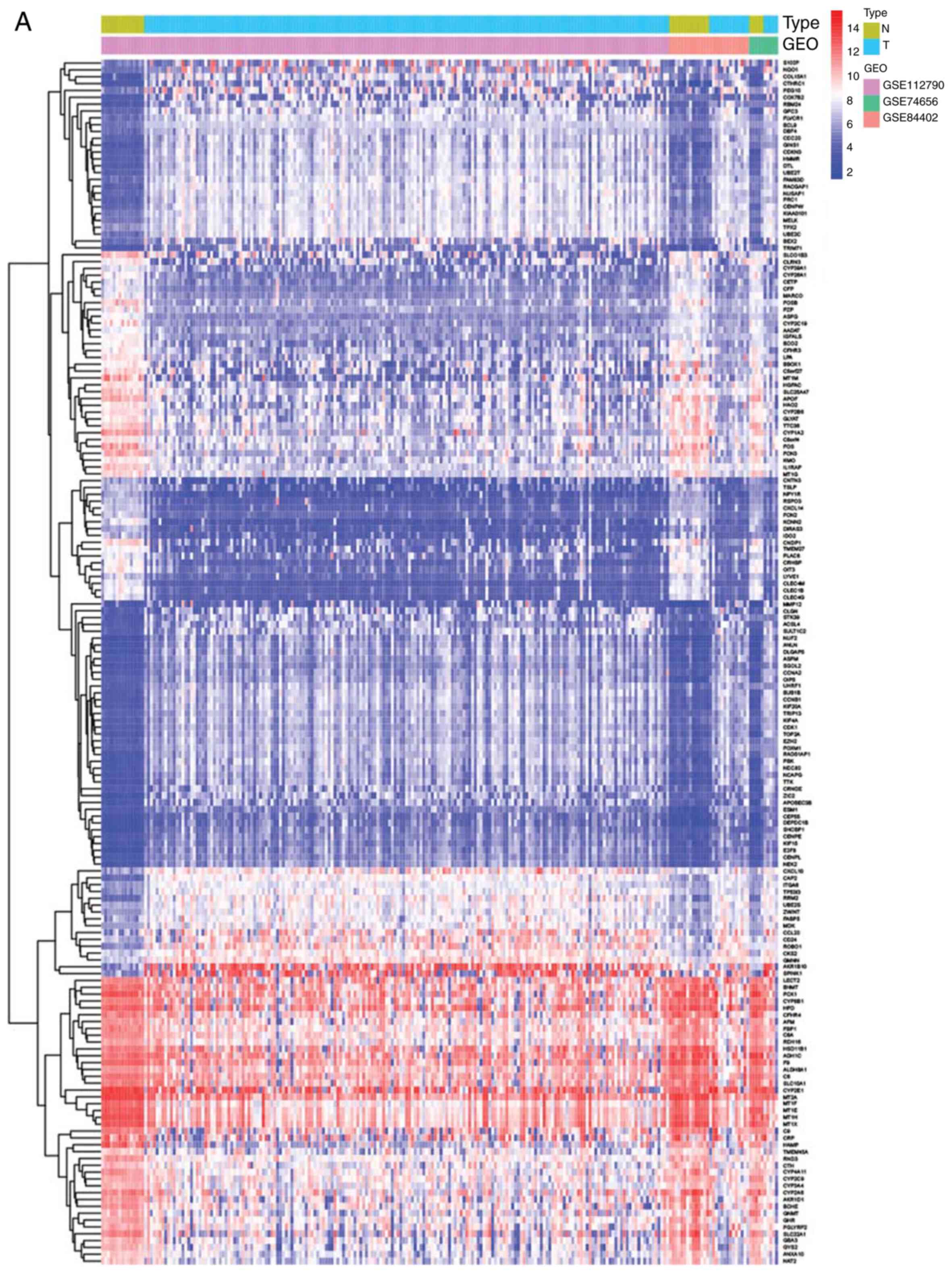
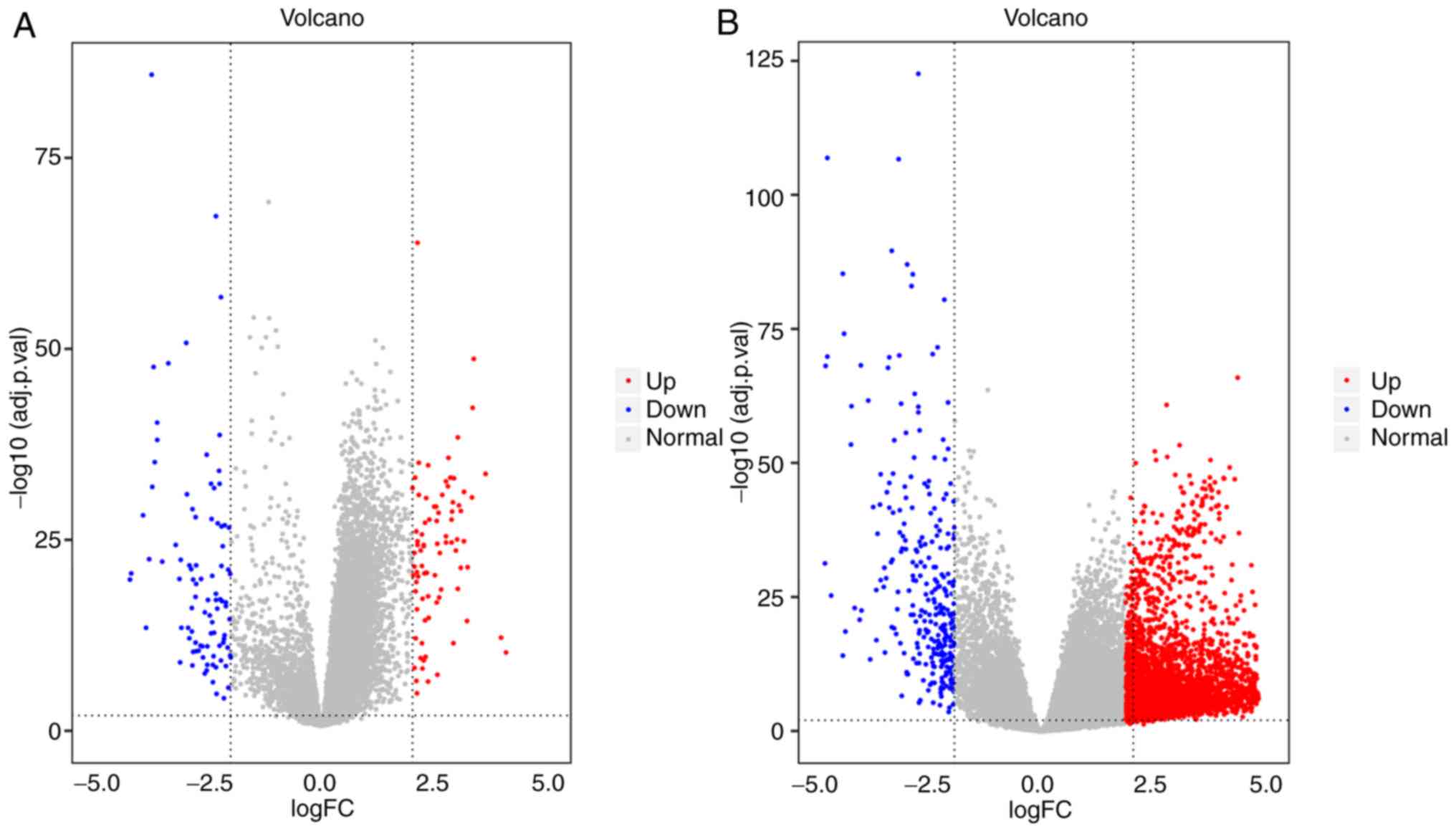
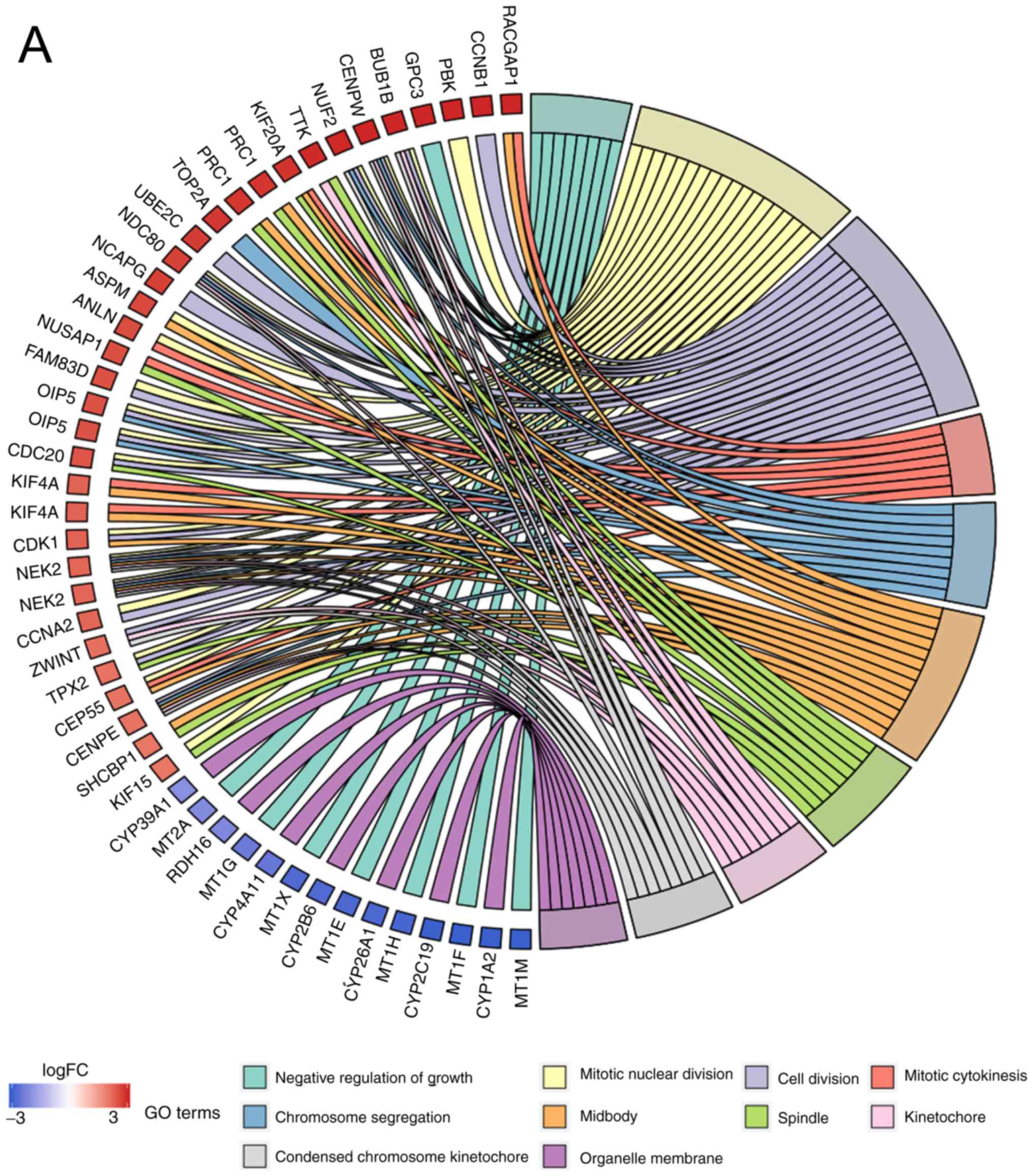
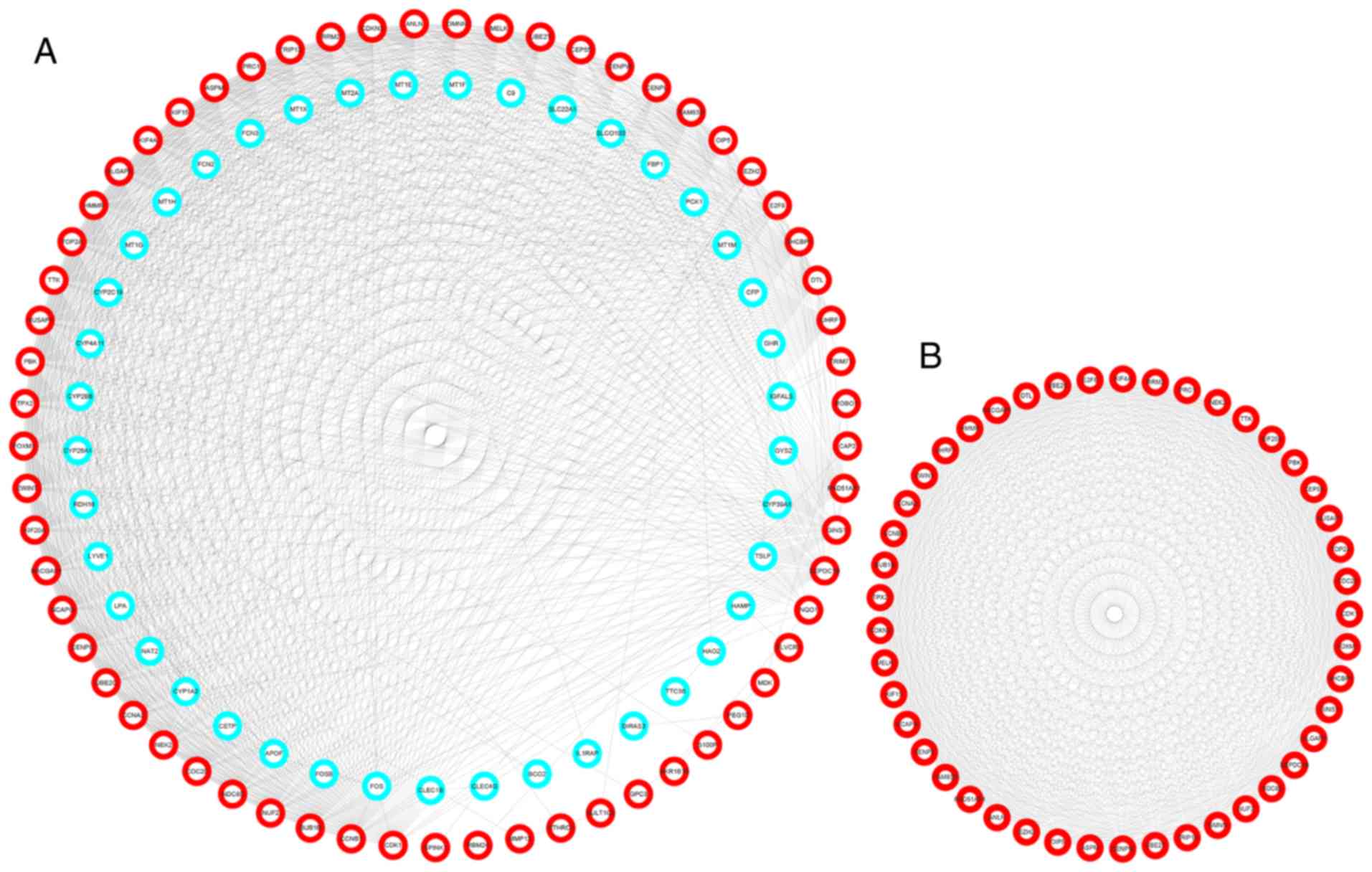
|
1
|
McGlynn KA, Petrick JL and El-Serag HB:
Epidemiology of hepatocellular carcinoma. Hepatology. Apr
22–2020.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maluccio M and Covey A: Recent progress in
understanding, diagnosing, and treating hepatocellular carcinoma.
CA Cancer J Clin. 62:394–399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
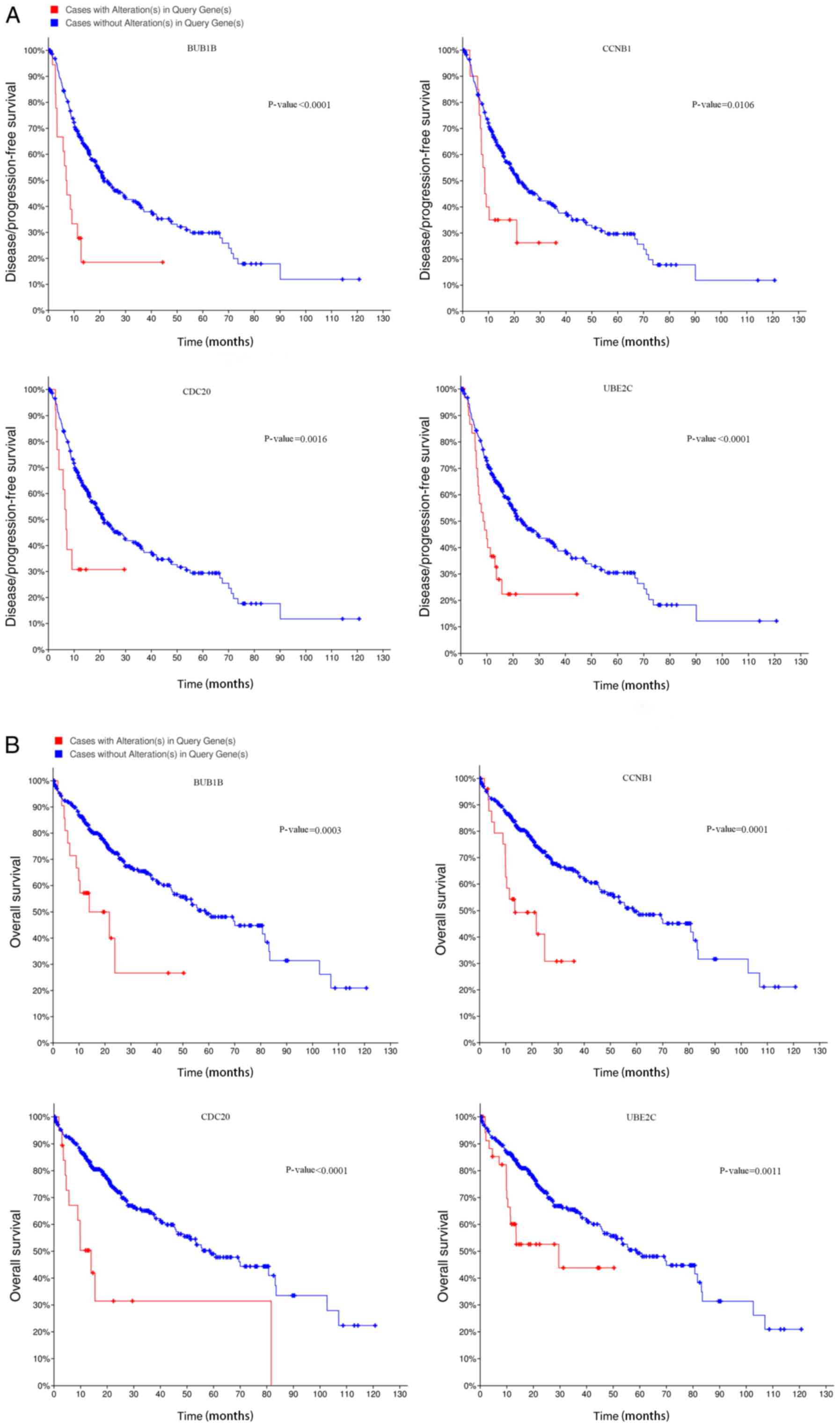
5
|
Parkin DM: The global health burden of
infection-associated cancers in the year 2002. Int J Cancer.
118:3030–3044. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rinella ME: Nonalcoholic fatty liver
disease: A systematic review. JAMA. 313:2263–2273. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mirizzi A, Franco I, Leone CM, Bonfiglio
C, Cozzolongo R, Notarnicola M, Giannuzzi V, Tutino V, De Nunzio V,
Bruno I, et al: Effects of some food components on non-alcoholic
fatty liver disease severity: Results from a cross-sectional study.
Nutrients. 11:27442019. View Article : Google Scholar
|
|
8
|
Dimitroulis D, Damaskos C, Valsami S,
Davakis S, Garmpis N, Spartalis E, Athanasiou A, Moris D,
Sakellariou S, Kykalos S, et al: From diagnosis to treatment of
hepatocellular carcinoma: An epidemic problem for both developed
and developing world. World J Gastroentero. 23:5282–5294. 2017.
View Article : Google Scholar
|
|
9
|
Fuks D, Dokmak S, Paradis V, Diouf M,
Durand F and Belghiti J: Benefit of initial resection of
hepatocellular carcinoma followed by transplantation in case of
recurrence: An intention-to-treat analysis. Hepatology. 55:132–140.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Han HS, Shehta A, Ahn S, Yoon YS, Cho JY
and Choi Y: Laparoscopic versus open liver resection for
hepatocellular carcinoma: Case-matched study with propensity score
matching. J Hepatol. 63:643–650. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hartke J, Johnson M and Ghabril M: The
diagnosis and treatment of hepatocellular carcinoma. Semin Diagn
Pathol. 34:153–159. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang P, Yang Y, Wen F, He X, Tang R, Du
Z, Zhou J, Zhang J and Li Q: Cost-effectiveness of sorafenib as a
first-line treatment for advanced hepatocellular carcinoma. Eur J
Gastroenterol Hepatol. 27:853–859. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li L, Lei Q, Zhang S, Kong L and Qin B:
Screening and identification of key biomarkers in hepatocellular
carcinoma: Evidence from bioinformatic analysis. Oncol Rep.
38:2607–2618. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang R, Ye J, Huang H and Du X: Mining
featured biomarkers associated with vascular invasion in HCC by
bioinformatics analysis with TCGA RNA sequencing data. Biomed
Pharmacother. 118:1092742019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang C, Peng L, Zhang Y, Liu Z, Li W,
Chen S and Li G: The identification of key genes and pathways in
hepatocellular carcinoma by bioinformatics analysis of
high-throughput data. Med Oncol. 34:1012017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shimada S, Mogushi K, Akiyama Y, Furuyama
T, Watanabe S, Ogura T, Ogawa K, Ono H, Mitsunori Y, Ban D, et al:
Comprehensive molecular and immunological characterization of
hepatocellular carcinoma. EBioMedicine. 40:457–470. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang H, Huo X, Yang XR, He J, Cheng L,
Wang N, Deng X, Jin H, Wang N, Wang C, et al: STAT3-mediated
upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer
metastasis by regulating SOX4. Mol Cancer. 16:1362017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Leek JT, Johnson WE, Parker HS, Jaffe AE
and Storey JD: The sva package for removing batch effects and other
unwanted variation in high-throughput experiments. Bioinformatics.
28:882–883. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Robinson MD, McCarthy DJ and Smyth GK:
EdgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Walter W, Sánchez-Cabo F and Ricote M:
GOplot: An R package for visually combining expression data with
functional analysis. Bioinformatics. 31:2912–2914. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Demchak B, Hull T, Reich M, Liefeld T,
Smoot M, Ideker T and Mesirov JP: Cytoscape: The network
visualization tool for GenomeSpace workflows. F1000Res. 3:1512014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li H, Han D, Hou Y, Chen H and Chen Z:
Statistical inference methods for two crossing survival curves: A
comparison of methods. PLoS One. 10:e01167742015. View Article : Google Scholar : PubMed/NCBI
|
|
30
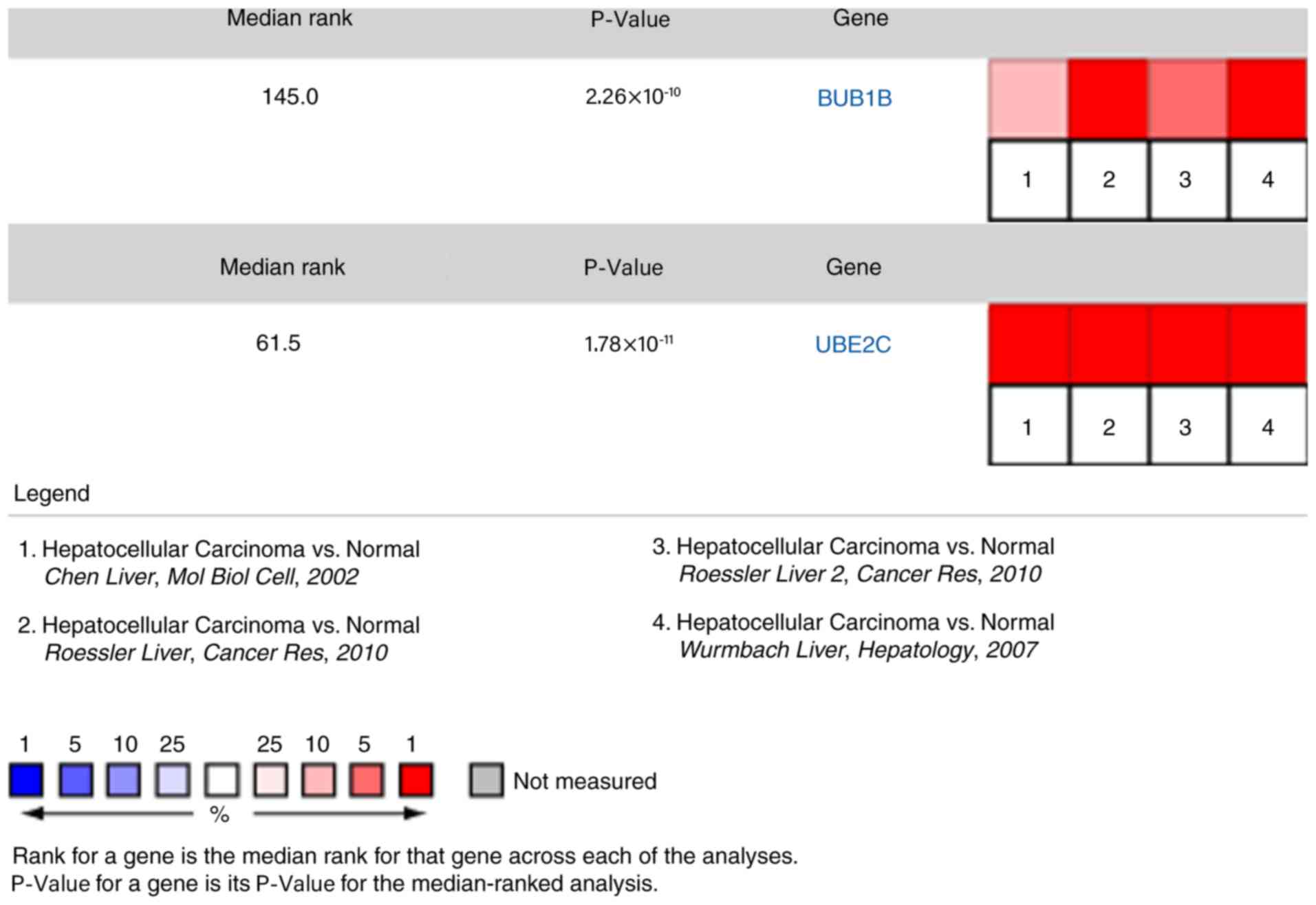
|
Rhodes DR, Kalyana-Sundaram S, Mahavisno
V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ,
Kincead-Beal C, Kulkarni P, et al: Oncomine 3.0: Genes, pathways,
and networks in a collection of 18,000 cancer gene expression
profiles. Neoplasia. 9:166–180. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wurmbach E, Chen YB, Khitrov G, Zhang W,
Roayaie S, Schwartz M, Fiel I, Thung S, Mazzaferro V, Bruix J, et
al: Genome-wide molecular profiles of HCV-induced dysplasia and
hepatocellular carcinoma. Hepatology. 45:938–947. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
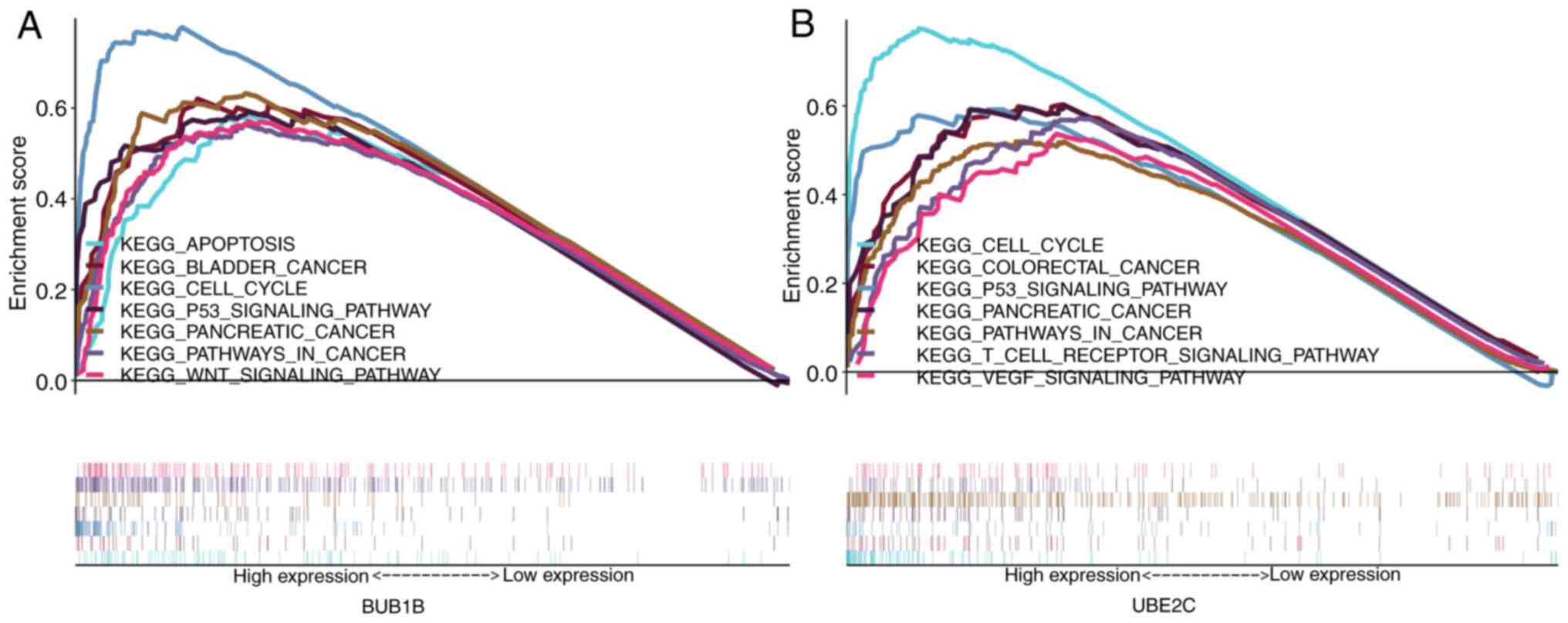
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liberzon A, Birger C, Thorvaldsdóttir H,
Ghandi M, Mesirov JP and Tamayo P: The Molecular Signatures
Database (MSigDB) hallmark gene set collection. Cell Syst.
1:417–425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
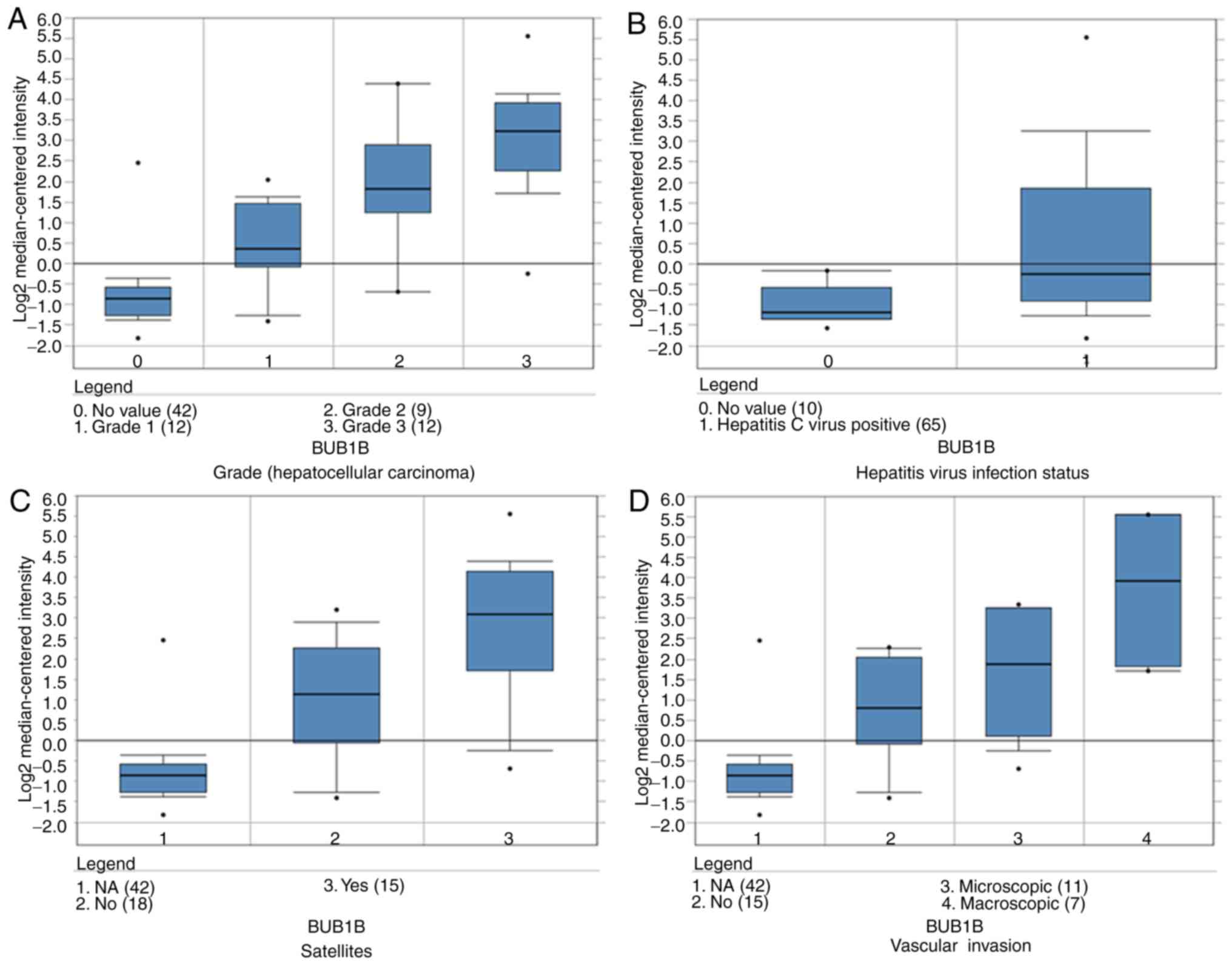
Zhou L, Du Y, Kong L, Zhang X and Chen Q:
Identification of molecular target genes and key pathways in
hepatocellular carcinoma by bioinformatics analysis. Onco Targets
Ther. 11:1861–1869. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dong S, Huang F, Zhang H and Chen Q:
Overexpression of BUB1B, CCNA2, CDC20 and CDK1 in tumor tissues
predicts poor survival in pancreatic ductal adenocarcinoma. Biosci
Rep. 39:BSR201823062019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang Q, Huang H, Liu A, Li J, Liu C, Sun
B, Chen L, Gao Y, Xu D and Su C: Cell division Cycle 20 (CDC20)
drives prostate cancer progression via stabilization of β-catenin
in cancer stem-like cells. EBioMedicine. 42:397–407. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cheng S, Castillo V and Sliva D: CDC20
associated with cancer metastasis and novel mushroom-derived CDC20
inhibitors with antimetastatic activity. Int J Oncol. 54:2250–2256.
2019.PubMed/NCBI
|
|
38
|
Shin HJ, Baek KH, Jeon AH, Park MT, Lee
SJ, Kang CM, Lee HS, Yoo SH, Chung DH, Sung YC, et al: Dual roles
of human BubR1, a mitotic checkpoint kinase, in the monitoring of
chromosomal instability. Cancer Cell. 4:483–497. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ando K, Kakeji Y, Kitao H, Iimori M, Zhao
Y, Yoshida R, Oki E, Yoshinaga K, Matumoto T, Morita M, et al: High
expression of BUBR1 is one of the factors for inducing DNA
aneuploidy and progression in gastric cancer. Cancer Sci.
101:639–645. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yamamoto Y, Matsuyama H, Chochi Y, Okuda
M, Kawauchi S, Inoue R, Furuya T, Oga A, Naito K and Sasaki K:
Overexpression of BUBR1 is associated with chromosomal instability
in bladder cancer. Cancer Genet Cytogen. 174:42–47. 2007.
View Article : Google Scholar
|
|
41
|
Liu AW, Cai J, Zhao XL, Xu AM, Fu HQ, Nian
H and Zhang SH: The clinicopathological significance of BUBR1
overexpression in hepatocellular carcinoma. J Clin Pathol.
62:1003–1008. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fu X, Chen G, Cai ZD, Wang C, Liu ZZ, Lin
ZY, Wu YD, Liang YX, Han ZD, Liu JC and Zhong WD: Overexpression of
BUB1B contributes to progression of prostate cancer and predicts
poor outcome in patients with prostate cancer. Onco Targets Ther.
9:2211–2220. 2016.PubMed/NCBI
|
|
43
|
Shichiri M, Yoshinaga K, Hisatomi H,
Sugihara K and Hirata Y: Genetic and epigenetic inactivation of
mitotic checkpoint genes hBUB1 and hBUBR1 and their relationship to
survival. Cancer Res. 62:13–17. 2002.PubMed/NCBI
|
|
44
|
Park HY, Jeon YK, Shin HJ, Kim IJ, Kang
HC, Jeong SJ, Chung DH and Lee CW: Differential promoter
methylation may be a key molecular mechanism in regulating BubR1
expression in cancer cells. Exp Mol Med. 39:195–204. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Williamson A, Wickliffe KE, Mellone BG,
Song L, Karpen GH and Rape M: Identification of a physiological E2
module for the human anaphase-promoting complex. Proc Natl Acad Sci
USA. 106:18213–18218. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang HQ, Zhao G, Ke B, Ma G, Liu GL,
Liang H, Liu LR and Hao XS: Overexpression of UBE2C correlates with
poor prognosis in gastric cancer patients. Eur Rev Med Pharmacol
Sci. 22:1665–1671. 2018.PubMed/NCBI
|
|
47
|
Mo CH, Gao L, Zhu XF, Wei KL, Zeng JJ,
Chen G and Feng ZB: The clinicopathological significance of UBE2C
in breast cancer: A study based on immunohistochemistry, microarray
and RNA-sequencing data. Cancer Cell Int. 17:832017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bavi P, Uddin S, Ahmed M, Jehan Z, Bu R,
Abubaker J, Sultana M, Al-Sanea N, Abduljabbar A, Ashari LH, et al:
Bortezomib stabilizes mitotic cyclins and prevents cell cycle
progression via inhibition of UBE2C in colorectal carcinoma. Am J
Pathol. 178:2109–2120. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Dastsooz H, Cereda M, Donna D and Oliviero
S: A comprehensive bioinformatics analysis of UBE2C in cancers. Int
J Mol Sci. 20:22282019. View Article : Google Scholar
|
|
50
|
Lin YT, Chen Y, Wu G and Lee WH: Hec1
sequentially recruits Zwint-1 and ZW10 to kinetochores for faithful
chromosome segregation and spindle checkpoint control. Oncogene.
25:6901–6914. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gao T, Han Y, Yu L, Ao S, Li Z and Ji J:
CCNA2 is a prognostic biomarker for ER+ breast cancer and tamoxifen
resistance. PLoS One. 9:e917712014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li CY, Xue C, Yang Q, Low BC and Liou YC:
NuSAP governs chromosome oscillation by facilitating the
Kid-generated polar ejection force. Nat Commun. 7:105972016.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Gordon CA, Gong X, Ganesh D and Brooks JD:
NUSAP1 promotes invasion and metastasis of prostate cancer.
Oncotarget. 8:29935–29950. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liu Z, Guan C, Lu C, Liu Y, Ni R, Xiao M
and Bian Z: High NUSAP1 expression predicts poor prognosis in colon
cancer. Pathol Res Pract. 214:968–973. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang Q, Su L, Liu N, Zhang L, Xu W and
Fang H: Cyclin Dependent Kinase 1 Inhibitors: A review of recent
progress. Curr Med Chem. 18:2025–2043. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Petri ET, Errico A, Escobedo L, Hunt T and
Basavappa R: The crystal structure of human cyclin B. Cell Cycle.
6:1342–1349. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhuang L, Yang Z and Meng Z: Upregulation
of BUB1B, CCNB1, CDC7, CDC20, and MCM3 in tumor tissues predicted
worse overall survival and disease-free survival in hepatocellular
carcinoma patients. Biomed Res Int. 2018:78973462018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Brown NR, Lowe ED, Petri E, Skamnaki V,
Antrobus R and Johnson LN: Cyclin B and Cyclin A confer different
substrate recognition properties on CDK2. Cell Cycle. 6:1350–1359.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Qiao M, Shapiro P, Fosbrink M, Rus H,
Kumar R and Passaniti A: Cell cycle-dependent phosphorylation of
the RUNX2 transcription factor by cdc2 regulates endothelial cell
proliferation. J Biol Chem. 281:7118–7128. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Pagano M, Pepperkok R, Verde F, Ansorge W
and Draetta G: Cyclin A is required at two points in the human cell
cycle. EMBO J. 11:961–971. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
David Y, Ziv T, Admon A and Navon A: The
E2 ubiquitin-conjugating enzymes direct polyubiquitination to
preferred lysines. J Biol Chem. 285:8595–8604. 2010. View Article : Google Scholar : PubMed/NCBI
|