|
1
|
Sánchez-Aranguren LC, Prada CE,
Riaño-Medina CE and Lopez M: Endothelial dysfunction and
preeclampsia: Role of oxidative stress. Front Physiol. 5:3722014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang Y, Zhang X, Cheng GM and Ren CC:
Expression of transforming growth factor-beta 1, vascular cell
adhesion molecule 1 and E-selectin in placenta of patients with
pre-clampsia. Zhonghua Fu Chan Ke Za Zhi. 41:514–517. 2006.(In
Chinese). PubMed/NCBI
|
|
3
|
Weissgerber TL and Mudd LM: Preeclampsia
and diabetes. Curr Diab Rep. 15:92015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Spradley FT: Metabolic abnormalities and
obesity's impact on the risk for developing preeclampsia. Am J
Physiol Regul Integr Comp Physiol. 312:R5–R12. 2017. View Article : Google Scholar : PubMed/NCBI
|
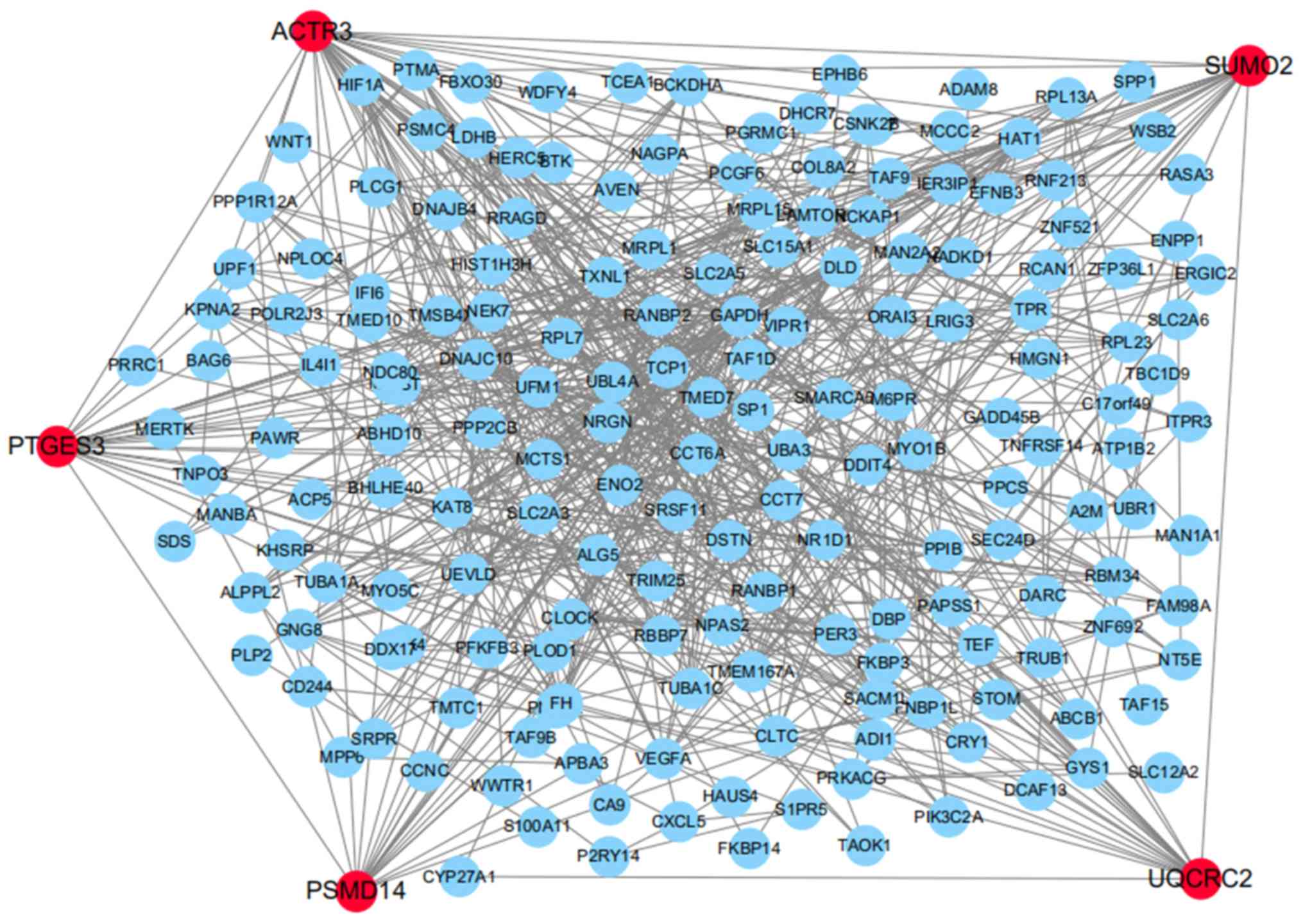
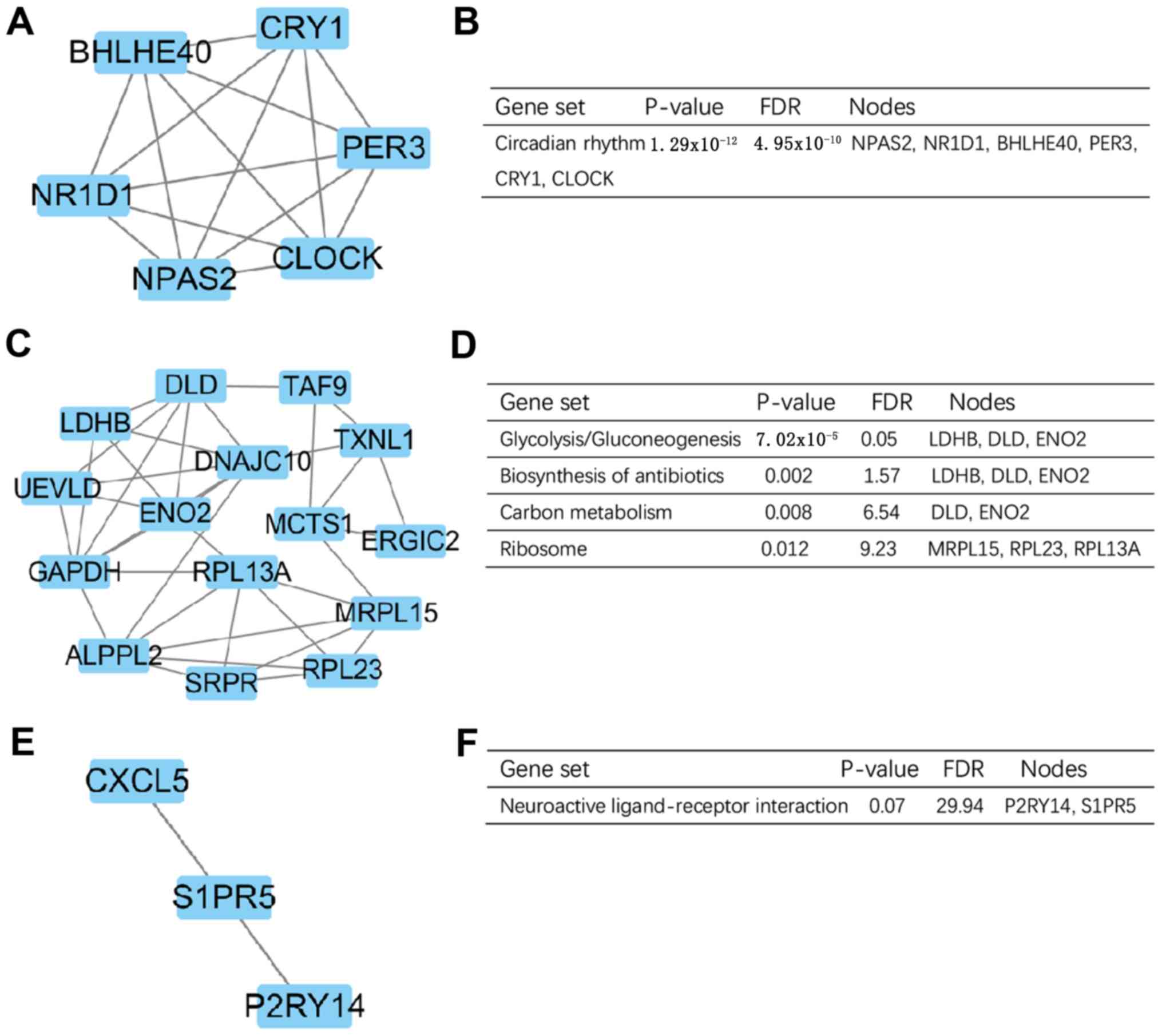
|
5
|
Karumanchi SA and Granger JP: Preeclampsia
and pregnancy-related hypertensive disorders. Hypertension.
67:238–242. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Facca TA, Kirsztajn GM and Sass N:
Preeclampsia (marker of chronic kidney disease): From genesis to
future risks. J Bras Nefrol. 34:87–93. 2012.(In English,
Portuguese). View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kumasawa K, Ikawa M, Kidoya H, Hasuwa H,
Saito-Fujita T, Morioka Y, Takakura N, Kimura T and Okabe M:
Pravastatin induces placental growth factor (PGF) and ameliorates
preeclampsia in a mouse model. Proc Natl Acad Sci USA.
108:1451–1455. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Filipek A and Jurewicz E: Preeclampsia - a
disease of pregnant women. Postepy Biochem. 64:232–229. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang J and Zhao ZM: LncRNA HOXD-AS1
promotes preeclampsia progression via MAPK pathway. Eur Rev Med
Pharmacol Sci. 22:8561–8568. 2018.PubMed/NCBI
|
|
10
|
Zhang XM, Xiong X, Tong C, Li Q, Huang S,
Li QS, Liu YM, Li HY, Baker P, Shan N and Qi HB: Down-regulation of
laminin (LN)-α5 is associated with preeclampsia and impairs
trophoblast cell viability and invasiveness through PI3K signaling
pathway. Cell Physiol Biochem. 51:2030–2040. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pan Q, Shai O, Lee LJ, Frey BJ and
Blencowe BJ: Deep surveying of alternative splicing complexity in
the human transcriptome by high-throughput sequencing. Nat Genet.
40:1413–1415. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Luo S, Cao N, Tang Y and Gu W:
Identification of key microRNAs and genes in preeclampsia by
bioinformatics analysis. PLoS One. 12:e01785492017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu X, Yang Y, Han T, Yin G, Gao P, Ni Y,
Su X, Liu Y and Yao Y: Suppression of microRNA-18a expression
inhibits invasion and promotes apoptosis of human trophoblast cells
by targeting the estrogen receptor alpha gene. Mol Med Rep.
12:2701–2706. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Barrett T, Troup DB, Wilhite SE, Ledoux P,
Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M,
Marshall KA, et al: NCBI GEO: Archive for high-throughput
functional genomic data. Nucleic Acids Res. 37:(Database Issue).
D885–D890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
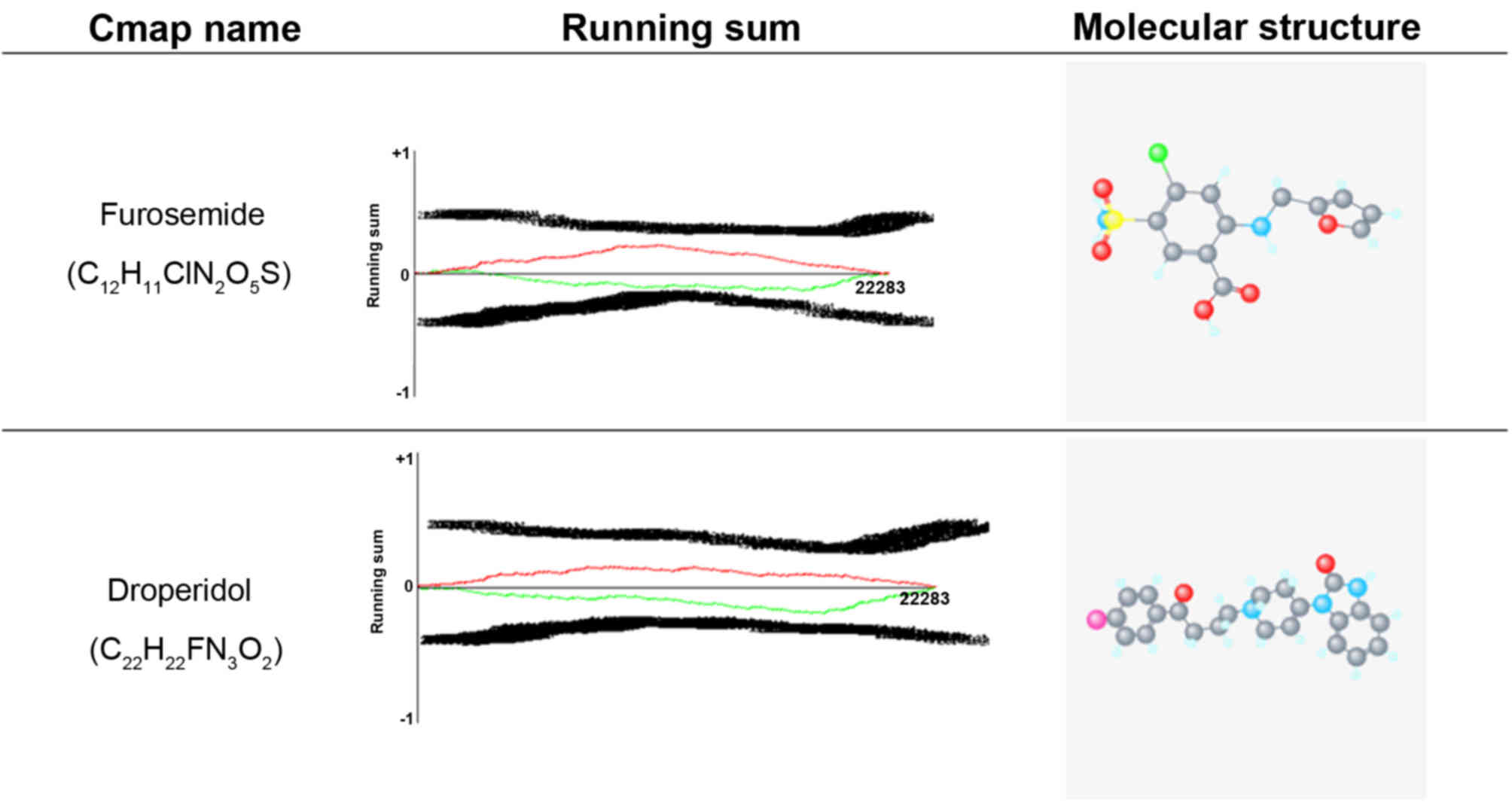
Lamb J, Crawford ED, Peck D, Modell JW,
Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et
al: The connectivitiy map: Using gene-expression signatures to
connect small molecules, genes, and disease. Scinece.
313:1929–1935. 2006. View Article : Google Scholar
|
|
17
|
Huang HL, Peng CY, Lai MJ, Chen CH, Lee
HY, Wang JC, Liou JP, Pan SL and Teng CM: Novel oral histone
deacetylase inhibitor, MPT0E028, displays potent growth-inhibitory
activity against human B-cell lymphoma in vitro and in vivo.
Oncotarget. 6:4976–4991. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wen Z, Wang Z, Wang S, Ravula R, Yang L,
Xu J, Wang C, Zuo Z, Chow MS, Shi L and Huang Y: Discovery of
molecular mechanisms of traditional Chinese medicinal formula
Si-Wu-Tang using gene expression microarray and connectivity map.
PLoS One. 6:e182782011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wei LL, Pan YS, Tang Q, Yang ZJ, Song WQ,
Gao YF, Li J, Zhang L and Liu SG: Decreased ALCAM expression and
promoter hypermethylation is associated with preeclampsia.
Hypertens Res. 43:13–22. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Q, Lu X, Li C, Zhang W, Lv Y, Wang L,
Wu L, Meng L, Fan Y, Ding H, et al: Down-regulated long non-coding
RNA PVT1 contributes to gestational diabetes mellitus and
preeclampsia via regulation of human trophoblast cells. Biomed
Pharmacother. 120:1095012019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hu S, Li J, Tong M, Li Q, Chen Y, Lu H,
Wang Y and Min L: MicroRNA-144-3p may participate in the
pathogenesis of preeclampsia by targeting Cox-2. Mol Med Rep.
19:4655–4662. 2019.PubMed/NCBI
|
|
22
|
Dong K, Zhang X, Ma L, Gao N, Tang H, Jian
F and Ma Y: Downregulations of circulating miR-31 and miR-21 are
associated with preeclampsia. Pregnancy Hypertens. 17:59–63. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Y, Cheng K, Zhou W, Liu H, Yang T,
Hou P and Li X: miR-141-5p regulate ATF2 via effecting MAPK1/ERK2
signaling to promote preeclampsia. Biomed Pharmacother.
115:1089532019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yong HE, Melton PE, Johnson MP, Freed KA,
Kalionis B, Murthi P, Brennecke SP, Keogh RJ and Moses EK:
Genome-wide transcriptome directed pathway analysis of maternal
pre-eclampsia susceptibility genes. PLoS One. 10:e01282302015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cao C, Chen J, Lyu C, Yu J, Zhao W, Wang Y
and Zou D: Bioinformatics analysis of the effects of tobacco smoke
on gene expression. PLoS One. 10:e01433772015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
29
|
Mootha VK, Lindgren CM, Eriksson KF,
Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E,
Ridderstråle M, Laurila E, et al: PGC-1alpha-responsive genes
involved in oxidative phosphorylation are coordinately
downregulated in human diabetes. Nat Genet. 34:267–273. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 4 (Suppl 4):S112014. View Article : Google Scholar
|
|
31
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lamb J, Crawford ED, Peck D, Modell JW,
Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et
al: The connectivity map: Using gene-expression signatures to
connect small molecules, genes, and disease. Science.
313:1929–1935. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Group of hypertensive disorders of
pregnancy, branch of Obstetrics and Gynecology, Chinese Medical
Association. Chin J Obst Gynecol. 10:2015.
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ditisheim AJ, Dibner C, Philippe J and
Pechère-Bertschi A: Biological rhythms and preeclampsia. Front
Endocrinol (Lausanne). 4:472013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Haelterman E, Marcoux S, Croteau A and
Dramaix M: Population-based study on occupational risk factors for
preeclampsia and gestational hypertension. Scand J Work Environ
Health. 33:304–317. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mozurkewich EL, Luke B, Avni M and Wolf
FM: Working conditions and adverse pregnancy outcome: A
meta-analysis. Obstet Gynecol. 95:623–635. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Munz W, Seufert R, Steiner E, Pollow K and
Brockerhoff P: Circadian blood pressure rhythm in preeclampsia as a
predictor of maternal and obstetrical outcome. Z Geburtshilfe
Neonatol. 207:132–136. 2003.(In German). PubMed/NCBI
|
|
39
|
van den Berg CB, Chaves I, Herzog EM,
Willemsen SP, van der Horst GTJ and Steegers-Theunissen RPM: Early-
and late-onset preeclampsia and the DNA methylation of circadian
clock and clock-controlled genes in placental and newborn tissues.
Chronobiol Int. 34:921–932. 2017. View Article : Google Scholar
|
|
40
|
Ding X, Yang Z, Han Y and Yu H: Fatty acid
oxidation changes and the correlation with oxidative stress in
different preeclampsia-like mouse models. PLoS One. 9:e1095542014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wata S, Lee JW, Okada K, Lee JK, Iwata M,
Rasmussen B, Link TA, Ramaswamy S and Jap BK: Complete structure of
the 11-subunit bovine mitochondrial cytochrome bc1 complex.
Science. 281:64–71. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Aouache R, Biquard L, Vaiman D and
Miralles F: Oxidative stress in preeclampsia and placental
diseases. Int J Mol Sci. 19(pii): E14962018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Elliot MG: Oxidative stress and the
evolutionary origins of preeclampsia. J Reprod Immunol. 114:75–80.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chiarello DI, Abad C, Rojas D, Toledo F,
Vázquez CM, Mate A, Sobrevia L and Marín R: Oxidative stress:
Normal pregnancy versus preeclampsia. Biochim Biophys Acta Mol
Basis Dis. 1866:1653542020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Heneweer C, Kruse LH, Kindhauser F,
Schmidt M, Jakobs KH, Denker HW and Thie M: Adhesiveness of human
uterine epithelial RL95-2 cells to trophoblast: Rho protein
regulation. Mol Hum Reprod. 8:1014–1022. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tamás P, Hantosi E, Farkas B, Ifi Z,
Betlehem J and Bódis J: Preliminary study of the effects of
furosemide on blood pressure during late-onset pre-eclampsia in
patients with high cardiac output. Int J Gynaecol Obstet.
136:87–90. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Adamantidis MM, Kerram P, Caron JF and
Dupuis BA: Droperidol exerts dual effects on repolarization and
induces early afterdepolarizations and triggered activity in rabbit
Purkinje fibers. J Pharmacol Exp Ther. 266:884–893. 1993.PubMed/NCBI
|
|
48
|
Gu ZC and Enenkel C: Proteasome assembly.
Cell Mol Life Sci. 71:4729–4745. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hass R and Sohn C: Increased oxidative
stress in pre-eclamptic placenta is associated with altered
proteasome activity and protein patterns. Placenta. 24:979–984.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
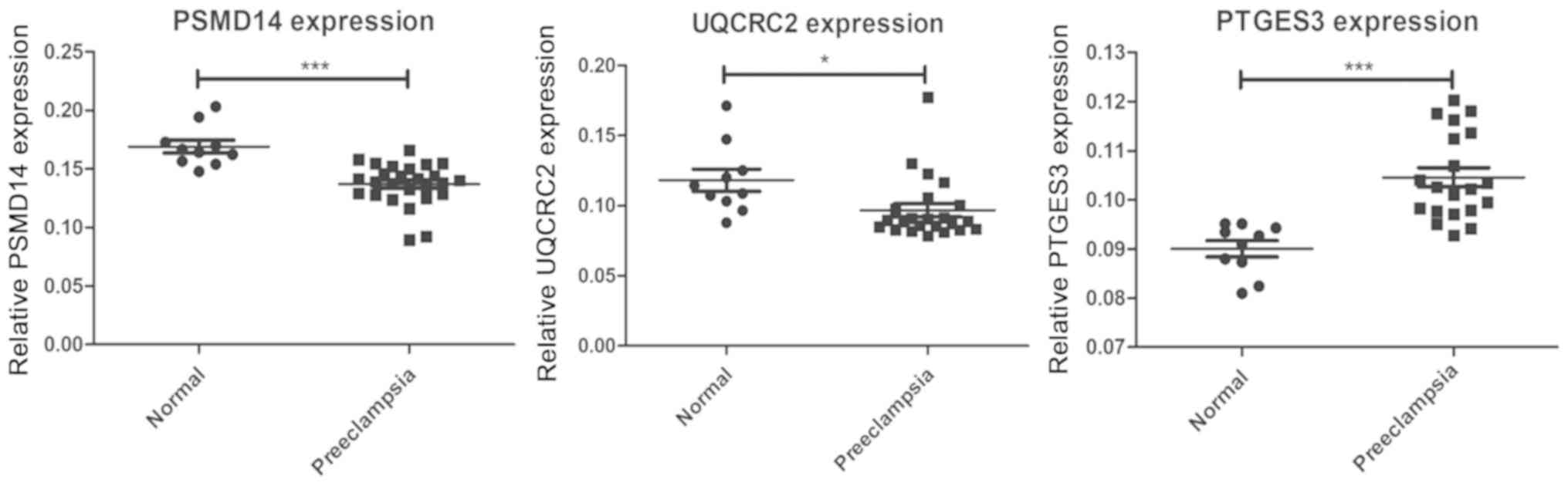
Zhou JF, Wang XY, Shangguan XJ, Gao ZM,
Zhang SM, Xiao WQ and Chen CG: Increased oxidative stress in women
with pregnancy-induced hypertension. Biomed Environ Sci.
18:419–426. 2005.PubMed/NCBI
|
|
51
|
Mistry HD, Wilso V, Ramsay MM, Symonds ME
and Broughton Pipkin F: Reduced selenium concentrations and
glutathione peroxidase activity in preeclamptic pregnancies.
Hypertension. 52:881–888. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Vanderlelie J, Venardos K, Clifton VL,
Gude NM, Clarke FM and Perkins AV: Increased biological oxidation
and reduced anti-oxidant enzyme activity in pre-eclamptic placenta.
Placenta. 26:53–58. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Miyake N, Yano S, Sakai C, Hatakeyama H,
Matsushima Y, Shiina M, Watanabe Y, Bartley J, Abdenur JE, Wang RY,
et al: Mitochondrial complex III deficiency caused by a homozygous
UQCRC2 mutation presenting with neonatal-onset recurrent metabolic
decompensation. Hum Mutat. 34:446–452. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Gaignard P, Eyer D, Lebigot E, Oliveira C,
Therond P, Boutron A and Slama A: UQCRC2 mutation in a patient with
mitochondrial complex III deficiency causing recurrent liver
failure, lactic acidosis and hypoglycemia. J Hum Genet. 62:729–731.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Freeman BC and Yamamoto KR: Disassembly of
transcriptional regulatory complexes by molecular chaperones.
Science. 296:2232–2235. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sinclair D, Fillman SG, Webster MJ and
Weickert CS: Dysregulation of glucocorticoid receptor co-factors
FKBP5, BAG1 and PTGES3 in prefrontal cortex in psychotic illness.
Sci Rep. 3:35392013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lv J, Zhang S, Wu H, Lu J, Lu Y, Wang F,
Zhao W, Zhan P, Lu J, Fang Q, et al: Deubiquitinase PSMD14 enhances
hepatocellular carcinoma growth and metastasis by stabilizing GRB2.
Cancer Lett. 469:22–34. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Song B, Du J, Feng Y, Gao YJ and Zhao JS:
Co-expressed differentially expressed genes and long non-coding
RNAs involved in the celecoxib treatment of gastric cancer: An RNA
sequencing analysis. Exp Ther Med. 12:2455–2468. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Gu Y, Chen G and Du Y: Screening of
prognosis-related genes in primary breast carcinoma using genomic
expression data. J Comput Biol. Nov 13–2019.(Epub ahead of print).
View Article : Google Scholar
|
|
60
|
Cao W, Wang X, Chen T, Zhu H, Xu W, Zhao
S, Cheng X and Xia L: The expression of Notch/Notch ligand, IL-35,
IL-17, and Th17/Treg in preeclampsia. Dis Markers. 2015:3161822015.
View Article : Google Scholar
|
|
61
|
Gu Y, Feng Y, Yu J, Yuan H, Yin Y, Ding J,
Zhao J, Xu Y, Xu J and Che H: Fasudil attenuates soluble fms-like
tyrosine kinase-1 (sFlt-1)-induced hypertension in pregnant mice
through RhoA/ROCK pathway. Oncotarget. 8:104104–104112. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zheng L, Shi L, Zhou Z, Chen X, Wang L, Lu
Z and Tang R: Placental expression of AChE, α7nAChR and NF-κB in
patients with preeclampsia. Ginekol Pol. 89:249–255. 2018.
View Article : Google Scholar : PubMed/NCBI
|