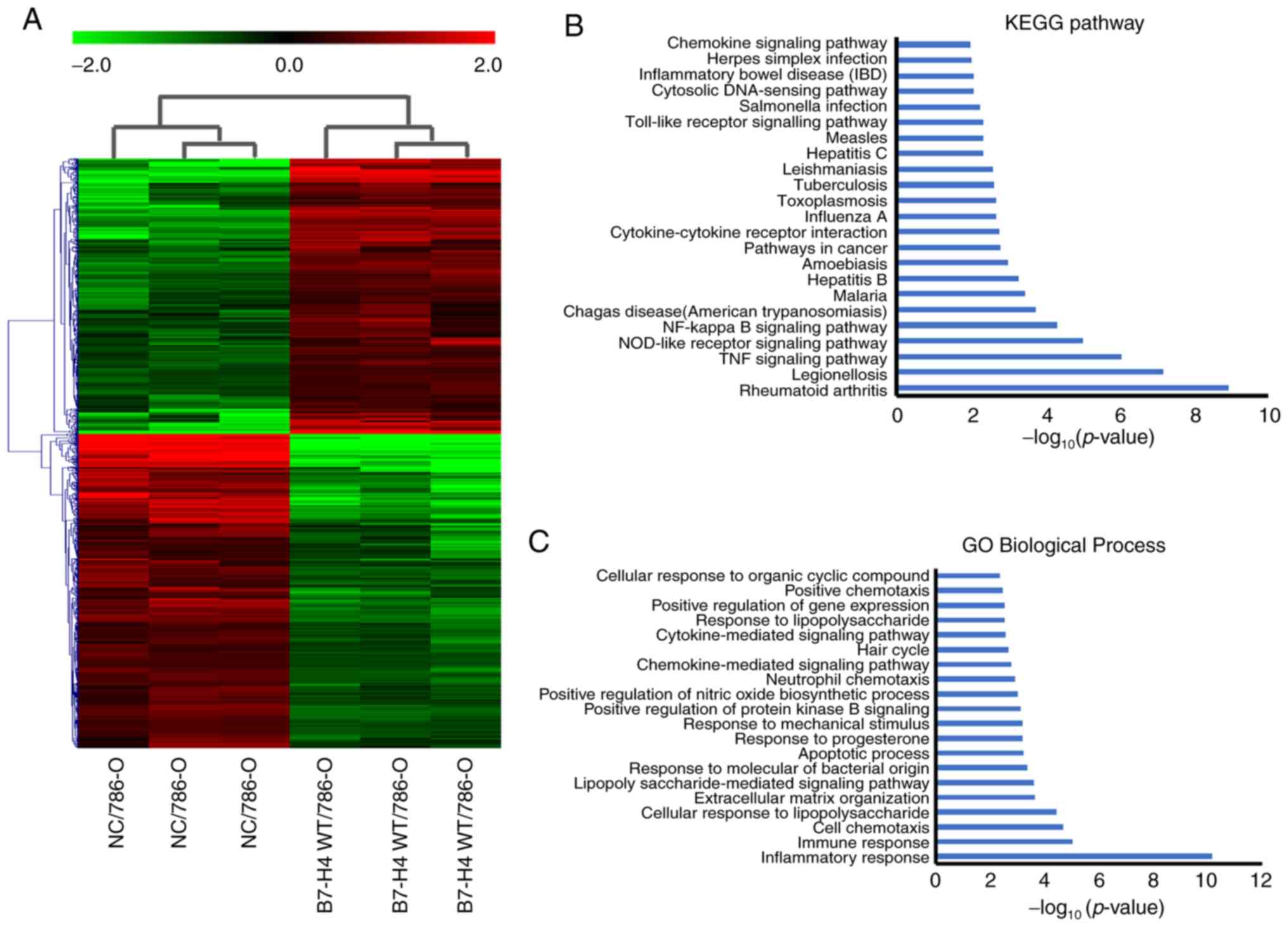 | Figure 1.Identification of DEGs between
NC/786-O and B7-H4/786-O cell lines by microarray analysis. (A)
Heatmap of DEGs between NC/786-O and B7-H4/786-O cells. (Red,
upregulated genes; green, downregulated genes and black, unchanged
genes). (B) Significant KEGG signaling pathways (P≤0.01) of the
upregulated genes are shown. No significant KEGG pathway was
identified for the downregulated genes. (C) GO terms enrichment
analysis of BPs. The top 20 BPs are shown. DEGs, differential
expressed genes; NC, negative control; B7-H4, B7 family member, H4;
KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, biological
process; GO, Gene Ontology; WT, wild-type. |
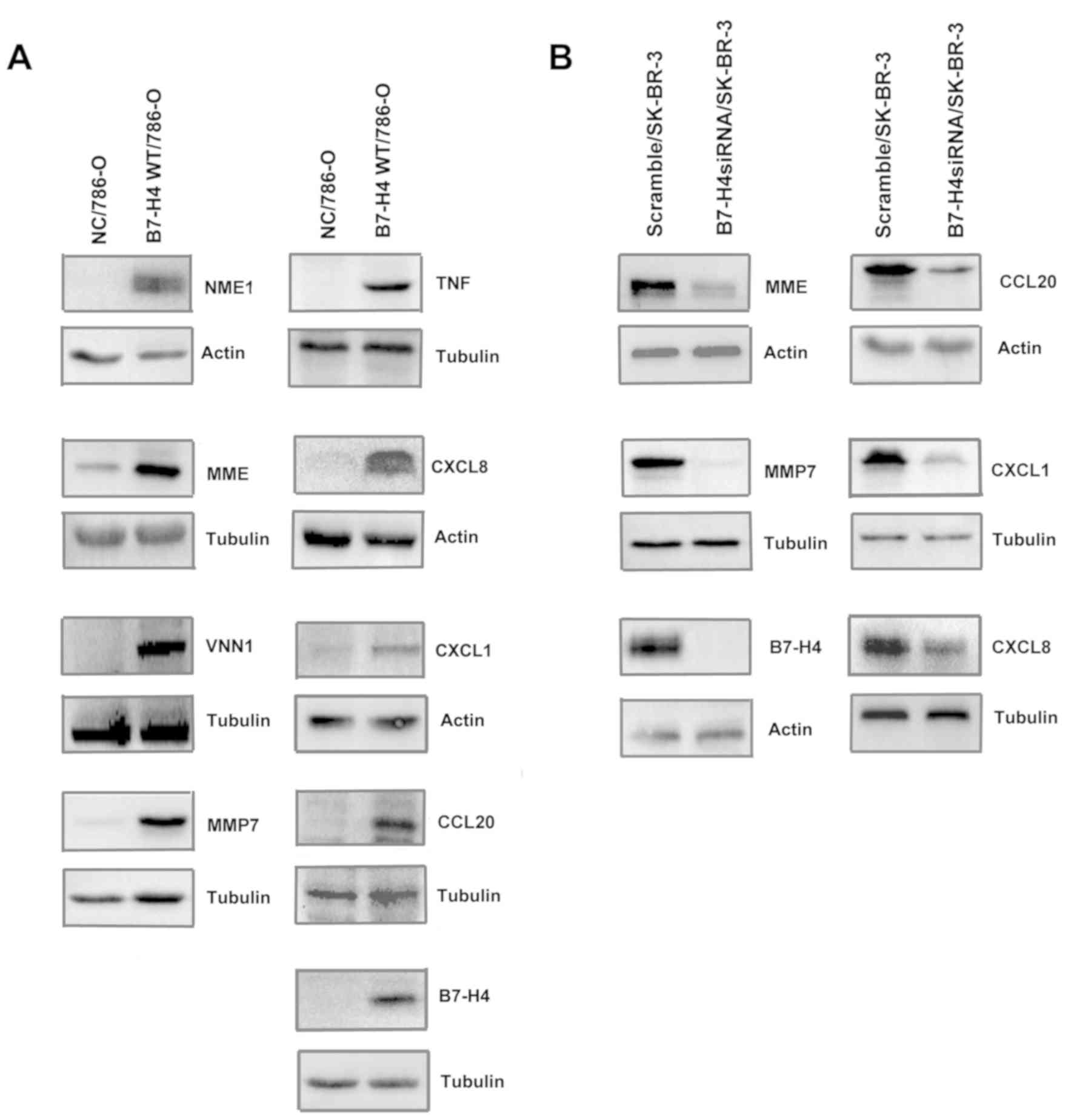
Verification of upregulated DEGs by
B7-H4 via western blotting
By GO BP analysis, the predominant BPs of the
upregulated DEGs were ‘inflammatory response’, ‘immune response’
and ‘cell chemotaxis’, all of which are associated with the immune
characteristics of tumor cells. Further analysis of GO revealed
that IL33, TLR2, CXCL1 and CXCL8 were involved in these BPs. The
upregulated DEGs involved in these BPs are listed in Table I. Next, the protein level of
upregulated DEGs in 786-O transfectants was verified using western
blot analysis. Fig. 2A revealed that
the protein levels of NME1, MME, VNN1, MMP7, TNF, CXCL8, CXCL1 and
CCL20 were upregulated in B7-H4/786-O cells compared with NC/786-O
cells. To further confirm the effect of B7-H4 on the expression of
these cytokines, the SK-BR-3 cell line was transfected with
scramble small interfering (si)RNA and B7-H4 siRNA. Western blot
analysis demonstrated that loss of B7-H4 in the SK-BR-3 cell line
resulted in decreased MME, MMP7, CXCL8, CXCL1 and CCL20 (Fig. 2B). Both overexpression and knocking
down of B7-H4 in cancer cell lines confirmed that B7-H4 expression
led to upregulation of the cytokines that were identified by
microarray analysis.
 | Table I.Upregulated differentially expressed
genes involved in the inflammatory response, immune response and
cell chemotaxis. |
Table I.
Upregulated differentially expressed
genes involved in the inflammatory response, immune response and
cell chemotaxis.
| Gene symbol | Regulation
(BW/NC) | FC (BW/NC) | Regulation
(BW/BM) | FC (BW/BM) |
|---|
| IL33 | Up | 379.026787 | Up | 258.11453 |
| TLR2 | Up | 34.398422 | Up | 2.3269036 |
| CXCL1 | Up | 34.114403 | Up | 104.74504 |
| CXCL8 | Up | 29.291115 | Up | 125.44275 |
| CSF2 | Up | 27.370817 | Up | 142.67291 |
| CCL20 | Up | 17.881693 | Up | 26.53977 |
| CXCL2 | Up | 15.134712 | Up | 29.041739 |
| TNF | Up | 11.655401 | Up | 14.347787 |
| NME5 | Up | 11.342079 | Up | 7.7392936 |
| VNN1 | Up | 10.856487 | Up | 10.219795 |
| IGF2 | Up | 10.503797 | Up | 7.646981 |
| MMP7 | Up | 10.202435 | Up | 3.5069153 |
| CXCL3 | Up | 9.860397 | Up | 24.532923 |
| PTGS2 | Up | 9.696702 | Up | 3.1078327 |
| MMP1 | Up | 8.226387 | Up | 12.556886 |
| CCL2 | Up | 5.6037107 | Up | 10.624764 |
| CXCL5 | Up | 4.761648 | Up | 5.3687253 |
| MME | Up | 3.3166819 | Up | 5.5750117 |
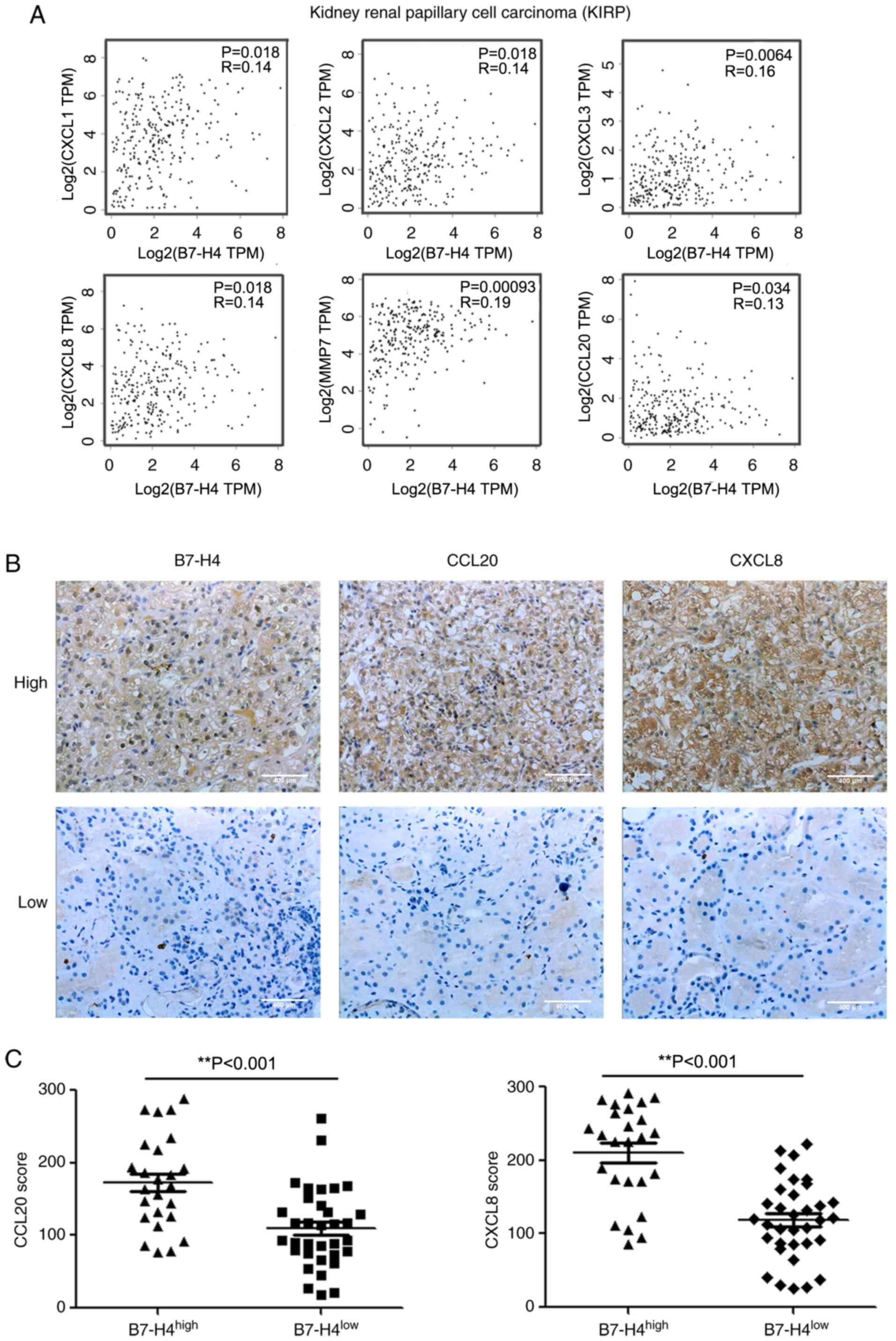
TCGA and IHC analyses reveal the
upregulation of chemokines by B7-H4 in clinical renal
carcinoma
To further confirm the upregulation of cytokines by
B7-H4 in cancer cell lines, the present study analyzed chemokine
gene expression in clinical kidney tumors. The RNA-sequencing data
of TCGA were analysed using the GEPIA website (10). As presented in Fig. 3A, B7-H4 mRNA was associated with
increased CXCL1, CXCL2, CXCL3, CXCL8, MMP7 and CCL20 mRNA levels in
renal papillary cell carcinoma. Although the correlations were not
strong according to the R values, they were significant. These data
further verified the results obtained in the cell lines. A total of
59 clinical renal carcinoma tissues were collected, and B7-H4,
CXCL8 and CCL20 expression was evaluated by IHC staining. The
patient characteristics are shown in Table II. B7-H4 high expression was
associated with tumor grade. As shown in Fig. 3B, representative specimens with high
B7-H4 expression exhibited high levels of CCL20 and CXCL8, while
specimens with low B7-H4 expression exhibited low levels of CCL20
and CXCL8. There was a positive association between CCL20 and CXCL8
levels with B7-H4 (Fig. 3C). These
data suggested that B7-H4 expression enhanced CCL20 and CXCL8
levels in clinical renal carcinoma.
 | Table II.Patient characteristics. |
Table II.
Patient characteristics.
|
| B7-H4 level |
|
|---|
|
|
|
|
|---|
| Clinical
variable |
B7-H4high (n=25) | B7-H4low
(n=34) |
P-valuea |
|---|
| Age, years (mean ±
SD) | 57.6±9.8 | 60.3±8.3 | 0.267 |
|
<60 | 14 | 14 | 0.260 |
|
≥60 | 11 | 20 |
|
| Sex |
|
| 0.891 |
|
Male | 15 | 21 |
|
|
Female | 10 | 13 |
|
| Grade |
|
| 0.010 |
|
G1-2 | 16 | 31 |
|
|
G3-4 | 9 | 3 |
|
| T stage |
|
| 0.200 |
|
T1-2 | 17 | 28 |
|
|
T3-4 | 8 | 6 |
|
B7-H4 promotes tumor proliferation and
infiltrating neutrophils by upregulating CXCL8
Since CXCL8 promoted tumor progression by recruiting
tumor-associated neutrophils (27),
the present study investigated the role of CXCL8 in the tumor
promotion role of B7-H4. NC/786-O and B7-H4/786-O cell lines were
inoculated into mice, and isotype IgG or anti-CXCL8 antibody were
administered. The tumor growth curve is shown in Fig. 4A. After 21 days, tumor tissues were
isolated and the tumor weight was evaluated. Fig. 4B revealed that overexpression of
B7-H4 significantly promoted tumor growth when isotype IgG was
administered. However, when administering anti-CXCL8 antibody,
there was no difference between NC/786-O and B7-H4/786-O,
suggesting that B7-H4 promoted tumor growth through CXCL8. The
fold-change in B7-H4/786-O (isotype/anti-CXCL8) decreased more
significantly than in NC/786-O (Fig.
4C). The gating strategy of tumor-infiltrating
CD11b+Ly6G+ neutrophils was shown in Fig. 4D. The results demonstrated that, in
the isotype IgG-treated group, the ratio of infiltrating
neutrophils in B7-H4/786-O was significantly higher than in
NC/786-O (Fig. 4E and F), but in the
anti-CXCL8-treated group, no difference in the number of
infiltrated neutrophils existed between the two cell lines. This
result suggested that overexpressed B7-H4 in tumors induced
neutrophil recruitment through CXCL8.
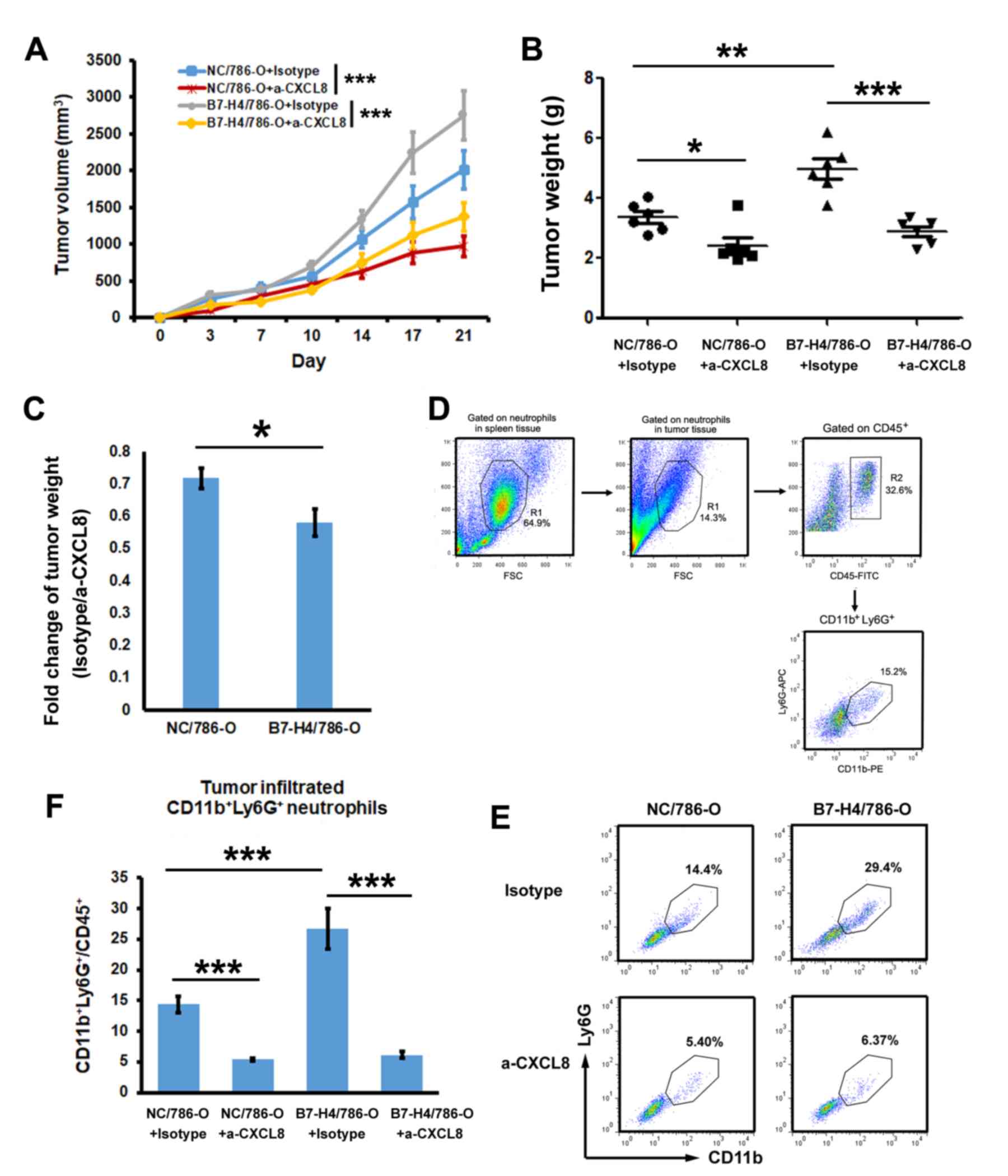 | Figure 4.Evaluation of the effects of CXCL8 on
the B7-H4-mediated promotion of tumor growth and tumor-infiltrating
neutrophils. 786-O transfectants were inoculated into non-obese
diabetic/severe combined immunodeficiency mice (n=6/group). (A)
Tumor growth curve was evaluated by assessing tumor volume. The
tumor volumes on day 21 showed significance, ***P<0.001. (B)
Tumor weight was evaluated after 21 days of treatment with isotype
IgG or anti-CXCL8 antibody (n=6). (C) Fold-change of tumor weight
between the isotype IgG and anti-CXCL8 antibody groups. (D) Gating
strategy of CD11b+Ly6G+ in tumor tissues is
shown. (E) Tumor-infiltrating neutrophils were analysed by flow
cytometry. Cells from tumor tissues were stained with fluorescein
conjugated CD45, CD11b and Ly6G. The ratio of
CD11b+Ly6G+ in CD45+ cells was
evaluated by flow cytometry. (F) Statistical analysis of the ratio
CD11b+Ly6G+/CD45+ (n=6). Multiple
comparisons among the groups were performed using the Tukey's post
hoc test. *P<0.05, **P<0.01, ***P<0.001, as indicated.
CXCL8, C-X-C motif chemokine ligand; B7-H4, B7 family member, H4;
CD, cluster of differentiation; Ly6G, lymphocyte antigen 6 complex,
locus G. |
Discussion
B7-H4 is an important negative co-stimulatory
molecule of the B7 family (8,9,17). B7-H4 protein expression is restricted
to activated T cells, B cells, DCs and macrophages (8). Additionally, it has been reported that
B7-H4 is upregulated in several types of cancer (28–31),
which suggests that B7-H4 has the potential to be used as a
biomarker or therapeutic target for tumors (19,31–33).
However, other studies revealed that B7-H4 promotes cell
proliferation (34,35), invasion and metastasis of tumor cells
(15,35,36),
enhances leukemia-initiating cell differentiation (23), and is correlated with carcinogenesis
and chemoresistance (34,37).
There are multiple mechanisms by which B7-H4 can
affect tumor cell biology. Wang et al (34) reported that silencing B7-H4 enhances
drug-induced apoptosis by inhibiting the phosphatase and tensin
homolog (PTEN)/PI3K/AKT signaling pathway, indicating the role of
B7-H4 in chemoresistance and suggesting that it may be an
attractive therapeutic target in triple-negative breast cancer. Xie
et al (15) demonstrated that
B7-H4 induced epithelial-mesenchymal transition, and promoted
invasion and metastasis of tumor cells by the activation of the
ERK1/2 signaling pathway. Furthermore, upregulated B7-H4 expression
was associated with downregulated Bax, upregulated Bcl-2 and
activation of caspase-3 (15). Qian
et al (38) analyzed the
microRNA (miRNA) expression profile following B7-H4 knockdown in
pancreatic cancer cell line L3.6p1 and noticed that the
differentially expressed miRNAs induced by B7-H4 siRNA were mainly
involved in the mitogen-activated protein kinase and PI3K/AKT
signaling pathways. Chen et al (37) demonstrated that B7-H4 expression is
positively correlated with IL6 expression and signal transducer and
activator of transcription 3 phosphorylation. Xia et al
(23) revealed that B7-H4 is one of
the highly expressed immune molecules on human acute myeloid
leukemia cells, and promotes the differentiation of
leukemia-initiating cells through the PTEN/AKT/hypoxia-inducible
factor-1α/REST corepressor 2/runt-related transcription factor 1
signaling pathway (23).
The present study constructed B7-H4 wild-type
overexpressing cells to investigate the specific DEGs induced by
B7-H4 wild-type in 786-O cells. The results revealed that there
were 704 upregulated and 804 downregulated DEGs. The upregulated
DEGs were associated with the inflammatory response, immune
response and cell chemotaxis. Of the upregulated DEGs obtained by
microarray, the upregulation of NME, MME, VNN1, MMP7, TNF, CXCL8,
CXCL1 and CCL20 were confirmed by western blot analysis. Since all
these molecules are involved in the inflammatory response, immune
response and cell chemotaxis, the current study further examined
the chemokine expression in clinical renal carcinoma by TCGA
dataset analysis and IHC staining in 59 clinical tumor tissues. The
results revealed that there was a positive correlation between
B7-H4 and CCL20 or CXCL8. Furthermore, B7-H4 increased
tumor-associated infiltrating neutrophils by upregulating CXCL8,
indicating another mechanism in the tumor promoting effect of
B7-H4. Similar the results obtained in the present study, Azuma
et al (39) revealed a
correlation between serum B7-H4 and neutrophil in peripheral blood
from the patients with clear cell renal cancer. The present study
demonstrated that B7-H4 expression increased tumor-infiltrating
neutrophils by upregulating CXCL8 and that blocking CXCL8 reversed
this increase. The present study revealed the molecular mechanism
underlying the B7-H4-associated increase in tumor-infiltrating
neutrophils, and suggested that B7-H4 and CXCL8 might serve as
therapeutic targets to remodel the tumor microenvironment.
Besides CXCL8, the chemokine CCL20 is also
upregulated in B7-H4-transfected cells. CCL20 is an 8-kDa protein
involved in the maintenance of immunological homeostasis (40). T cells, natural killer (NK) cells, B
cells and immature DCs are recruited to the tumor by the
interaction of CCL20 with CCR6 (41–43). As
CCL20 recruits both anti-tumor leukocytes and pro-tumor leukocytes
(regulatory T cells, myeloid dendritic cells and NK cells), the
role of CCL20 in tumor progression is complex (44). Tumor cells, macrophages and
neutrophils produce CXCL1 and recruit myeloid-derived suppressor
cells, which suppress the activity of CD8+ T effector
cells to prevent tumor cell killing by CD8+ T cells
(44,45). Thus, upregulated CXCL1 expression by
B7-H4 expression may contribute to tumor progression (44). Of the upregulated DEGs identified in
the present study, MMP7 exhibited the largest fold-change
difference. MMPs are a family of enzymes responsible for the
degradation of a wide spectrum of extracellular matrix and
non-matrix proteins (46). During
carcinogenesis, MMPs can regulate the microenvironment and
contribute to several critical steps in cancer development via
their involvement in cell proliferation, differentiation,
apoptosis, invasion, migration and immune surveillance (46–48).
Thus, upregulated MMP7 expression by B7-H4 may serve an important
role in tumor progression.
In conclusion, the results of the present study
revealed that in renal cell carcinoma, B7-H4 may upregulate CXCL1,
CXCL8, CCL20 and MMP7 and thus recruit tumor-associated
neutrophils.
Acknowledgements
The authors would like to thank Dr Chuanyang Sun
(The Department of Urology, The Second Affiliated Hospital of
Soochow University, Suzhou, China) for his valuable assistance in
collecting clinical samples.
Funding
The present study was supported by the National
Nature Science Foundation of China (grant nos. 31370872, 81402381
and 81502454).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LZ designed the study wrote the manuscript. AL
performed the cellular experiments. NZ performed the experiments on
the clinical samples. ZZ performed the bioinformatics and
statistical analyses. AL and YC performed the animal experiments.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved and guided by the
Ethics Committee of Soochow University (Suzhou, China). All
patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rooney MS, Shukla SA, Wu CJ, Getz G and
Hacohen N: Molecular and genetic properties of tumors associated
with local immune cytolytic activity. Cell. 160:48–61. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Santoni M, Berardi R, Amantini C,
Burattini L, Santini D, Santoni G and Cascinu S: Role of natural
and adaptive immunity in renal cell carcinoma response to
VEGFR-TKIs and mTOR inhibitor. Int J Cancer. 134:2772–2777. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Motzer RJ, Hutson TE, Tomczak P,
Michaelson MD, Bukowski RM, Rixe O, Oudard S, Negrier S, Szczylik
C, Kim ST, et al: Sunitinib versus interferon alfa in metastatic
renal-cell carcinoma. N Engl J Med. 356:115–124. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Motzer RJ, Escudier B, McDermott DF,
George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G,
Plimack ER, et al: Nivolumab versus everolimus in advanced
renal-cell carcinoma. N Engl J Med. 373:1803–1813. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gill DM, Hahn AW, Hale P and Maughan BL:
Overview of current and future first-line systemic therapy for
metastatic clear cell renal cell carcinoma. Curr Treat Options
Oncol. 19:62018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kawashima A, Uemura M and Nonomura N:
Importance of multiparametric evaluation of immune-related T-cell
markers in renal-cell carcinoma. Clin Genitourin Cancer.
17:e1147–e1152. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sica GL, Choi IH, Zhu G, Tamada K, Wang
SD, Tamura H, Chapoval AI, Flies DB, Bajorath J and Chen L: B7-H4,
a molecule of the B7 family, negatively regulates T cell immunity.
Immunity. 18:849–861. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zang X, Loke P, Kim J, Murphy K, Waitz R
and Allison JP: B7×: A widely expressed B7 family member that
inhibits T cell activation. Proc Natl Acad Sci USA.
100:10388–10392. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li C, Zhan Y, Ma X, Fang H and Gai X:
B7-H4 facilitates proliferation and metastasis of colorectal
carcinoma cell through PI3K/Akt/mTOR signaling pathway. Clin Exp
Med. Oct 29–2019.doi: 10.1007/s10238-019-00590-7 (Epub ahead of
print).
|
|
12
|
Genova C, Boccardo S, Mora M, Rijavec E,
Biello F, Rossi G, Tagliamento M, Dal Bello MG, Coco S, Alama A, et
al: Correlation between B7-H4 and survival of non-small-cell lung
cancer patients treated with nivolumab. J Clin Med. 8(pii):
E15662019. View Article : Google Scholar
|
|
13
|
Gruosso T, Gigoux M, Manem VSK, Bertos N,
Zuo D, Perlitch I, Saleh SMI, Zhao H, Souleimanova M, Johnson RM,
et al: Spatially distinct tumor immune microenvironments stratify
triple-negative breast cancers. J Clin Invest. 129:1785–1800. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Krambeck AE, Thompson RH, Dong H, Lohse
CM, Park ES, Kuntz SM, Leibovich BC, Blute ML, Cheville JC, Kwon
ED, et al: B7-H4 expression in renal cell carcinoma and tumor
vasculature: Associations with cancer progression and survival.
Proc Natl Acad Sci USA. 103:10391–10396. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xie N, Cai JB, Zhang L, Zhang PF, Shen YH,
Yang X, Lu JC, Gao DM, Kang Q, Liu LX, et al: Upregulation of B7-H4
promotes tumor progression of intrahepatic cholangiocarcinoma. Cell
Death Dis. 8:32052017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu J, Chu BF, Yang YP, Zhang SL, Zhuang
M, Lu WJ and Liu YB: B7-H4 expression is associated with cancer
progression and predicts patient survival in human thyroid cancer.
Asian Pac J Cancer Prev. 14:3011–3015. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Prasad DV, Richards S, Mai XM and Dong C:
B7S1, a novel B7 family member that negatively regulates T cell
activation. Immunity. 18:863–873. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li J, Lee Y, Li Y, Jiang Y, Lu H, Zang W,
Zhao X, Liu L, Chen Y, Tan H, et al: Co-inhibitory molecule B7
superfamily member 1 expressed by tumor-infiltrating myeloid cells
induces dysfunction of Anti-tumor CD8(+) T cells. Immunity.
48:773–786.e5. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen C, Qu QX, Xie F, Zhu WD, Zhu YH and
Huang JA: Analysis of B7-H4 expression in metastatic pleural
adenocarcinoma and therapeutic potential of its antagonists. BMC
Cancer. 17:6522017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen C, Zhu WD, Xie F and Huang JA:
Nuclear localization of B7-H4 in pulmonary adenocarcinomas
presenting as a solitary pulmonary nodule. Oncotarget.
7:58563–58568. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang L, Wu H, Lu D, Li G, Sun C, Song H,
Li J, Zhai T, Huang L, Hou C, et al: The costimulatory molecule
B7-H4 promote tumor progression and cell proliferation through
translocating into nucleus. Oncogene. 32:5347–5358. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jeon YK, Park SG, Choi IW, Lee SW, Lee SM
and Choi I: Cancer cell-associated cytoplasmic B7-H4 is induced by
hypoxia through hypoxia-inducible factor-1a and promotes cancer
cell proliferation. Biochem Biophys Res Commun. 459:277–283. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xia F, Zhang Y, Xie L, Jiang H, Zeng H,
Chen C, Liu L, He X, Hao X, Fang X, et al: B7-H4 enhances the
differentiation of murine leukemia-initiating cells via the
PTEN/AKT/RCOR2/RUNX1 pathways. Leukemia. 31:2260–2264. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tang Z, Li C, Kang B, Gao G and Zhang Z:
GEPIA: A web server for cancer and normal gene expression profiling
and interactive analyses. Nucleic Acids Res. 45:W98–W102. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Allred DC, Clark GM, Elledge R, Fuqua SA,
Brown RW, Chamness GC, Osborne CK and McGuire WL: Association of
p53 protein expression with tumor cell proliferation rate and
clinical outcome in node-negative breast cancer. J Natl Cancer
Inst. 85:200–206. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang DH, Lee HS, Yoon D, Berry G, Wheeler
TM, Sugarbaker DJ, Kheradmand F, Engleman E and Burt BM:
Progression of EGFR-Mutant lung adenocarcinoma is driven by
alveolar macrophages. Clin Cancer Res. 23:778–788. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ogawa R, Yamamoto T, Hirai H, Hanada K,
Kiyasu Y, Nishikawa G, Mizuno R, Inamoto S, Itatani Y, Sakai Y and
Kawada K: Loss of SMAD4 promotes colorectal cancer progression by
recruiting tumor-associated neutrophils via CXCL1/8-CXCR2 axis.
Clin Cancer Res. 25:2287–2899. 2019. View Article : Google Scholar
|
|
28
|
Wang L, Heng X, Lu Y, Cai Z, Yi Q and Che
F: Could B7-H4 serve as a target to activate anti-cancer immunity?
Int Immunopharmacol. 38:97–103. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bregar A, Deshpande A, Grange C, Zi T,
Stall J, Hirsch H, Reeves J, Sathyanarayanan S, Growdon WB and
Rueda BR: Characterization of immune regulatory molecules B7-H4 and
PD-L1 in low and high grade endometrial tumors. Gynecol Oncol.
145:446–452. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Han S, Wang Y, Shi X, Zong L, Liu L, Zhang
J, Qian Q, Jin J, Ma Y, Cui B, et al: Negative roles of B7-H3 and
B7-H4 in the microenvironment of cervical cancer. Exp Cell Res.
371:222–230. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang H, Li C and Ren G: Clinical
significance of the B7-H4 as a novel prognostic marker in breast
cancer. Gene. 623:24–28. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
MacGregor HL and Ohashi PS: Molecular
pathways: Evaluating the potential for B7-H4 as an immunoregulatory
target. Clin Cancer Res. 23:2934–2941. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tan Z and Shen W: Prognostic role of B7-H4
in patients with non-small cell lung cancer: A meta-analysis.
Oncotarget. 8:27137–27144. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang L, Yang C, Liu XB, Wang L and Kang
FB: B7-H4 overexpression contributes to poor prognosis and
drug-resistance in triple-negative breast cancer. Cancer Cell Int.
18:1002018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Qian Y, Sang Y, Wang FX, Hong B, Wang Q,
Zhou X, Weng T, Wu Z, Zheng M, Zhang H and Yao H: Prognostic
significance of B7-H4 expression in matched primary pancreatic
cancer and liver metastases. Oncotarget. 7:72242–72249. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Han S, Li Y, Zhang J, Liu L, Chen Q, Qian
Q, Li S and Zhang Y: Roles of immune inhibitory molecule B7-H4 in
cervical cancer. Oncology Rep. 37:2308–2316. 2017. View Article : Google Scholar
|
|
37
|
Chen X, Wang W, Man H, Li P and Shan B:
Increased B7-H4 expression during esophageal squamous cell
carcinogenesis is associated with IL-6/STAT3 signaling pathway
activation in mice. Oncol Lett. 13:2207–2215. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Qian Y, Feng L, Wu W, Weng T, Hu C, Hong
B, Wang FXC, Shen L, Wang Q, Jin X and Yao H: MicroRNA expression
profiling of pancreatic cancer cell line L3.6p1 following B7-H4
Knockdown. Cell Physiol Biochem. 44:494–504. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Azuma T, Sato Y, Ohno T, Azuma M and Kume
H: Serum soluble B7-H4 is a prognostic marker for patients with
non-metastatic clear cell renal cell carcinoma. PLoS One.
13:e01997192018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jacquelot N, Duong CPM, Belz GT and
Zitvogel L: Targeting chemokines and chemokine receptors in
melanoma and other cancers. Front Immunol. 9:24802018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Y, Roth TL, Gray EE, Chen H, Rodda
LB, Liang Y, Ventura P, Villeda S, Crocker PR and Cyster JG:
Migratory and adhesive cues controlling innate-like lymphocyte
surveillance of the pathogen-exposed surface of the lymph node.
Elife. 5(pii): e181562016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ramirez-Valle F, Gray EE and Cyster JG:
Inflammation induces dermal Vγ4+ γδT17 memory-like cells that
travel to distant skin and accelerate secondary IL-17-driven
responses. Proc Natl Acad Sci USA. 112:8046–8051. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hartwig T, Pantelyushin S, Croxford AL,
Kulig P and Becher B: Dermal IL-17-producing γδ T cells establish
long-lived memory in the skin. Eur J Immunol. 45:3022–3033. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Vilgelm AE and Richmond A: Chemokines
modulate immune surveillance in tumorigenesis, metastasis, and
response to immunotherapy. Front Immunol. 10:3332019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Susek KH, Karvouni M, Alici E and
Lundqvist A: The role of CXC chemokine receptors 1–4 on immune
cells in the tumor microenvironment. Front Immunol. 9:21592018.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Young D, Das N, Anowai A and Dufour A:
Matrix metalloproteases as influencers of the Cells' social media.
Int J Mol Sci. 20(pii): E38472019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liao CH, Chang WS, Hu PS, Wu HC, Hsu SW,
Liu YF, Liu SP, Hung HS, Bau DT and Tsai CW: The Contribution of
MMP-7 promoter polymorphisms in renal cell carcinoma. In Vivo.
31:631–635. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang Z, Dong T, Fu Y, Zhou W, Tian X,
Chen G and Liu S: MMP-11 promotes papillary thyroid cell
proliferation and invasion via the NF-κB pathway. J Cell Biochem.
Sep 1–2018.doi: 10.1002/jcb.27500 (Epub ahead of print).
|